Abstract
Annexins (ANXs) are a large group of calcium-binding proteins participating in diverse important biological processes. ANXA10 is the least expressed new member of unknown function. We showed that ANXA10 mRNA was expressed in adult liver and hepatocellular carcinoma (HCC), but not in multiple adult and fetal tissues, cholangiocarcinoma, and several other common carcinomas. Of 182 unifocal primary HCCs, ANXA10 mRNA was dramatically reduced in 121 (66%), and the down-regulation correlated with p53 mutation (P = 0.024), early intrahepatic tumor recurrence (P = 0.0007), and lower 4-year survival (P = 0.0014). Down-regulation of ANXA10 was twofold more frequent in large than small HCCs (P = 0.0012), in grade II to III than grade I HCC (P < 0.00001), and in stage IIIA to IV than stage I to II HCC (P < 0.00001). Moreover, ANXA10 down-regulation and p53 mutation acted synergistically toward high-grade (P < 0.00001), high-stage HCC (P < 0.00001), and poorer prognosis (P = 0.0025). Our results indicate that the expression of the tissue- and tumor-restricted ANXA10 is a marker of liver cell differentiation and growth arrest, and its down-regulation associated with malignant phenotype of hepatocytes, vascular invasion, and progression of HCC, leading to poor prognosis. Thus, ANXA10 might serve as a new potential target of gene therapy for HCC.
The annexin (ANX) family is the largest single category of eukaryotic calcium-binding proteins without EF hands (the helix-loop-helix calcium-binding motif). 1 The ANXs play an important role in a broad range of physiological processes, including anti-coagulation, 2 endocytosis, exocytosis, 3,4 immunosuppression, 5 differentiation, 6-8 proliferation, 9,10 and inhibition of calcium channels, phospholipase A2, and protein kinase C. 7,11-14 There are at least 13 human ANX members, designated A1, A2, A3, A4, A5, 5′-A6, 3′-A6, A7, A8, A9, A10, A11, and A13. 15 The ANXs share a similar structure characterized by the presence of four or eight repeats of a 70-amino acid motif and a highly variable N-terminal end. 1,14,16 Unlike the conserved repeats, the amino-terminal domains of ANXs are highly variable, and may mediate the specific functions of individual ANXs. 1
Several ANXs are involved in tumorigenesis. ANXA1 is overexpressed in breast cancer and hepatocellular carcinoma (HCC), 17-19 ANXA2 in brain glial tumors and pancreatic cancer, 20-23 and ANXA8 in acute promyelocytic leukemia. 13,24 Overexpression of ANXA1 correlates positively with metastatic potential of breast cancer, 19 and increased growth of tumor cells inoculated in nude mice. 25 In contrast, ANXA6 has tumor suppressive activity in squamous cell carcinoma, 26 and a decrease in ANXA6 and ANXA7 proteins is associated with malignant phenotype and the metastatic potential of melanoma. 27,28 Moreover, ANXA4 plays a role in chemoresistance. 29 Hence, there are at least two categories of ANXs implicated in tumor progression and tumor suppression, and different ANXs play important and complex roles in cancer.
ANXA10, a novel member of the ANX family, has several distinct features, including rare expression, codon deletion in conserved repeat 3, and an unusual ablation of the type II calcium-binding sites in tetrad core repeats. 15 The loss of type II calcium-binding sites is a fundamental deviation from ANX structure conservation and undoubtedly has functional consequences for membrane association and aggregation properties. 30 ANXA10 is mapped to chromosome 4q33. 31 Chromosome 4q is one of the most common regions with a high frequency of allelic loss in HCC. 32-35 Introduction of normal human chromosome 4 leads to suppressed tumorigenicity of teratocarcinoma PA-1 cells in nude mice, suggestive of a putative tumor suppressor gene on chromosome 4. 35 The biological function of ANXA10 and its role in HCC are not known. Using differential display, we found a cDNA clone identical to ANXA1015 that was expressed in liver and often down-regulated in HCC. In this study, we correlate the mRNA expression of ANXA10 in HCC with clinicopathological features. We show that ANXA10 might be a potential tumor suppressor gene because its down-regulation is associated with malignant phenotype of liver cells, and correlates with vascular invasion and progression of HCC.
Materials and Methods
Study Group
Between 1987 and 1997, 909 surgically resected primary and 127 recurrent HCCs were pathologically assessed at the National Taiwan University Hospital. The tissue samples were immediately cut into small pieces, snap-frozen in liquid nitrogen, and stored in deep freezers. Of these, 187 patients who already had RNA samples taken from resected HCCs were analyzed for ANXA10 mRNA levels, including 182 unifocal and 5 multifocal primary HCCs. To elucidate the clinical implication of ANXA10 expression, the 182 cases of unifocal primary HCC, as previously defined, 36-39 were selected for clinicopathological analysis. Multifocal HCC was excluded from this correlation because of incomplete sampling of the tumor nodules for analysis and their variation in pathological features. To study the tissue and tumor distribution, we also examined the mRNA expression of ANXA10 in multiple adult tissues and several common tumor tissues obtained from surgical pathology specimens, and multiple fetal tissues obtained from autopsy.
The intrahepatic tumor recurrence (IHTR) was based on imaging diagnosis with ultrasonography and/or computed tomography, supplemented with elevated serum α-fetoprotein (AFP). Until the end of October 2001, tumor recurrence was detected in 113 patients. Among the 182 patients studied, 156 were eligible for the evaluation of early IHTR (≤1 year). Twenty-six patients who died within 1 year after resection and had no information about or were negative for IHTR were excluded from the evaluation of early recurrence.
