Abstract
A relatively crude preparation of herpes simplex virus was rapidly visualized by atomic force microscopy after exposure to conditions that produced gradual degradation of the virions. Images were obtained of 1) the intact, enveloped virus; 2) the underlying capsid with associated tegument proteins along with fragments of the membrane; 3) the capsomeres composing the capsid and their surface arrangement; 4) damaged and partially degraded capsids with missing capsomeres; and 5) the DNA extruded from damaged virions. These images provide a unique perspective on the structures of individual virus particles. Atomic force microscopy can thus be used as a diagnostic tool to provide a rapid way to obtain high-resolution images of human pathogens from crude preparations. It is a useful technique that complements X-ray-based structure determination, cryo-electron microscopy techniques, and optical microscopies in the field of molecular pathogenesis.
A key to the understanding of pathogenesis on the molecular level is knowledge of the three-dimensional structure of the macromolecules involved. In the case of viral infections, this involves detailed knowledge of the virion composition and architecture. However, the structures of complex human and animal viruses are difficult to determine in detail. They are fragile, often structurally heterogeneous, are very difficult to produce in sufficient quantities for crystallization, and in addition, their sheer size often places them outside the range of X-ray crystallography. The only approach, using X-rays as the probe, may be to address the protein components, expressed in some type of cells, individually, and by some means reconstructing the various parts to create the virion. Even this is problematic, however, considering the membrane that envelops the virus and the hydrophobic proteins that populate the membranes.
An alternative that has proven effective, although at significantly lower resolution, is cryo-electron microscopy (cryo-EM), often coupled with image reconstruction (IR) techniques. This approach has yielded excellent images of large animal viruses 1 at resolutions ranging from 8 Å upwards. A limitation of this method, however, is that it benefits greatly from high particle symmetry, such as icosahedral symmetry, and is far less powerful for nonsymmetrical macromolecular ensembles.
Both X-ray diffraction and cryo-EM/IR directly provide three-dimensional structural information, and they allow visualization of the interior of the virus as well as its surface. A limitation that some might consider of minor consequence, is that both techniques are averaging techniques. That is, they present a time and space average model of the entire population of particles found in the crystal or on the EM grid. They produce rather little information (although some) regarding the characteristics of the individuals of the population that distinguish them from the average. For this reason, the beautifully symmetrical, and apparently perfect, models of larger viruses derived from X-ray crystallography and cryo-EM may be somewhat deceptive, and not entirely representative of the entire population. Therefore, imaging and interpretation of individual virus particles should always be a part of initial virus characterization.
Another limitation to both X-ray and EM techniques is that preparation of samples can be time-consuming and potentially damaging to the specimens. Especially in the case of X-ray structure determination, the entire cycle of crystallization (if even possible), data collection, and calculations can consume months. Preparation, data collection, and calculations for EM/IR also can take substantial time, albeit generally less. There is a need for a rapid, relatively simple and reasonably inexpensive technique that is still capable of high-resolution imaging.
Atomic force microscopy (AFM) 2 has different virtues (and different problems) and it introduces an effective complement to the techniques above.
For applications in pathology, a key benefit is the ability to rapidly examine the architectures of single virus particles or a collection of distinct individuals that originate from a relatively crude preparation. The capability to study individual particles enables us to see variations (such as defects or different reactions to the addition of detergents) within a population of a certain type of virus. The crude preparation that suffices for this makes the whole process relatively easy and fast. Imaging may be performed at a resolution comparable to or slightly lower than conventional EM methods. One important advantage of AFM is that it can be performed into many different environments: in air, vacuum, and most importantly to biological applications, in solution. Usually, this requires no staining, freezing, fixing, labeling, or other preparations. Normally, samples are either dried and imaged in air or imaged in solution, enabling the study of live samples.
This method has already been used for imaging capsid structures of plant viruses in crystals. 3,4 With future development of AFM technology, which is now rapid in the area of biological imaging, AFM may maintain pace with conventional EM in resolution. Since its development less than two decades ago, AFM has matured into a easy-to-use, cross-disciplinary and relatively cheap way to study a multitude of samples to a high resolution.
