Abstract
Aberrant crypt foci (ACF) are postulated to be the earliest precursor lesion in colorectal carcinogenesis, and CpG island methylation has been described as an important molecular pathway. We therefore studied methylation in ACF from patients with familial adenomatous polyposis (FAP) or sporadic colorectal cancer. We assessed methylation status of the p16 tumor suppressor gene, MINT1 (methylated in tumor 1), MINT2, MINT31, O6-methylguanine-DNA methyltransferase gene, and hMLH1 mismatch repair gene. We compared methylation to ACF histopathology, K-ras proto-oncogene mutation, loss of heterozygosity at chromosome 1p, and microsatellite instability. Methylation was present in 34% (21 of 61) of ACF, including both FAP and sporadic types, but was more frequent in sporadic ACF [53% (18 of 34) versus 11% (3 of 27), P = 0.002], especially dysplastic sporadic ACF [75% (3 of 4) versus 8% (2 of 24), P = 0.004]. MINT31 was more frequently methylated in heteroplastic ACF than dysplastic ACF [35% (11 of 31) versus 7% (2 of 30), P = 0.01]. Strong associations of ACF methylation with K-ras mutation (P = 0.007) and with loss of chromosome 1p (P = 0.04) were observed, but methylation was the only molecular abnormality identified in 16% (10 of 61) of ACF. Our findings suggest that methylation in ACF is an early event in the pathogenesis of a subset of colorectal carcinomas, and that ACF from FAP patients and patients with sporadic colorectal cancer have distinct epigenetic changes that reflect differences in molecular pathogenesis.
Aberrant crypt foci (ACF) in colorectal mucosa are the earliest known morphological precursors to colorectal cancer (CRC). 1-6 A role for ACF in colorectal carcinogenesis is supported by the presence of histopathological intraepithelial neoplasia (dysplasia) in some ACF, and the expression of markers of proliferation and of tumor-associated antigens and lectin-binding moieties. 1,2 This role is further corroborated by the presence in some ACF of genetic alterations that are present in colorectal carcinomas, such as alterations in the adenomatous polyposis coli (APC) tumor suppressor gene, K-ras proto-oncogene mutations, and microsatellite instability (MSI). 1-6
The histopathology of human ACF is variable but can be subclassified into two broad categories: dysplastic and heteroplastic. 1 Dysplastic ACF resemble adenomas and are more common in familial adenomatous polyposis (FAP), which is because of germline mutation of the APC gene, than in patients with sporadic colorectal neoplasia. In addition to dysplasia, these ACF are characterized by abnormal epithelial proliferation in the upper aspects of the crypts, lack of K-ras mutations, and presence of APC mutations in dysplastic ACF from FAP patients but not patients with sporadic CRCs. 5-6 In contrast, heteroplastic ACF resemble hyperplastic polyps histopathologically, lack dysplasia, have proliferation mainly in the lower aspects of the crypts, have frequent K-ras mutations, and lack APC mutations. 1,5,6 Thus, ACF are phenotypically and genotypically diverse, but reflect their molecular pathogenesis.
Recent studies have shown that methylation of CpG islands is a molecular defect common in CRC. 7 CpG islands are 0.5- to 2-kb regions rich in cytosine-guanine dinucleotides and are present in the 5′ region of approximately half of all human genes. 8 CpG island methylation (CIM) is a mechanism for suppression of transcription of genes in physiological and pathological settings including neoplasia. 9 The recently discovered CpG island methylator phenotype (CIMP) is a novel pathway characterized by methylation of multiple CpG islands in colorectal carcinomas and adenomas, including genes known to be important in tumorigenesis such as the p16 tumor suppressor gene and hMLH1 mismatch repair gene. 10,11 In addition, CIMP is associated with K-ras mutations through methylation of MGMT (O6-methylguanine-DNA methyltransferase). 12
Dysplastic ACF are recognized precursors to CRC, but the relationship of heteroplastic ACF is less certain. 1-6 However, recent studies have proposed a hyperplastic polyp/serrated adenoma-carcinoma sequence as an alternative pathway to the usual adenoma-carcinoma sequence. 13-17 In addition, CIMP is present in hyperplastic polyps from patients with hyperplastic polyposis and colorectal neoplasia. 18 Therefore in the present study, we characterized ACF for the presence of CIM and compared the findings with histopathological and molecular alterations of importance in colorectal tumorigenesis, including K-ras mutation, loss of heterozygosity (LOH) of chromosome 1p, and MSI.