Histological Study and Tumor Staging
The tumor grade was simply divided into three groups: well (grade I), moderately (grade II), and poorly differentiated HCC (grades III and IV). Resected unifocal HCC was staged as stage I, II, IIIA, IIIB, and IV as previously described. 40 Briefly, stage I HCC is encapsulated and without parenchymal invasion. Stage II HCC has liver invasion, but no vascular invasion. Stage IIIA HCC has invasion of small vessels in the tumor capsule. Stage IIIB and IV HCCs have invasion of portal vein branches. The pathological stage of HCC correlates closely with survival. 40
Differential Display Analysis
Differential display was performed according to the recommendation of the manufacturer using the RNAimage kit (GenHunter Corp., Nashville, TN). Total RNA (0.2 μg) was reverse-transcribed in a mixture containing 100 U MMLV reverse transcriptase, 20 μmol/L dNTP, and 0.2 μmol/L anchor primer H-T11C (5′-AAGCT11C-3′). The reverse transcription products (2 μl) were subjected to polymerase chain reaction (PCR) amplification in a PCR mixture containing 2 μmol/L dNTP, 0.2 μmol/L H-T11C (5′-AAGCT11C-3′), 25 mCi/mmol [α-33P]dATP (Amersham, Buckinghamshire, England), 1 U Vent DNA polymerase (New England Biolabs, Inc. Beverly, MA), 0.2 μmol/L arbitrary primer HAP35 (5′-AAGCTTCAGGGCA-3′). PCR was done in Perkin-Elmer’s 9600 thermocycler (94°C for 30 seconds, 40°C for 2 minutes, 72°C for 1 minute for 40 cycles, and a final elongation at 72°C for 5 minutes). The amplification products were separated by 6% denaturing polyacrylamide gel electrophoresis. The dried gel was then exposed by film autoradiography. The differentially expressed band was cut, eluted, and sequenced.
Reverse Transcriptase (RT)-PCR)
RT-PCR assays were used for large-scale quantitative measurements for ANXA10 mRNA levels, as previously described, 41 using S26 ribosomal protein mRNA as internal control. 42 Briefly, 2 μl of RT product, 1.25 U Pro Taq DNA polymerase (Protech Technology Enterprise Co, Taipei, Taiwan), Pro Taq buffer, and 200 μmol/L each dATP, dTTP, dGTP, and dCTP were mixed with primer pairs for ANXA10 and S26 in a total volume of 30 μl. The PCR was performed in an automated DNA thermal cycler 480 (Perkin-Elmer/Cetus). The PCR was stopped at the exponential phases, 29 cycles for ANXA10 and 22 cycles for S26. These experiments were repeated when the S26 loading showed inappropriate template amount for PCR. The primers used included the sense primer ANXA10-A: 5′-TTGTTCTCTGTGTTCGAGAGAAACC-3′, and anti-sense primer ANXA10-F: 5′-GTAGGCAAATTCAGGATAGTAGGC-3′.
The products were electrophoresed on 2% agarose gel. The ANXA10 mRNA expression level was determined by the ratio of signal intensity of ANXA10 to that of S26 measured by 1D Image Analysis Software (Kodak Digital Science, Rochester, NY), and scored as high (ratio ≥ 1.0), intermediate (0.5 ≤ ratio < 1.0), trace (ratio < 0.5), and negative (or immeasurable). An ANXA10/S26 ratio <0.5 was regarded as down-regulation, because ANXA10 mRNA level in liver was rarely less than a ratio of 0.5.
Analysis of p53 Mutation
The analysis of p53 mutations was done as described. 37,39
Follow-Up Observation
To minimize the influence of second and third recurrent primary HCC, the endpoint for follow-up was set at the 4th year. Of the 182 cases studied, 179 (98%) had been followed for >4 years or until death.
Statistics
The analyses were performed using the Statistica for Windows software (Statsoft, Inc., Chicago, IL). Two-tailed chi-square and Fisher exact tests were used for univariate analysis. The cumulative survival after tumor removal was calculated with the log-rank test. P values <0.05 were considered statistically significant.
Results
Cloning and Expression of ANXA10 mRNA
We used differential display for the analysis of genes aberrantly expressed in HCC, and cloned a 192-bp cDNA fragment that was consistently expressed in liver and often down-regulated in HCC. This cDNA clone was identical to ANXA1015 (GenBank GI no. 9625245). A nearly full-length cDNA of 1268-bp long was subsequently cloned from Hep3B cell line by RT-PCR amplification; the sequence was identical to ANXA10. 15 The preferential down-regulation of ANXA10 in HCC was verified by Northern blotting (Figure 1) ▶ . Using RT-PCR for large-scale analysis, ANXA10 mRNA was expressed abundantly in liver, and in low level in only 12 (6.4%) of 176 livers harboring benign (focal nodular hyperplasia) and malignant tumors (HCC, cholangiocarcinoma, metastatic carcinoma). In contrast, ANXA10 mRNA was dramatically decreased in 121 of 182 unifocal primary HCCs (66%) as compared to paired nontumor livers (Table 1 ▶ ; Figure 2 ▶ ).
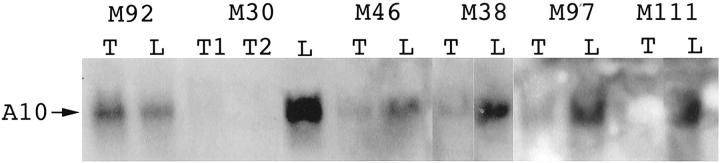
Figure 1.
Northern blot for ANXA10 using a cDNA fragment cloned from differential display as probe. ANXA10 mRNA (A10) of 1.4 kb was detected in the livers (L) and dramatically decreased in HCC (T) in five cases.