Although AFM produces images only of surfaces, as we show here, that may be sufficient to reveal most of the interior detail. If the native virus particles are subjected to systematic degradation, with enzymes or chemical agents, for example, then layers of structure may be sequentially peeled away to reveal those below. In addition, particles in different states of disrepair reveal a variety of features that can provide a more comprehensive image of broader scope.
The virus that we have chosen to test the capabilities of AFM in molecular pathogenesis is herpes simplex virus-1 (HSV-1). From a methodological point of view, the most important reason for this is that HSV-1 is a well-studied virus, of which the capsid structure is known to the details, because of EM and especially cryo-EM combined with three-dimensional IR. Furthermore, it is of reasonable large size, enabling to study it in detail by AFM. Also, HSV-1 is an enveloped virus; hence we could test out procedures for its removal. All of these make HSV-1 a perfect measuring stick to judge the capabilities of AFM.
From a pathological point of view, the most important reason we chose Herpes is because it is one of the most widespread human viruses, making it important to study its structure and interaction with cells. Because in our lab we have the ability to infect HSV-1 into cells, this will be the next stage of our HSV-1 studies.
HSV-1 consists of an icosahedral capsid of protein containing the double-stranded DNA genome, and which is itself surrounded by a lipid envelope with embedded proteins. Between the membrane and the capsid are tegument proteins of various sizes. The intact virion is ∼200 nm in diameter, but from cryo-EM, we know that the capsid shell is ∼125 nm in diameter and ∼15-nm thick. The main components of the icosahedral capsid of triangulation number T = 16, as determined by EM, are 12 pentameric capsomeres (pentons) and 150 hexameric capsomeres (hexons). These capsomeres are interconnected by 320 smaller protein complexes known as triplexes. 5 Together, these units construct a capsid consisting of 20 equivalent triangles, with pentons on the corners, a row of three hexons between them, and three more hexons in the center of each triangle (Figure 1) ▶ . Pentons (hexons) consist of five (six) copies of the 150-kd VP5 capsid protein, 5 and each hexon also contains six copies of 12-kd VP26, forming a ring on top of the VP55. 5,6 Each triplex consists of two copies of 33-kd VP23 and one 53-kd VP19 protein. 7
Figure 1.
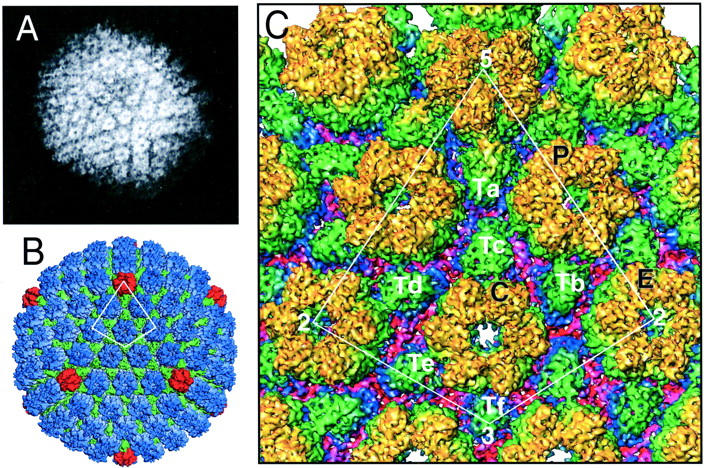
HSV-1 capsid structure as seen by EM. A: Single HSV-1 capsid as seen by conventional EM using negative staining. 9 B and C: HSV-1 capsid at 8.5 Å resolution, reconstructed by merging of cryo-EM images of 5860 particles. 8 B: The entire capsid, with pentons in red, hexons in blue, and triplexes in green. The asymmetric unit, indicated by the white line, is enlarged in C. C: Asymmetric unit. The penton is labeled 5; three slightly distinct hexons are denoted P, C, and E; the six types of triplexes are labeled Ta, Tb, Tc, Td, and Tf; and the twofold, threefold, and fivefold icosahedral axes are denoted by 2, 3, and 5. Here, color coding is according to the distance from the virion core, yellow indicating the most extreme features and red indicating the most proximal. Figure A: Reprinted with permission from http://www.uct.ac.za/depts/mmi/stannard/linda.html. Copyright Linda M. Stannard, 1995. Figures B and C: Reprinted with permission from ZH Zhou et al, Seeing the herpesvirus capsid at 8.5Ȧ, Science 2000, 288:877–880. Copyright 2000 American Association for the Advancement of Science.