Materials and Methods
Specimens and Patients
ACF were isolated from the grossly normal mucosa in 10 colectomy specimens from patients with sporadic CRCs and from the nonpolyploid mucosa in 2 colectomy specimens from FAP patients with numerous polyps but no cancer. The specimens were collected in surgical pathology at the M. D. Anderson Cancer Center. The colons or rectums were opened longitudinally and rinsed of their luminal contents. The mucosa was dissected from the underlying muscularis propria, and the entire mucosal sheet was pinned flat on wax and fixed in formalin for 2 to 4 hours. The fixed colonic sheet was stained with 0.2% methylene blue for several minutes, and the ACF were visualized using an Olympus dissecting microscope (Olympus Optical Company, Ltd., Tokyo, Japan). ACF were marked with India ink, excised from the mucosal sheet in small strips, and submitted for routine histological processing. The mucosal strips were embedded longitudinally in paraffin blocks, sectioned onto glass slides, and stained with hematoxylin and eosin (H&E).
The H&E-stained ACF were characterized by light microscopy (Figure 1) ▶ of coded specimens by two gastrointestinal pathologists (RB and AR) unaware of the source of the specimens (Figure 1) ▶ . The ACF were classified as dysplastic, heteroplastic, or mixed (features of both dysplastic and heteroplastic ACF). 1 The size of ACF was measured from the H&E-stained slides.
Figure 1.

Histopathology of ACF. Heteroplastic ACF (top two panels), mixed heteroplastic and dysplastic ACF (third panel), and dysplastic ACF (bottom panel).
DNA Extraction
Lesional and nonlesional tissues were microdissected from the H&E-stained sections of ACF and mucosa without coverslip. Genomic DNA was extracted and prepared from the microdissected ACF as described previously. 19
Bisulfite Treatment of DNA and Methylation-Specific PCR
The methylation status of the p16 tumor suppressor gene, MINT1 (methylated in tumor 1), MINT2, MINT31, MGMT gene, and hMLH1 mismatch repair gene was determined by bisulfite treatment of DNA followed by methylation-specific PCR as described. 12,20 The six loci selected for methylation analysis are unmethylated (<1% methylation) in normal tissues. 7 MINT1 and MINT2 correspond to CpG islands that are in the 5′ region of cDNAs with open reading frames that have no known protein homology (JP Issa, unpublished data). MINT31 is 2-kb upstream of the CACNA1G, a T-type calcium channel gene. 21
In brief, 2 μg of microdissected genomic DNA were denatured with 2 mol/L of NaOH at 37°C for 10 minutes, followed by incubation with 3 mol/L of sodium bisulfite (pH 5.0) at 50°C for 16 hours in darkness. After treatment, DNA was purified using the DNA Cleanup Kit (Promega, Madison, WI) as recommended by the manufacturer, incubated with 3 mol/L of NaOH at room temperature for 5 minutes, precipitated with 10 mol/L ammonium acetate and 100% ethanol, washed with 70% ethanol, and finally resuspended in 20 μl of distilled water.
The primers and polymerase chain reaction (PCR) conditions for p16 were the same as reported by Herman and colleagues. 20 The primers and PCR conditions for the MINT loci, MGMT, and hMLH1 are listed in Table 1 ▶ . In brief, 2 μl of bisulfite-treated DNA was used as template for PCR reactions using primers specific for methylated and unmethylated alleles. DNA from the RKO colon cancer cell line (American Type Culture Collection, Manassas, VA) and water were used as positive and negative controls, respectively. PCR products from methylated and unmethylated reactions were electrophoresed on 6% acrylamide gels and visualized by ethidium bromide staining.
Table 1.