Table 1.
ANXA10 mRNA Expression in Benign and Malignant Human Tumors
| Tumor type | ANXA10 mRNA expression | |
|---|---|---|
| Expressed | Not expressed or dramatically decreased | |
| Hepatocellular carcinoma (n = 182) | 61 | 121 (66%) |
| Hepatoblastoma (n = 5) | 0 | 5 (100%) |
| Cholangiocarcinoma (n = 2) | 0 | 2 |
| Metastatic colorectal carcinoma (n = 10)* | 0 | 10 (100%) |
| Focal nodular hyperplasia (n = 9) | 8 | 1 (11%) |
| Ovarian carcinoma (n = 16) | 0 | 16 (100%) |
| Lung carcinoma (n = 3) | 0 | 3 |
| Breast carcinoma (n = 10) | 0 | 10 (100%) |
*Includes one ovarian and one breast, and eight metastatic colorectal carcinomas.
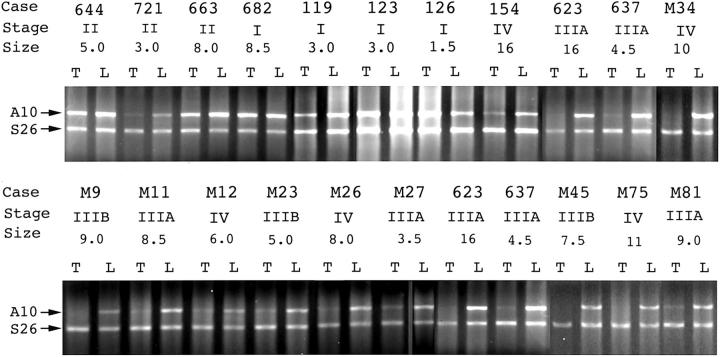
Figure 2.
RT-PCR of ANXA10 mRNA in paired HCC and liver. ANXA10 mRNA (A10) was dramatically decreased in HCC (T) in 16 of 22 representative cases. Stage, tumor stage (I to IV); size, tumor size (cm).
Tissue-Specific Expression of ANXA10 mRNA in Fetal and Adult Tissues
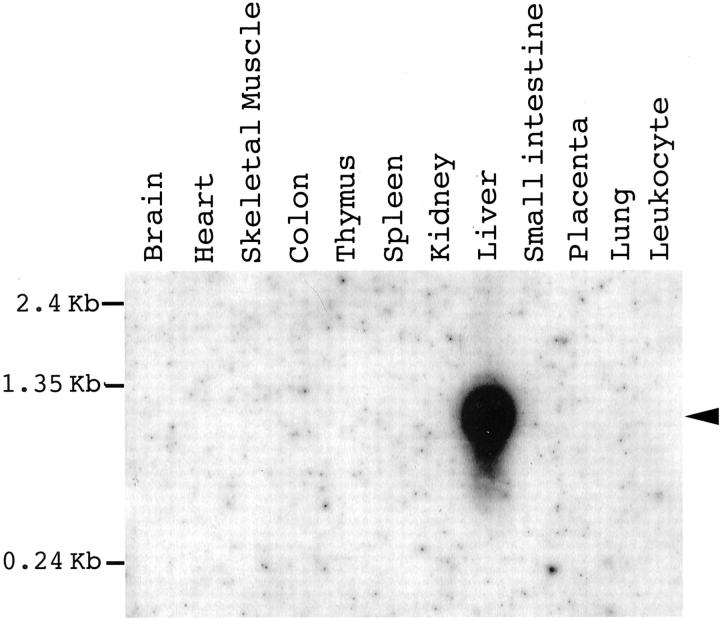
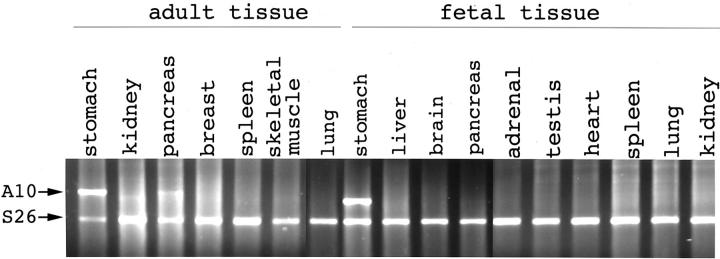
To elucidate the tissue-specific expression of the ANXA10, RNA samples taken from multiple fetal and adult tissues were analyzed. Using polyA+ RNA blot obtained from multiple adult tissues (RPN4800; Amersham Pharmacia Biotech, Buckinghamshire, UK), a single ANXA10 transcript of ∼1.3 kb was identified in adult liver alone (Figure 3) ▶ . By RT-PCR analysis, ANXA10 mRNA was expressed only in a very limited number of adult tissues, abundant in liver and stomach, low in pancreas, and not expressed in other adult tissues, such as lung, heart, breast, kidney, and spleen (Figure 4) ▶ . ANXA10 mRNA was also undetectable in most fetal tissues, such as brain, thyroid, lung, heart, liver, kidney, adrenal gland, and testes. ANXA10 mRNA was expressed abundantly in fetal stomach (Figure 4) ▶ . Among three fetal livers taken from fetuses less than gestation age of 22 weeks and without congenital anomalies, ANXA10 mRNA was expressed in low level in one and not detected in two (Figure 4) ▶ .
Figure 3.