The 8.5-Å cryo-EM model on which much of our current knowledge of the structure of HSV-1 rests 8 provides a standard against which the AFM studies presented here can be compared. The AFM images and the cryo-EM images are very different in character, and different as well in content, and both present separate verities of a common structure.
Materials and Methods
Cells and Viruses
Virus was grown in rabbit skin fibroblasts grown on minimal essential medium with Earle’s salts (Life Technologies, Inc., Grand Island, NY), supplemented with 292 μg ml−1 glutamine, 100 U of penicillin, 100 μg ml−1 streptomycin, and 5% newborn calf serum (Atlanta Biologicals, Inc., Norcross, GA). For a typical stock of virus, 8 × 107 cells were infected at a multiplicity of 0.01 plaque-forming units/cell of HSV strain 17syn+. After a 1-hour adsorption period, cells were overlain in minimal essential medium with 5% fetal calf serum (Atlanta Biologicals), and incubated at 37°C for 48 hours under 5% CO2.
Partially purified cell-free virus was then prepared as follows. The infected cells were shaken off into media, the suspension centrifuged at 1000 × g for 5 minutes in a Beckman tabletop centrifuge at room temperature. The clarified supernatant was then centrifuged at 14,000 × g for 2.5 hours at 10°C using a Beckman 35 rotor. The resultant pellet was drained thoroughly and resuspended in 350 μl of sterile TE buffer (0.01 mol/L Tris, 0.001 mol/L ethylenediaminetetraacetic acid, pH 7.6) at 4°C overnight. The virus suspension was then gently and thoroughly mixed, and stored at −55°C until use. The concentration was ∼1010 virions/ml.
AFM
The frozen virus suspension was thawed and either first mixed 1:1 with 0.2% Triton X-100 detergent or directly deposited on freshly cleaved mica. After a few minutes, samples were gently rinsed with double-distilled water and quickly dried with a flow of nitrogen gas. Imaging was performed using a Nanoscope IIIa AFM (Digital Instruments, Santa Barbara, CA), operated in tapping mode, 9,10 and equipped with phase-imaging hardware. Most images presented here are phase images, because they display the greatest amount of contrast and details. A small number of height images are also included to show topological differences. Commercially available tapping mode tips (TESP, RTESP; Digital Instruments), as well as extra oxide-sharpened nanotips (kindly made available by Ami Chand, Digital Instruments) were used.
AFM Imaging of HSV-1
Envelope
Before treatment with any agents, the experiments revealed enveloped virions such as that presented in Figure 2A ▶ . In this image, a number of features are visible on the surface of the lipid envelope. The largest of them, some of which are indicated with arrows, have lengths of ∼25 nm. The envelope of HSV contains several types of viral glycoproteins projecting from the virion envelope. These glycoproteins are essential for virion infectivity and penetration into the host cell. The lengths of spikes formed by different glycoproteins vary in the range of 8 to 24 nm. 11 Thus the observed surface features may correspond to the virus-encoded glycoproteins that are incorporated into the virion envelope.
Figure 2.