Primers and PCR Conditions for MINT 1, MINT 2, MINT 31, MGMT, and hMLH1
| Locus | Allele | Sense primers | Antisense primers | Cycling conditions |
|---|---|---|---|---|
| MINT1 | Methylated | 5′-AATTTTTTTATATATATTTTCGAAGC-3′ | 5′-AAAAACCTCAACCCCGCG-3′ | 95°C for 10 minutes, 37 cycles of |
| Unmethylated | 5′-AATTTTTTTATATATATTTTTGAAGTGT-3′ | 5′-AACAAAAAACCTCAACCCCACA-3′ | 95°C for 30 seconds, 55°C for 45 seconds | |
| MINT2 | Methylated | 5′-TTGTTAAAGTGTTGAGTTCGTC-3′ | 5′-AATAACGACGATTCCGTACG-3′ | 95°C for 10 minutes, 40 cycles of |
| Unmethylated | 5′-GATTTTGTTAAAGTGTTGAGTTTGTT-3′ | 5′-CAAAATAATAACAACAATTCCATACA-3′ | 95°C for 30 seconds, 60°C for 45 seconds | |
| MINT31 | Methylated | 5′-TGTTGGGGAAGTGTTTTTCGGC-3′ | 5′-CGAAAACGAAACGCCGCG-3′ | 95°C for 10 minutes, 38 cycles of |
| Unmethylated | 5′-TAGATGTTGGGGAAGTGTTTTTTGGT-3′ | 5′-TAAATACCCAAAAACAAAACACCACA-3′ | 95°C for 30 seconds, 60°C for 45 seconds | |
| MGMT | Methylated | 5′-GGTCGTTTGTACGTTCGC-3′ | 5′-GACCGATACAAACCGAACG-3′ | 95°C for 10 minutes, 38 cycles of |
| Unmethylated | 5′-GTAGGTTGTTTGTATGTTTGT-3′ | 5′-AACCAATACAAACCAAACA-3′ | 95°C for 30 seconds, 55°C for 45 seconds | |
| hMLH1 | Methylated | 5′-GATAGCGATTTTTAACGC-3′ | 5′-TCTATAAATTACTAAATCTCTTCG-3′ | 95°C for 10 minutes, 40 cycles of 95°C for 30 seconds, 53°C for 45 seconds |
| Unmethylated | 5′-AGAGTGGATAGTGATTTTTAATGT-3′ | 5′-ACTCTATAAATTACTAAATCTCTTCA-3′ |
Mutation of K-ras Proto-Oncogene
The first exon of K-ras was amplified in 50-μl volumes using 2 μl of genomic DNA, 10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L potassium chloride, 2 mmol/L magnesium chloride, 0.8 mmol/L dNTP mix, 2.25 U Ampli Taq Gold (Applied Biosystems, Foster City, CA), 0.125 U pfu DNA Polymerase (Stratagene, La Jolla, CA), and 20 pmol of forward and reverse primers (5′-GGCCGGTAGTGTATTAACCTTATG TGTGACAT-3′ and 5′-CCGCGGCCGGCGGCCAAAACAAGATTTACCTCTATTGTTGG-3′; Life Technologies, Rockville, MD). PCR reactions were performed using the following cycling conditions: denaturation at 95°C for 10 minutes; 14 cycles of 95°C for 20 seconds, 59°C to 52°C in 0.5°C decrements/cycle for 60 seconds, and 72°C for 60 seconds; 25 cycles of 95°C for 20 seconds, 52°C for 60 seconds, and 72°C for 60 seconds; and extension at 72°C for 10 minutes on a GeneAmp PCR System 9700 (Applied Biosystems).
PCR products were purified using 10 U of exonuclease I and 2 U of shrimp alkaline phosphatase (Amersham Life Science, Indianapolis, IN), incubated at 37°C for 15 minutes, and inactivated by incubating at 80°C for 15 minutes. DNA sequencing was performed in 20-μl volumes comprised of 2 μl of purified PCR product, 8 μl ABI Prism BigDye Terminator Cycle Sequencing Kit (Applied Biosystems), and 5 pmol of forward primer using the following cycling conditions: 25 cycles of 95°C for 20 seconds, 52°C for 60 seconds, and 72°C for 60 seconds. After spin-column purification (Princeton Separations, Adelphia, NJ), the reaction products were sequenced by capillary electrophoresis using an ABI Prism 3700 DNA Analyzer (Applied Biosystems). Mutations were confirmed by sequencing using the reverse primer.
Fluorescently Labeled PCR Amplification
LOH and MSI were determined by fluorescently labeled PCR amplification using fluorescent dye-labeled and unlabeled primers (Applied Biosystems). The 5′ oligonucleotide was end-labeled with 6-FAM (BAT-25, D1S199, and D1S507), HEX (BAT-26, D1S468, and D1S255), or NED (D1S214) fluorescent dye. PCR was performed in 15-μl reaction volumes containing 40 ng of DNA, 9 μl ABI Prism True Allele PCR Premix (Applied Biosystems), and 5 pmol of each primer. PCR was performed using the following cycling conditions: denaturation at 95°C for 12 minutes; 10 cycles (94°C for 15 seconds, 55°C for 15 seconds, 72°C for 30 seconds), 32 cycles (89°C for 15 seconds, 55°C for 15 seconds, 72°C for 30 seconds), and extension at 72°C for 10 minutes. A 1.0-μl aliquot of each fluorescent-labeled PCR product was combined with 12 μl of formamide and 0.5 μl of Genescan 400HD (ROX) size standard (Applied Biosystems). The samples were then subjected to capillary electrophoresis on an ABI 3700 DNA Analyzer using Genescan Analysis software (Applied Biosystems).