Northern blot analysis of ANXA10 in multiple adult tissues. The commercially obtained Hybond Northern blot (Amersham) containing polyA+ RNAs was hybridized with a 1.2-kb cDNA fragment containing the whole coding region and 3′-UTR of ANXA10. A single ANXA10 transcript of ∼1.3 kb (arrowhead) was identified in adult liver alone.
Figure 4.
Expression of ANXA10 in multiple adult and fetal tissues. ANXA10 mRNA (A10) was expressed in abundance in adult and fetal stomach, in low level in adult pancreas, but undetectable in most adult tissues and fetal tissues.
Expression of Annexin A10 in Benign and Malignant Liver Tumors
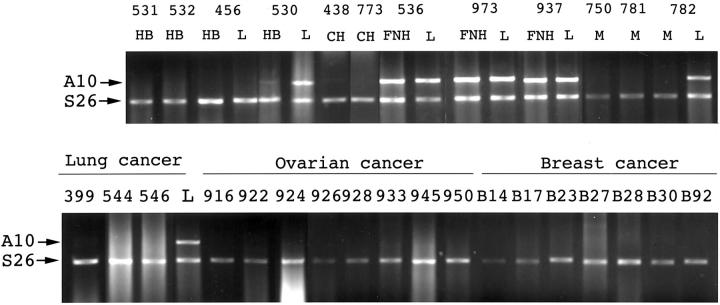
ANXA10 mRNA was expressed in benign liver lesions such as focal nodular hyperplasia, and decreased dramatically in hepatoblastoma (Table 1) ▶ . To clarify the tumor-specific distribution, we analyzed the ANXA10 mRNA expression in several common carcinomas of other anatomical sites. As shown in Table 1 ▶ , ANXA10 mRNA was undetectable or weakly expressed in carcinomas of the ovary, breast, colon, and lung, and metastatic carcinomas of the liver (Figure 5) ▶ .
Figure 5.
Expression of ANXA10 in other malignant tumors. ANXA10 mRNA (A10) was detected in focal nodular hyperplasia (FNH), but not in hepatoblastoma (HB), cholangiocarcinoma (CH), metastatic colon carcinoma (M), lung carcinoma (399, adenosquamous cell carcinoma; 544, squamous cell carcinoma; 546, spindle cell cancer), breast carcinoma, and ovarian carcinoma (926, 950, endometrioid carcinoma; 916, serous cystadenocarcinoma; 922, 928, adenocarcinoma; 924, yolk sac tumor; 945, mucinous cystadenocarcinoma; 933, clear cell carcinoma).
Clinicopathological Correlations of ANXA10 mRNA Expression
To elucidate the biological significance of its expression, we correlated the ANXA10 expression with clinicopathological features. As shown in Table 2 ▶ , the decreased ANXA10 expression was closely associated with serum AFP elevation (P < 0.00001). The ANXA10 expression did not correlate with age, gender, hepatitis B virus (HBV) infection, and liver cirrhosis (data not shown). Histopathologically, down-regulation of ANXA10 was found more often in large HCCs than in HCCs <3 cm (P = 0.0012), in grade II and III than in grade I HCCs (P < 0.00001), and in HCCs with vascular invasion (stage IIIA to IV) than in stage I to II HCCs that has no vascular invasion (P < 0.00001).
Table 2.
Correlation of ANXA10 mRNA Expression with Clinicopathological Features in 182 Patients with Unifocal Primary Hepatocellular Carcinoma
| ANXA10 mRNA expression | |||
|---|---|---|---|
| Normal expression (n = 61) | Down-regulation (n = 121) | P value | |
| AFP | |||
| <320 ng/ml (n = 97) | 51 | 46 (47%) | <0.00001 |
| ≥320 ng/ml (n = 85) | 10 | 75 (88%) | |
| p53 mutation | |||
| Yes (n = 77) | 19 | 58 (75%) | 0.024 |
| No (n = 79) | 33 | 46 (58%) | |
| Tumor size | |||
| ≤3 cm (n = 45) | 24 | 21 (47%) | 0.0012 |
| >3 cm (n = 137) | 37 | 100 (73%) | |
| Tumor grade | |||
| I (n = 39) | 25 | 14 (36%) | <0.00001 |
| II–III (n = 142) | 36 | 106 (75%) | |
| Tumor stage | |||
| I–II (n = 83) | 42 | 41 (49%) | <0.00001 |
| IIIA–IV (n = 99) | 19 | 80 (81%) | |
| Early recurrence (≤1-yr)* | 14/52 (27%) | 58/104 (56%) | 0.0007 |
*Tumor recurrence in the liver was detected ≤12 months after resection of the primary HCC.
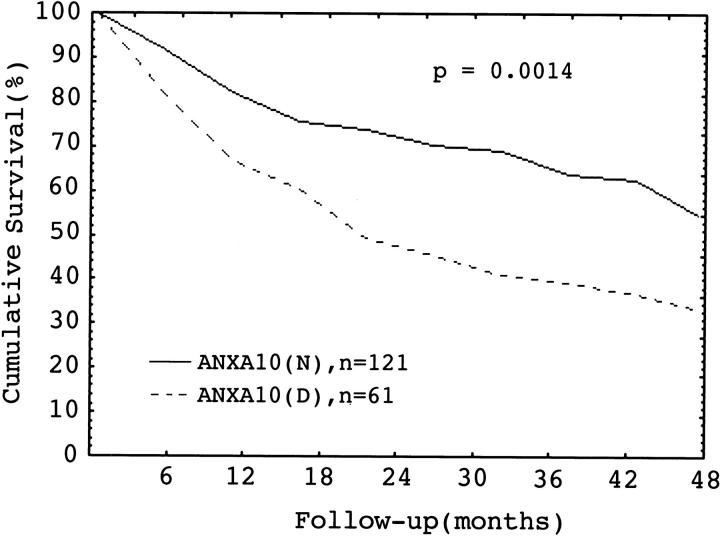
Down-regulation of ANXA10 was associated with higher frequency of early intrahepatic tumor recurrence (P = 0.0007). Furthermore, HCC with down-regulation of ANXA10 was associated with a lower 4-year patient survival rate than HCC with normal ANXA10 expression, 54% versus 33%, P = 0.0014 (Figure 6) ▶ .