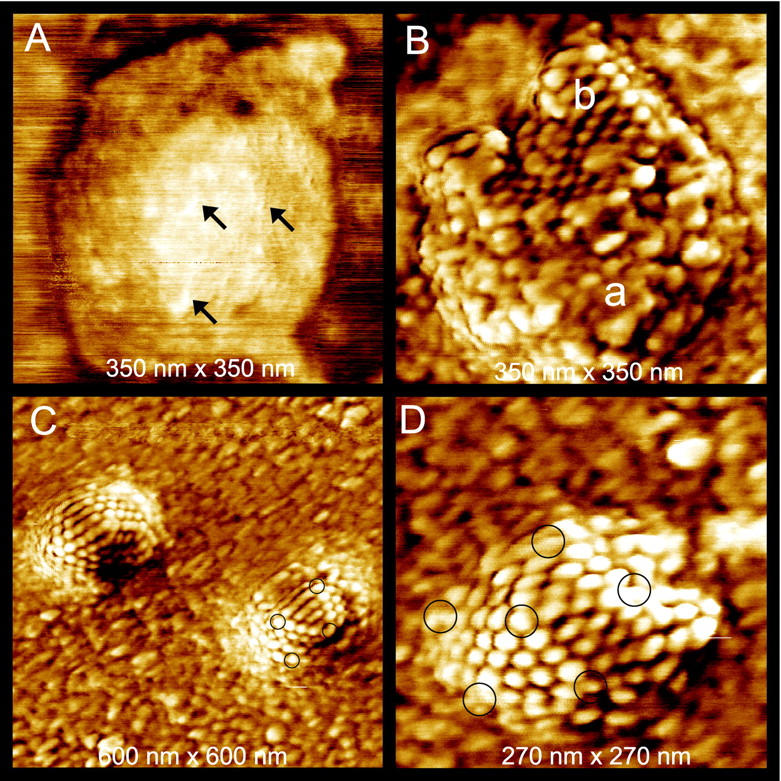
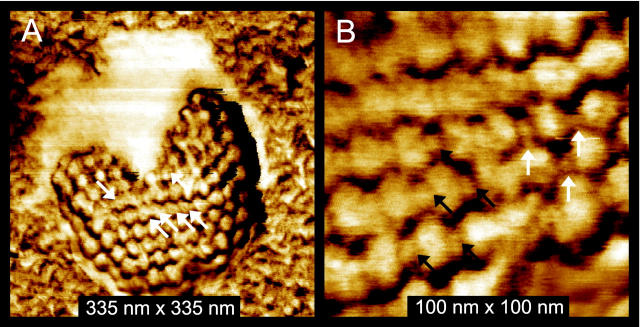
HSV-1 virions in different conformations adsorbed on mica and imaged with tapping-mode AFM in air. A: The capsid is still covered by the lipid envelope. Some elongated features of sizes 10 to 25 nm are denoted by arrows. These may correspond to glycoproteins. B–D: Addition of 0.2% Triton X-100 removes the lipid envelope. B: Most of this capsid is covered by an irregular collection of particles of ∼10 nm in size (area a) that corresponds to tegument proteins. In a smaller region the underlying, highly regular capsid is exposed (area b). C: Two particles with lipid bilayer and tegument completely removed. Black circles denote pentons in one of the capsids. D: A completely stripped capsid at higher magnification. Again, black circles denote the pentons.
Tegument
Mixing of the virus solution with 0.2% Triton X-100 results in the removal of the lipid envelope from most virions, which then renders the tegument and capsomere structure visible. In Figure 2B ▶ , a capsid that is mostly covered with an irregular coating of particles of various sizes up to 10 nm is seen, whereas on the upper part of the virus capsid, the underlying, highly regular packing of the proteins composing the virus capsid can be recognized as well. It was shown earlier 12 that the tegument has a complex nonicosahedrally ordered arrangement and contains at least 18 different viral proteins, the largest of which, VP 1-3, has a predicted size of 336 kd. 13 This corresponds well to the observed particles of ∼10 nm and smaller.