LOH of Chromosome 1p
Loss of chromosome 1p was determined by using five dinucleotide-repeat microsatellite markers on the short arm of chromosome 1 (in order from centromere to telomere D1S255, D1S199, D1S507, D1S214, and D1S468). Complete or partial loss of chromosome 1p was based on the pattern of loss of the five dinucleotide microsatellite markers evaluated. Loss of a marker was considered to be present when the PCR assay showed absence or decrease in intensity by more than 50% of a band from a tumor sample as compared with the paired control nonneoplastic sample.
MSI
MSI status was determined by using the five markers on the short arm of chromosome 1p and two mononucleotide-repeat microsatellite markers, BAT 25 and BAT 26. 13 Specimens with high levels of MSI (MSI-high) were defined by shifts of bands as compared to control DNA in at least 30% of evaluable markers, and low levels of MSI (MSI-low) by shifts in less than 30% of evaluable microsatellite markers, as in previous studies. 13
Statistical Analysis
Fisher’s exact test and chi-square test were used for comparing associations among genetic alterations and between genetic alterations and clinicopathological factors. Student’s t-test was used to compare the means of size, age, and methylation.
Results
The patient demographic data in relationship to ACF characteristics are summarized in Table 2 ▶ . Twenty-seven ACF were identified from two FAP patients and 34 ACF from 10 patients with sporadic colorectal carcinomas. The histopathological type of ACF was strongly associated with the patient groups: 89% (24 of 27) of ACF from FAP patients were dysplastic, and only 11% (3 of 27) were heteroplastic. In contrast, 82% (28 of 34) of ACF from the patients with sporadic CRC were heteroplastic, only 12% (4 of 34) were dysplastic, and 6% (2 of 34) were mixed having both dysplastic and heteroplastic regions (P = 0.000001 for sporadic versus FAP patients).
Table 2.
Demographic Data and Characteristics of Patients with ACF
| Patient | Age | Sex | Site of ACF | Histology of ACF, no. | Other clinical associations | ||
|---|---|---|---|---|---|---|---|
| Heteroplastic | Dysplastic | Mixed | |||||
| 1 | 50 | F | L | 2 | 7 | 0 | FAP |
| 2 | 17 | M | R | 1 | 17 | 0 | FAP |
| 3 | 49 | M | L | 7 | 0 | 0 | Sporadic CRC |
| 4 | 79 | F | R | 1 | 0 | 0 | Sporadic CRC |
| 5 | 69 | M | R | 0 | 0 | 1 | Sporadic CRC |
| 6 | 61 | M | L | 8 | 0 | 1 | Sporadic CRC |
| 7 | 64 | F | L | 3 | 1 | 0 | Sporadic CRC |
| 8 | 65 | F | L | 2 | 1 | 0 | Sporadic CRC |
| 9 | 37 | F | L | 2 | 0 | 0 | Sporadic CRC |
| 10 | 45 | F | L | 5 | 0 | 0 | Sporadic CRC |
| 11 | 60 | F | L | 0 | 1 | 0 | Sporadic CRC |
| 12 | 58 | M | R | 0 | 1 | 0 | Sporadic CRC |
F, female; M, male; L, left, colorectum; R, right, colon.