Figure 6.
Cumulative survival in HCC in relation to ANXA10 mRNA expression. A decreased ANXA10 (D) expression had a lower 4-year survival than normal expression (N), P = 0.0014 (log-rank test).
Correlation and Synergy between ANXA10 Expression and p53 Mutations
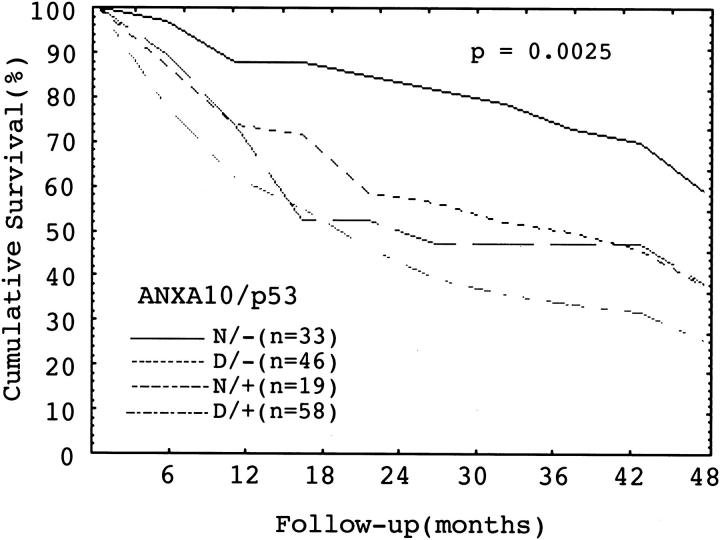
Because p53 mutation correlates closely with tumor aggressiveness and unfavorable prognosis of HCC, 37,39 we analyzed the potential interplay between these two important unfavorable prognostic factors. In this study, p53 mutation was found in 77 (49%) of 156 HCCs examined. Down-regulation of ANXA10 correlated with p53 mutation, P = 0.024 (Table 2) ▶ . HCC with both ANXA10 down-regulation and p53 mutation had the highest frequencies of grade II to III and stage IIIA to IV tumors than other groups, P < 0.00001 and P < 0.00001, respectively (Table 3) ▶ . Moreover, HCCs with both ANXA10 down-regulation and p53 mutation also had the most frequent early intrahepatic tumor recurrence, P = 0.0001 (Table 3) ▶ , and the worst 4-year survival, P = 0.0025 (Figure 7) ▶ .
Table 3.
Synergistic Effect between ANXA10 Down-Regulation and p53 Mutation in Unifocal Hepatocellular Carcinoma
| Feature | Down-Regulation of ANXA10/p53 mutation | P value | |||
|---|---|---|---|---|---|
| No/No n = 33 | Yes/No n = 46 | No/Yes n = 19 | Yes/Yes n = 58 | ||
| Tumor grade | |||||
| I | 16 (48%) | 9 (20%) | 4 (21%) | 4 (7%) | <0.00001 |
| II–III | 17 | 37 | 15 | 54 | |
| Tumor stage | |||||
| I–II | 26 (79%) | 23 (50%) | 10 (53%) | 13 (22%) | <0.00001 |
| III–IV | 8 | 23 | 9 | 45 | |
| Early intrahepatic tumor recurrence* | 7/30 (23%) | 15/38 (39%) | 6/15 (40%) | 36/53 (68%) | 0.0001 |
*Tumor recurs in the liver ≤12 months after resection of the primary HCC.
Figure 7.
Cumulative survival in HCC in relation to normal (N) or decreased (D) expression of ANXA10 and presence (+) or absence (−) of p53 mutation. HCC with decreased ANXA10 expression and p53 mutation had the lowest 4-year survival, P = 0.0025 (log-rank test).
Discussion
Using differential display analysis in paired HCC and liver tissue, a cDNA band preferentially decreased in HCC was cloned. The partial cDNA sequence was identical to liver ANXA10, a novel member of the ANX family with unique loss of the calcium-binding site and of unknown function. In this study, we demonstrated that ANXA10 mRNA was expressed in a tissue-restricted and developmentally regulated pattern. In contrast to fetal liver, ANXA10 mRNA was expressed abundantly in adult liver. Among the more than 13 ANX family members, different ANXs have unique tissue and cell distribution in the vertebrates. ANXA2, -A5, -A6, and -A11 are ubiquitous, whereas ANXA3, -A8, and -A13 have more restricted tissue distribution. 1,43,44 These findings indicate their functions are tissue-specific. 45,46 Although the function of ANXA10 remains to be clarified, our results indicate that ANXA10 is developmentally regulated in liver, and may serve as a potential marker of differentiation, maturation, or growth arrest of hepatocytes. This suggestion is strengthened by the finding that ANXA10 mRNA was expressed abundantly in benign liver lesions such as focal nodular hyperplasia. ANXA10 mRNA was also expressed in HCC and hepatoblastoma, although often decreased. In contrast, ANXA10 mRNA was not detected in cholangiocarcinoma, and in several common carcinomas of other anatomical sites, where ANXA10 mRNA was also not expressed in their normal tissues, such as colorectal, breast, and ovarian carcinomas. These findings suggest that ANXA10 mRNA expression is a marker of hepatocellular origin. The ANXA10 mRNA expression in adult liver and malignant liver cell tumors seems paradoxical. However, we found a close association of normal ANXA10 expression with grade I HCC, 64% versus 25%, P < 0.00001. This finding supports the suggestion that ANXA10 expression indeed might represent tumor cell differentiation and growth arrest in HCC.