Capsid
More vigorous treatment of virions with detergent results in the complete loss of all envelope and tegument proteins and reveals the icosahedral capsid structure of the virions as seen at relatively low (Figure 2C) ▶ and higher (Figure 2D) ▶ magnification. In Figure 1B ▶ each penton is surrounded by five hexons, whereas each hexon is surrounded by six other capsomeres. Based on the cryo-EM structure of HSV, the pentameric and hexameric capsomeres can be discriminated from one another in AFM images as seen in Figure 2D ▶ , because of their different environments. The distortion in the symmetry of the capsomere packing in the capsid is chiefly because of the attachment of virions to the mica substrate and sample drying, which results in some flattening of the capsids.
In Figure 3A ▶ , a capsid that is stripped of its lipid envelope (white layer) is seen, leaving the capsid fully exposed. In AFM phase images, the lipid envelope contrasts strongly with both capsids and the thin layer of background debris, which consists of small biological particles from within the HSV-1 solution, such as tegument protein, that adheres to the mica surface together with the virions. The reason for this contrast is that the lipid envelope has a different chemical composition, giving it a different interaction with the AFM tip, and thus a distinct phase delay. The simultaneously recorded height image (Figure 3B) ▶ shows that the lipid envelope consists of three different areas, with heights of 2, 6, and 12 nm. Because lipid bilayers have a typical thickness of ∼6 nm, 14 these plateaus could correspond to half, single, and double bilayers, respectively. A higher magnification image of this layer (Figure 3C) ▶ reveals many particles of 4 to 6 nm in size (some of which are denoted by circles), which may well be membrane proteins.
Figure 3.
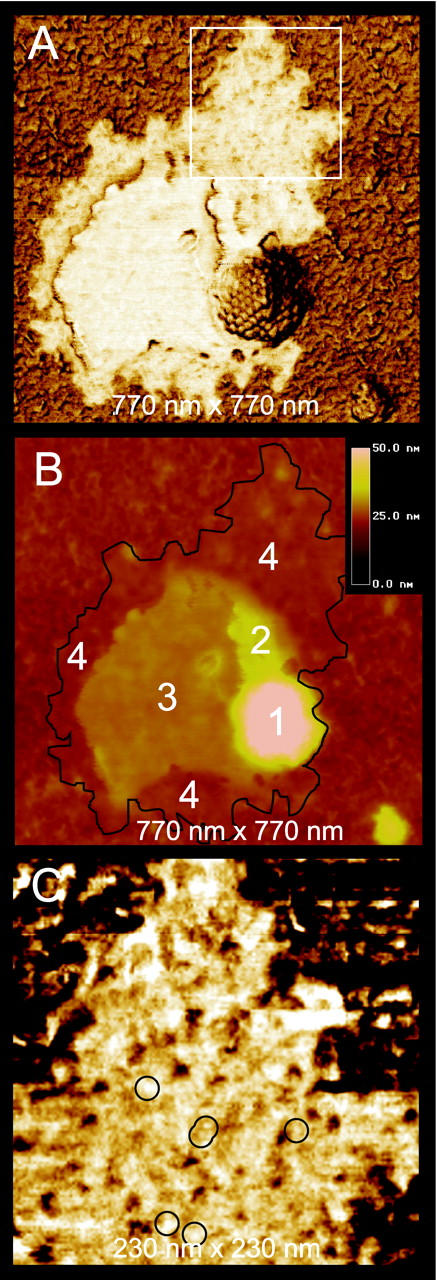
A: AFM phase image of an exposed capsid with attached lipid envelope adsorbed on mica. The white square is enlarged in C. B: Simultaneously acquired height image of the same capsid. Black line denotes the circumference of the envelope as seen in A. Numbers correspond to the capsid (1), lipid envelope layers of height 12 nm (2), 6 nm (3), and 2 nm (4), respectively. C: Lipid envelope at higher resolution. Circles indicate 4- to 6-nm-sized particles.