CpG Island Methylation in ACF
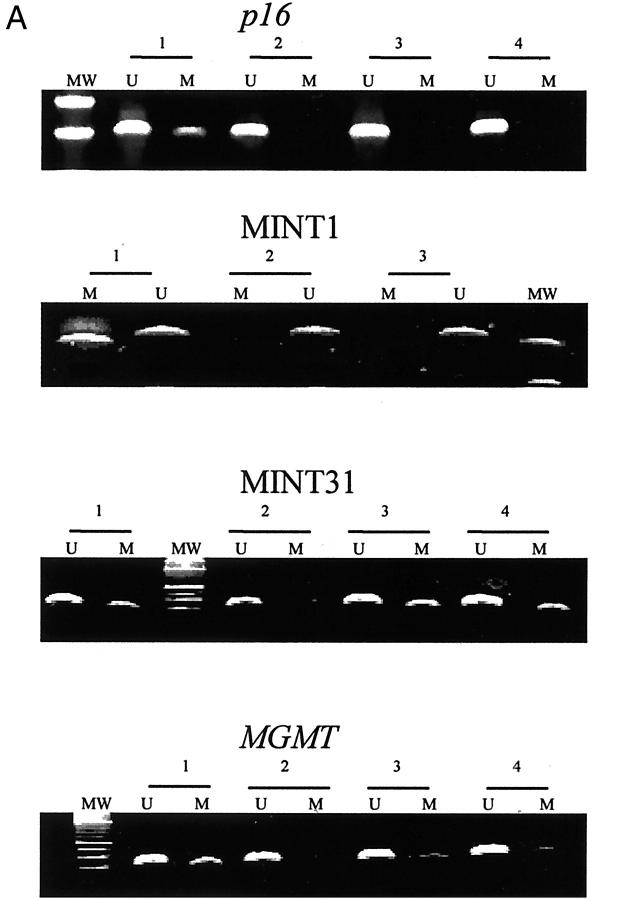
Methylation of the p16 gene, MINT31, MINT2, MINT1, MGMT gene, and hMLH1 gene was present in 4% (2 of 56), 21% (13 of 61), 5% (3 of 59), 8% (5 of 59), 12% (6 of 51), and 3% (2 of 61) of the ACF, respectively (examples in Figure 2A ▶ and summarized in Figure 3 ▶ ). Thirty-four percent (21 of 61) of the ACF were methylated in at least one locus: 2% (1 of 61) of ACF were methylated at three loci, 13% (8 of 61) at two loci, and 20% (12 of 61) at one locus. The p16 gene was methylated in two ACF, the hMLH1 gene in two ACF, and the MGMT gene in six ACF (Figure 3) ▶ . Six of eight ACF with methylation of p16, hMLH1, or MGMT had methylation of at least one additional locus.
Figure 2.
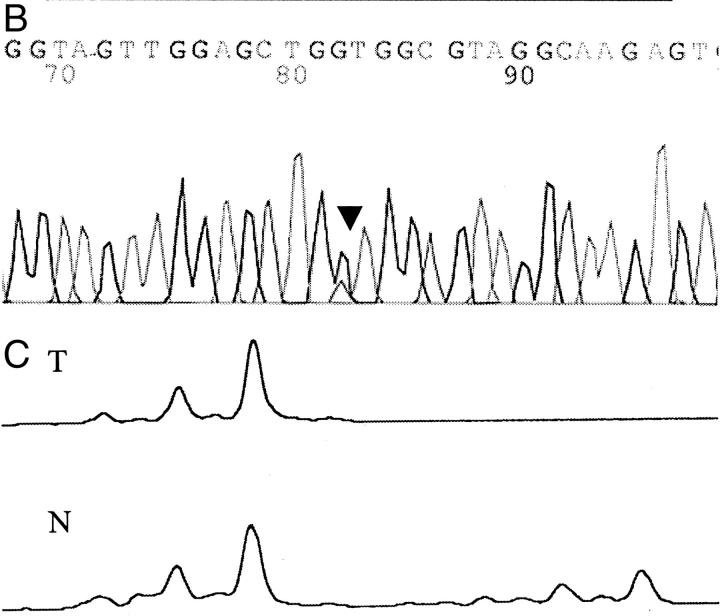
A: Methylation analysis of CpG islands in ACF. Examples of methylation of p16, MINT1, MINT31, and MGMT are shown. Methylation-specific PCR using primers for methylated (M) and unmethylated (U) alleles of bisulfite-treated DNA was performed. Loci examined and ACF numbers are indicated above each gel. MW represents lane with molecular weight marker. B: Nucleotide sequencing of the K-ras gene in ACF. A G-to-A mutation of codon 12 is indicated by the arrowhead. C: Allelic loss of chromosome 1p in ACF. The lane from normal DNA (N) has two alleles at marker D1S199 and the lane from the tumor DNA (T) shows loss of one of the alleles.
Figure 3.