Human ANXA10 is mapped to chromosome 4q33. 31 Chromosome 4q is one of the most common regions with a high frequency of allelic loss in HCC. 33-35,47 Chromosomal alterations at 4q are of great interest because it is a unique trait for HCC, 32,33 and chromosome 4q contains genes encoding growth factors or genes expressed predominantly in the liver. These include albumin, alcohol dehydrogenase (ADH3), fibrinogen, AFP, and UDP-glucuronyl-transferase. In this study, we showed that ANXA10, a new member of genes at 4q, was expressed predominantly in liver. To elucidate its role in HCC, we demonstrated for the first time that HCC with ANXA10 mRNA expression was dramatically down-regulated in 121 (66%) of 182 unifocal primary HCCs. The down-regulation was associated with more frequent AFP elevation (P < 0.00001), and grade II and III tumors (P < 0.00001). These results are in accord with our previous observation that poor differentiation of HCC correlates with AFP mRNA expression and serum AFP elevation. 38 During the tumor progression, HCC tends to spread throughout the liver via the portal vein branches even at the advanced stage, and the extent of portal vein invasion is the most crucial factor for tumor staging and prognosis. 40 However, the molecular mechanisms for portal vein spread remain primarily unknown. In this study, down-regulation of ANXA10 was nearly twofold more frequent in large than in small HCCs ≤ 3 cm (P = 0.0012), in grade II to III than in grade I HCC (P < 0.00001), and in stage IIIA to IV HCC that has various extents of vascular invasion than in stage I to II HCC that has no vascular invasion (P < 0.00001). These results suggest that down-regulation of ANXA10 contributes to the progression and metastasis of HCC. Furthermore, down-regulation of ANXA10 was associated with early IHTR (P = 0.0007), which is mostly because of true recurrence of HCC and associated with more unfavorable prognosis. 48 Indeed, HCC with ANXA10 down-regulation had a worse 4-year survival (P = 0.0014). Hence, down-regulation of ANXA10 may serve as a potential molecular marker to identify a subset of HCCs with high risk for early IHTR. In this study, our observations provide evidence that ANXA10 is a new member of ANXs that may possess metastasis suppression activity, 26-28 However, the molecular mechanisms for the metastasis-suppressive activity of ANXA10 in HCC warrants more studies for better understanding.
Mutation of p53 occurs frequently in HCC and is associated with tumor invasion and unfavorable outcome. 37,39 In this study, down-regulation of ANXA10 correlated with p53 mutation, P = 0.024. The molecular mechanism for this association is currently unknown, but the potential interplay between these two factors in HCC needs to be clarified. We showed that HCC with both ANXA10 down-regulation and p53 mutation had the highest frequencies of grade II to III tumor and vascular invasion (stage IIIA to IV HCC), significantly higher than other groups (P < 0.00001 and P < 0.00001, respectively). Moreover, HCC with ANXA10 down-regulation and p53 mutation had more frequent early IHTR (P = 0.0001), and worse 4-year survival (P = 0.0025). These results indicate that combined alterations of the two molecular markers act synergistically toward poorly differentiated and more aggressive HCC. Our results also suggest that a combinatorial analysis of multiple genetic alterations may help to identify subsets of HCC with different biological course and prognosis. The tissue- and tumor-restricted expression makes ANXA10 a potential ideal target of gene therapy for HCC.
Footnotes
Address reprint requests to Professor Dr. Hey-Chi Hsu, Department of Pathology, National Taiwan University Hospital, 7 Chung-Shan South Rd., Taipei, Taiwan. E-mail: heychi@ha.mc.ntu.edu.tw.
Supported by the National Health Research Institute, Department of Health of the Republic of China, Taiwan (DOH88-HR-701; NHRI-GT-EX89B701L to H. C. H.).