In most cases, the acuity of commercially available Si AFM tips used in this work allowed lateral resolution of ∼4 to 5 nm, which is insufficient to visualize the substructure of capsomeres, but is adequate to discriminate the individual capsomeres. Their diameters of ∼15 nm are accurately reflected by AFM. However, when Si AFM tips that were additionally oxide sharpened (see Materials and Methods) were used, substructure of individual capsomeres emerged (Figure 4) ▶ . Many capsomeres show a hole (Figure 4A ▶ , arrows) in the middle consistent with the structure of the capsid determined from EM (Figure 1) ▶ . In addition, smaller clusters that link adjacent capsomeres (Figure 4B ▶ , arrows) can also be visualized. We believe that these correspond to triplexes that are a characteristic features of the capsid (indicated as Ta, Tb, Tc, Td, Te, and Tf in Figure 1C ▶ ) 8 ) and are formed by three capsid proteins. Some of these clusters are situated in the cavities between three capsomeres (Figure 4B ▶ , white arrows), as expected from cryo-EM (Figure 1) ▶ , whereas other units seem positioned between two, rather than three, capsomeres (black arrows). Possibly, there has been some conformational distortion on adsorption of the capsids to the mica.
Figure 4.
Tapping-mode AFM images recorded with sharp tips yielding 2-nm resolution. A: A capsid that flattened on absorption onto mica partly covered with (white) lipid envelope. White arrows point at capsomeres with visible hollow channels in their center. B: Center region of the same capsid as in A. In addition to the channels, units between the hexons and pentons are also visible. These are the triplexes, consistent with the cryo-EM images in Figure 1 ▶ . EM shows these triplexes to be positioned in the cavities between three capsomeres. AFM images show triplexes both in these cavities (white arrows) as well as in the space between two capsomeres (black arrows).
DNA
AFM can probe the structures of large macromolecular assemblies as well as dynamic processes involving the assemblies. Thus, for example, we were able to observe disassembly of viruses as seen in Figure 5 ▶ . Investigations of the extrusion of HSV DNA can be important for understanding DNA packing in the capsid shell, and for simulating DNA exit from the capsid in vivo.
Figure 5.
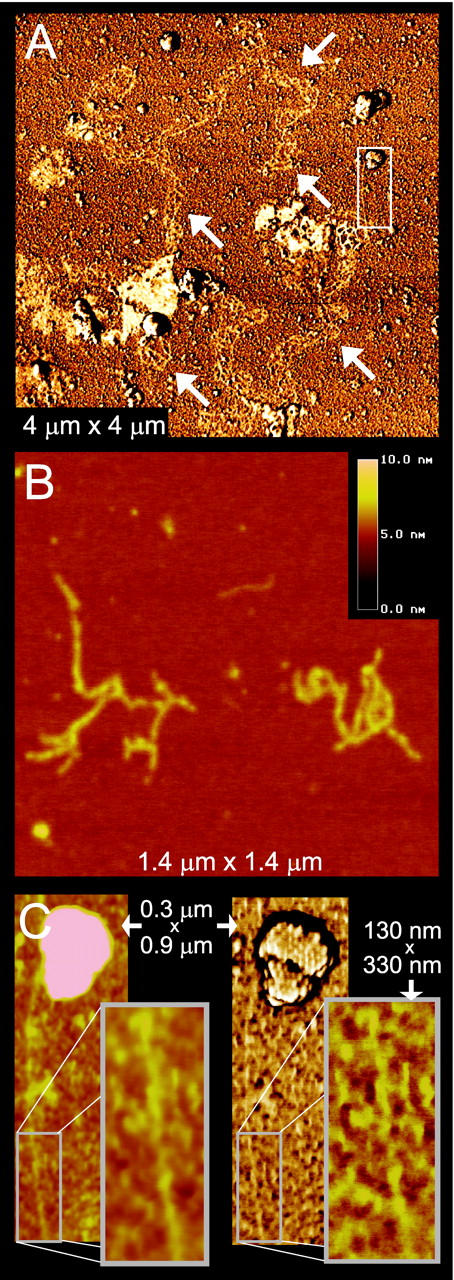
DNA escapes from the capsids after treatment with 0.5% SDS. A: Bundle of several DNA chains (white arrows). The area in the white rectangle is enlarged in C. B: Height image of two fragments of double-stranded DNA. C: Height image (left) and corresponding phase image (right) of double-stranded DNA escaping from a capsid. In the inset, this DNA chain is seen to have some helical sections, with a pitch of ∼60 nm.
In Figure 5, A to C ▶ , virions treated with 0.5% sodium dodecyl sulfate are seen in the presence of thin fibers resembling DNA. The form of these fibers varies from what seems a complex bundle of many intertwined DNA chains (Figure 5A) ▶ to what appear to be individual double-stranded DNA molecules (Figure 5, B and C) ▶ . These thin DNA fibers were found to have diameters of 1.5 to 3 nm, which corresponds well to the 2- to 3-nm diameter of a double-stranded DNA double helix. The chain of Figure 5C ▶ , furthermore, appears to originate from the capsid at the top of the image, also indicating that the fibers are indeed DNA strands emerging from the HSV capsids.
Discussion
Using AFM, the major components of HSV-1, membrane, membrane proteins, tegument proteins, capsid, and DNA were directly visualized for single particles. Virions that were damaged or partially degraded were not detrimental, but provided additional perspectives on the virus structure. Using phase imaging, the membrane and its embedded proteins, despite the structural fragility and diverse composition, were readily seen. The tegument proteins were revealed as a fluid aggregate of protein molecules having little apparent order. Unfortunately, AFM cannot unambiguously identify the exact nature of the observed membrane components and tegument proteins that are observed, because it records topography alone. To accomplish this, more elaborate studies involving specific antibodies must be conducted.
The capsids seen in the AFM images do not exhibit the level of perfection implied by the cryo-EM/IR structure, although all are consistent, in a general sense, with that model. The adsorbed virions reveal the range of eccentricities that characterize the individual members of the purified population. Not only does AFM permit us to visualize the aperiodic features of the virions, it shows us the natural variation about the mean structure. When compared to EM single images (Figure 1A) ▶ , the resolution is similar or only slightly lower.
As the virions are degraded, additional features appear. Entire hexameric capsomeres are lost from the capsid surface, apparently as discrete units, and distinct holes remain in the capsids. As the effects of the detergent become more severe, extrusion of double-stranded DNA occurs, and it too can be visualized on the substrate surface.
Obtaining high AFM resolution on very flat crystal surfaces (mica, graphite, silicon, ionic crystals) is not critically dependent on the tip radius, because only a very small part of the tip is in contact with the sample surface, and that part alone determines the resolution. In the case of a surface with greater height variation, such as a virus capsid surface, a larger portion of the tip contacts the sample. Here, overall tip sharpness becomes more important in seeing details such as the capsomere channels and the deeper lying triplexes. Based on recent improvements in the fabrication of ultrasharp tips having a tip radius of ∼1 nm, as a result of tip sharpening (Nanosensors, Norderfriedrichskoog, Germany: superSharpSilicon Tips; http://www.nanosensors.com/newprod.htm; 2001) and nanotubes, 15-17 we expect that AFM resolution will considerably improve in the very near future. This may make AFM resolution comparable to that of conventional EM.
Pathogen diagnosis may depend, in general, on several techniques that agree on different aspects of the pathogen. These include immunological tests, enzyme tests, staining characteristics, cell and tissue morphology, and identification by some means of the infectious agent. It is in this last regard that AFM is particularly relevant to pathology, and the most important point here is that relatively crude cellular or tissue extracts are not essential, nor is purification, with the often attendant artifacts lengthy procedures introduce.
In addition to high-resolution imaging of pathogens themselves, of which we presented some results here, AFM potentially may be used to image immunoreactions. Its high-resolution capacity combined with its capability of imaging in aqueous environments make AFM well suited for this, and will further increase its use in the field of molecular pathogenesis. The same capacities make AFM suitable for the study of cell infection by viruses, which is currently performed in our laboratory.
Conclusions
The primary features of single particles of HSV originating from a relatively crude preparation were rapidly imaged with significant detail through the use of AFM. Although an overall resemblance to the capsid structure revealed by cryo-EM was seen, deviations from this inherent symmetric structure were observed as well.