References
- 1.Smith PD, Moss SE: Structural evolution of the annexin supergene family. Trends Genet 1994, 10:241-246 [DOI] [PubMed] [Google Scholar]
- 2.Tait JF, Sakata M, Mcmullen BA, Miao CH, Funakoshi T, Hendrickson LE, Fujikawa K: Placental anticoagulant proteins: isolation and comparative characterization of four members of the lipocortin family. Biochemistry 1998, 27:6268-6276 [DOI] [PubMed] [Google Scholar]
- 3.Creutz CE: The annexins and exocytosis. Science 1992, 258:924-931 [DOI] [PubMed] [Google Scholar]
- 4.Emans N, Gorvel JP, Walter C, Gerke V, Kellner R, Griffiths G, Gruenberg J: Annexin II is a major component of fusogenic endosomal vesicles. J Cell Biol 1993, 120:1357-1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakata T, Iwagami S, Tsuruta Y, Suzuki S, Suzuki R: Study of natural lipocortin I. A potent mediator for macrophage-mediated immunosuppression in tumor-bearing mice. J Immunol 1993, 151:4964-4972 [PubMed] [Google Scholar]
- 6.Ohnishi M, Tokuda M, Masaki M, Fujimura T, Tai Y, Matsui H, Itano T, Ishida T, Takahara J, Konishi R, Hatase O: Changes in annexin I and II levels during the postnatal development of rat pancreatic islets. J Cell Sci 1994, 107:2117-2125 [DOI] [PubMed] [Google Scholar]
- 7.Violette SM, King I, Browning JL, Pepinsky RB, Wallner BP, Sartorelli AC: Role of lipocortin I in the glucocorticoid induction of the terminal differentiation of a human squamous carcinoma. J Cell Physiol 1990, 142:70-77 [DOI] [PubMed] [Google Scholar]
- 8.Horlick KR, Ganjianpour M, Frost SC, Nick HS: Annexin-I regulation in response to suckling and rat mammary cell differentiation. Endocrinology 1990, 128:1574-1579 [DOI] [PubMed] [Google Scholar]
- 9.Schlaepfer DD, Haigler HT: Expression of annexins as a function of cellular growth state. J Cell Biol 1990, 111:229-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masaki M, Tokuda M, Fujiwara T, Ohnishi M, Tai Y, Miyamoto K, Itano T, Matsui H, Watanabe S, Sogawa K, Yamada T, Konishi R, Nishioka M, Hatase O: Involvement of annexin I and annexin II in hepatocyte proliferation: can annexins I and II be markers for proliferative hepatocytes? Hepatology 1994, 20:425-435 [PubMed] [Google Scholar]
- 11.Schlaepfer DD, Jones J, Haigler HT: Inhibition of protein kinase C by annexin V. Biochemistry 1993, 31:1886-1891 [DOI] [PubMed] [Google Scholar]
- 12.Rand JH: Annexinopathies—a new class of diseases. N Engl J Med 1999, 340:1035-1036 [DOI] [PubMed] [Google Scholar]
- 13.Sarkar A, Yang P, Fan YH, Mu ZM, Hauptmann R, Adolf GR, Stass SA, Chang KS: Regulation of the expression of annexin VIII in acute promyelocytic leukemia. Blood 1994, 84:279-286 [PubMed] [Google Scholar]
- 14.Dubois T, Oudinet JP, Mira JP, Russo-Marie F: Annexins and protein kinase C. Biochim Biophys Acta 1996, 1313:290-294 [DOI] [PubMed] [Google Scholar]
- 15.Morgan RO, Jenkins NA, Gilbert DJ, Copeland NG, Balsara BR, Testa JR, Fernandez MP: Novel human and mouse annexin A10 are linked to the genome duplications during early chordate evolution. Genomics 1999, 60:40-49 [DOI] [PubMed] [Google Scholar]
- 16.Benz J, Hofmann A: Annexins: from structure to function. Biol Chem 1997, 78:177-183 [PubMed] [Google Scholar]
- 17.Masaki T, Tokuda M, Ohnishi M, Watanabe S, Fujimura T, Miyamoto K, Itano T, Matsui H, Arima K, Shirai M, Maeba T, Sogawa K, Konishi R, Taniguchi K, Hatanaka Y, Hatase O, Nishioka M: Enhanced expression of the protein kinase substrate annexin I in human hepatocellular carcinoma. Hepatology 1996, 24:72-81 [DOI] [PubMed] [Google Scholar]
- 18.Ahn SH, Sawada H, Ro JY, Nicolson GL: Differential expression of annexin I in human mammary ductal epithelial cells in normal and benign and malignant breast tissues. Clin Exp Metastasis 1997, 15:151-156 [DOI] [PubMed] [Google Scholar]
- 19.Pencil SD, Toh Y, Nicolson GL: Candidate metastasis-associated genes of the rat 13762NF mammary adenocarcinoma. Breast Cancer Res Treat 1993, 25:165-174 [DOI] [PubMed] [Google Scholar]
- 20.Vishwanatha JK, Chiang Y, Kumble KD, Hollingsworth MA, Pour PM: Enhanced expression of annexin II in human pancreatic carcinoma cells and primary pancreatic cancers. Carcinogenesis 1993, 14:2575-2579 [DOI] [PubMed] [Google Scholar]
- 21.Roseman BJ, Bollen A, Hsu J, Lamborn K, Israel MA: Annexin II marks astrocytic brain tumors of high histologic grade. Oncol Res 1994, 6:561-567 [PubMed] [Google Scholar]
- 22.Nygaard SJ, Haugland HK, Kristoffersen EK, Lund-Johansen M, Laerum OD, Tysnes OB: Expression of annexin II in glioma cell lines and in brain tumor biopsies. J Neurooncol 1998, 38:11-18 [DOI] [PubMed] [Google Scholar]
- 23.Paciucci R, Tora M, Diaz VM, Real FX: The plasminogen activator system in pancreas cancer: role of t-PA in the invasive potential in vitro. Oncogene 1998, 16:625-633 [DOI] [PubMed] [Google Scholar]
- 24.Chang KS, Wang G, Freireich EJ, Daly M, Naylor SL, Trujillo JM, Stass SA: Specific expression of the annexin VIII gene in acute promyelocytic leukemia. Blood 1992, 79:1802-1810 [PubMed] [Google Scholar]
- 25.Frey BM, Reber BFX, Vishwanath B, Escher G, Frey FJ: Annexin I modulates cell functions by controlling intracellular calcium release. FASEB J 1999, 13:2235-2245 [DOI] [PubMed] [Google Scholar]