The procedures and techniques applied here to image single herpesvirus particles are not in any way unique to that system. They can equally well be applied to other viruses. 4 With ongoing rapid advances in AFM tip fabrication, increased resolution is expected. These developments, together with AFM’s capability of visualizing biological samples in situ, may lead to increased use of AFM in the field of molecular pathology.
Footnotes
Address reprint requests to Alexander J. Malkin, Department of Molecular Biology and Biochemistry, University of California, Irvine, CA 92697-3900. E-mail: amalkin@uci.edu.
Supported by the National Aeronautics and Space Administration (grant NAG8-1569) and the Lawrence Livermore National Laboratory (grant MI-01-004).
References
- 1.Baker TS, Olson NH, Fuller SD: Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev 1999, 63:862-922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binnig G, Quate CF, Gerber CH: Atomic force microscope. Phys Rev Lett 1986, 56:930-933 [DOI] [PubMed] [Google Scholar]
- 3.Malkin AJ, Kuznetsov YUG, Lucas RW, McPherson A: Surface processes in the crystallization of turnip yellow mosaic virus visualized by atomic force microscopy. J Struct Biol 1999, 127:35-43 [DOI] [PubMed] [Google Scholar]
- 4.Kuznetsov YUG, Malkin AJ, Lucas RW, Plomp M, McPherson A: Imaging of viruses by atomic force microscopy. J Gen Virol 2001, 82:2025-2034 [DOI] [PubMed] [Google Scholar]
- 5.Zhou ZH, Prasad BVV, Jakana J, Rixon FJ, Chiu W: Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol 1994, 242:456-469 [DOI] [PubMed] [Google Scholar]
- 6.Booy FP, Trus BL, Newcomb WW, Brown JC, Conway JF, Steven AC: Finding a needle in a haystack: detection of a small protein (the12-kDa VP26) in a large complex (the 200 MDa capsid of herpes simplex virus). Proc Natl Acad Sci USA 1994, 91:5652-5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newcomb WW, Brown JC, Booy FP, Steven AC, Wall JS, Brown JC: Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol 1993, 232:499-511 [DOI] [PubMed] [Google Scholar]
- 8.Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W: Seeing the herpesvirus capsid at 8.5 Å. Science 2000, 288:877-880 [DOI] [PubMed] [Google Scholar]
- 9.Zhong Q, Inniss D, Kjoller K, Elings VB: Fractured polymer/silica fiber surface studied by tapping mode atomic force microscopy. Surf Sci 1993, 290:L688-L692 [Google Scholar]
- 10.Magonov SN: Encyclopedia of Analytical Chemistry. Meyers RA eds. 2000:p 7432 John Wiley & Sons, Chichester
- 11.Stannard LM, Fuller AO, Spear PG: Herpes simplex virus glycoproteins associated with different morphological entities projecting from the virion envelope. J Gen Virol 1986, 68:715-725 [DOI] [PubMed] [Google Scholar]
- 12.Steven AC, Spear PG: Structural Biology of Viruses. Burnett RM Garcea R eds. 1997:pp 312-351 Oxford University Press, New York
- 13.Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W: Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol 1999, 73:3210-3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voet D, Voet JG: Biochemistry. Voet D D Voet JG eds. 1995:p 286 John Wiley, New York
- 15.Hafner JH, Cheung CL, Lieber CM: Growth of nanotube for probe microscopy tips. Nature 1999, 398:761-762 [Google Scholar]
- 16.Hafner JH, Cheung CL, Lieber CM: Direct growth of single-walled carbon nanotube scanning probe microscopy tips. J Am Chem Soc 1999, 121:9750-9751 [Google Scholar]
- 17.Nishijima H, Kamo S, Akita S, Nakayama Y, Hohmura KI, Yoshimura SH, Takeyasu K: Carbon-nanotube tips for scanning probe microscopy: preparation by a controlled process and observation of deoxyribonucleic acid. Appl Phys Lett 1999, 74:4061-4063 [Google Scholar]