- 26.Theobald J, Hanby A, Patel K, Moss SE: Annexin VI has tumor-suppressor activity inhuman A431 squamous epithelial carcinoma cells. Br J Cancer 1995, 71:786-788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francia G, Mitchell SD, Moss SE, Hanby AM, Marshall JF, Hart IR: Identification by differential display of annexin-VI, a gene differentially expressed during melanoma progression. Cancer Res 1996, 56:3855-3858 [PubMed] [Google Scholar]
- 28.Kataoka TR, Ito A, Asada H, Watabe K, Nishiyama K, Nakamoto K, Itami S, Yoshikawa K, Ito M, Nojima H, Kitamura Y: Annexin VII as a novel marker for invasive phenotype of malignant melanoma. Jpn J Cancer Res 2000, 91:75-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han EKH, Tahir SK, Cherian SP, Collins N, Ng SC: Modulation of pacitaxel resistance by annexin IV in human cancer cell lines. Br J Cancer 2000, 83:83-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bitto E, Cho W: Roles of individual domains of annexin I in its vesicle binding and vesicle aggregation: a comprehensive study. Biochemistry 1998, 37:10231-10237 [DOI] [PubMed] [Google Scholar]
- 31.Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tomé P, Hui L, Matise TC, McKusick KB, Beckman JS, et al: A physical map of 30,000 human genes. Science 1998, 282:744-746 [DOI] [PubMed] [Google Scholar]
- 32.Yeh SH, Chen PJ, Lai MY, Chen DS: Allelic loss on chromosomes 4q and 16q in hepatocellular carcinoma: association with elevated alpha-fetoprotein production. Gastroenterology 1996, 110:184-192 [DOI] [PubMed] [Google Scholar]
- 33.Nagai H, Pineau P, Tiollais P, Buendia MA, Dejean A: Comprehensive allelotyping of human hepatocellular carcinoma. Oncogene 1997, 14:2927-2933 [DOI] [PubMed] [Google Scholar]
- 34.Bando K, Nagai H, Matsumoto S, Koyama M, Kawamura N, Onda M, Emi M: Identification of a 1-cM region of common deletion on 4q35 associated with progression of hepatocellular carcinoma. Genes Chromosom Cancer 1999, 25:284-289 [PubMed] [Google Scholar]
- 35.Wong N, Lai P, Lee SW, Fan S, Pang E, Liew CT, Sheng Z, Lau JW, Johnson PJ: Assessment of genetic changes in hepatocellular carcinoma detected by comparative genomic hybridization: relationship to disease stage, tumor size, and cirrhosis. Am J Pathol 1999, 154:37-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu HC, Chiou TJ, Chen ZY, Lee CS, Lee PH, Peng SY: Clonality and clonal evolution of hepatocellular carcinoma with multiple nodules. Hepatology 1991, 13:923-928 [PubMed] [Google Scholar]
- 37.Hsu HC, Tseng HJ, Lai PL, Lee PH, Chen DS, Peng SY: Expression of p53 gene in 184 unifocal hepatocellular carcinomas (HCC): association with tumor growth and invasiveness. Cancer Res 1993, 53:4691-4694 [PubMed] [Google Scholar]
- 38.Peng SY, Lai PL, Chu JS, Lee PH, Tsung PT, Chen DS, Hsu HC: Expression and hypomethylation of alpha-fetoprotein gene in unicentric and multicentric human hepatocellular carcinomas. Hepatology 1993, 17:35-41 [PubMed] [Google Scholar]
- 39.Hsu HC, Peng SY, Lai PL, Chu JS, Lee PH: Mutations of p53 gene in hepatocellular carcinoma (HCC) correlate with tumor progression and patient prognosis: a study of 138 patients with unifocal HCC. Int J Oncol 1994, 4:1341-1347 [DOI] [PubMed] [Google Scholar]
- 40.Hsu HC, Jeng YM, Mao TL, Chu JS, Lai PL, Peng SY: β-Catenin mutations are associated with a subset of low stage hepatocellular carcinoma negative for hepatitis B virus and with favorable prognosis. Am J Pathol 2000, 157:763-770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng SY, Chou SP, Hsu HC: Association of downregulation of cyclin D1 and of overexpression of cyclin E with p53 mutation, high tumor grade and poor prognosis in hepatocellular carcinoma. J Hepatol 1998, 29:281-289 [DOI] [PubMed] [Google Scholar]
- 42.Vincent S, Marty L, Fort P: S26 ribosomal protein RNA: an invariant control for gene regulation experiments in eucaryotic cells and tissues. Nucleic Acids Res 1993, 21:1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raynal P, Pollard HB: Annexins: the problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim Biophys Acta 1994, 1197:63-93 [DOI] [PubMed] [Google Scholar]
- 44.Morgan RO, Fernandez MP: Annexin gene structures and molecular evolutionary genetics. Cell Mol Life Sci 1997, 53:508-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pepinsky RB, Tizard R, Mattaliano RJ, Sinclair LK, Miller GT, Browning JL, Chow EP, Burne C, Huang KS, Pratt D, Wachter L, Hession C, Frey AZ, Wallner BP: Five distinct calcium and phospholipid binding proteins share homology with lipocortin I. J Biol Chem 1988, 263:10799-10811 [PubMed] [Google Scholar]
- 46.Kaetzel MA, Hazarika P, Dedman JR: Differential tissue expression of three 35-kDa annexin calcium-dependent phospholipid-binding proteins. J Biol Chem 1989, 264:14463-14470 [PubMed] [Google Scholar]
- 47.McGowan-Jordan IJ, Speevak MD, Blakey D, Cevrette M: Suppression of tumorigenicity in human teratocarcinoma cell line PA-1 by introduction of chromosome 4. Cancer Res 1994, 54:2568-2572 [PubMed] [Google Scholar]
- 48.Poon RTP, Fan ST, Ng IOL, Lo CM, Liu CL, Wong J: Different risk factors and prognosis for early and late intrahepatic recurrence after resection of hepatocellular carcinoma. Cancer 2000, 89:500-507 [PubMed] [Google Scholar]