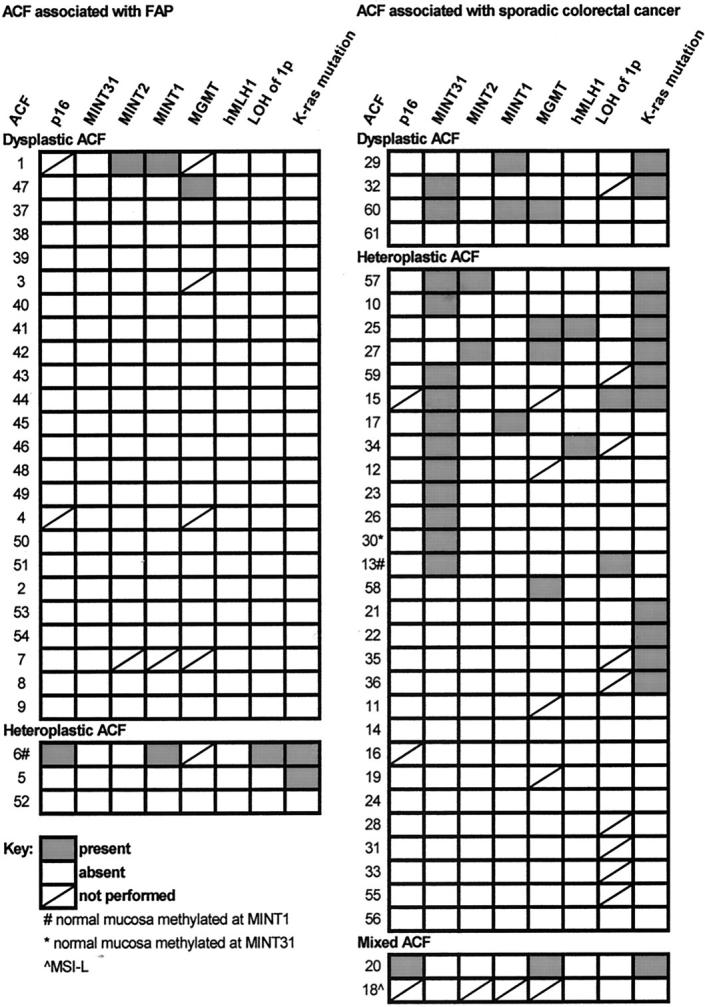
CIM, loss of chromosome 1p, K-ras mutation, and histopathology of ACF from FAP patients and patients with sporadic CRC.
CpG Island Methylation in Mucosa
Methylation of the six markers was analyzed in 36 samples of mucosa adjacent to the ACF (Figure 3) ▶ . Methylation at one locus was found in only three mucosal samples. Two of these samples were methylated at MINT31 and MINT1, respectively, and the adjacent ACF were concordantly methylated for the same loci. The third sample of mucosa was methylated at MINT1, and was discordant with the methylation status of the adjacent ACF.
K-ras Mutation, Loss of Chromosome 1p, and MSI in ACF
K-ras mutations were present in 25% (15 of 61) of ACF (examples in Figure 2B ▶ and summarized in Figure 3 ▶ ). All 15 mutations were present at codon 12, and none were found in codon 13. G-to-A transition at the second nucleotide of codon 12 was present in seven ACF, G-to-T transversion at the second nucleotide in seven ACF, and G-to-T transversion at the first nucleotide in one ACF.
Chromosome 1p loss was present in 6% (3 of 52) of ACF (examples in Figure 2C ▶ and summarized in Figure 3 ▶ ). Chromosome 1p loss was present at locus 1p35 (marker D1S199) in one ACF, and at locus 1p35-36 (markers D1S468 and D1S507, and markers D1S199 and D1S468, respectively) in the other two ACF.
Allelic shift was present in only one ACF, which was classified as MSI-low. No ACF had MSI-high.
Associations of CpG Island Methylation with Clinicopathological and Molecular Characteristics
CIM occurred in ACF from FAP and sporadic patients but was more characteristic of sporadic ACF: methylation was present in 53% (18 of 34) of ACF from patients with sporadic colorectal carcinomas but only 11% (3 of 27) of ACF from FAP patients (P = 0.002, Figure 3 ▶ ). This difference in methylation frequency was most apparent in dysplastic ACF, which had a higher frequency of methylation in sporadic patients than in FAP patients [75% (3 of 4) versus 8% (2 of 24), P = 0.004]. In contrast, the frequency of methylation was similar in heteroplastic ACF from FAP and sporadic patients [33% (1 of 3) versus 50% (14 of 28); P = 0.9, not significant].
When individual markers were considered, MINT31 was more frequently methylated in heteroplastic ACF than dysplastic ACF [35% (11 of 31) versus 7% (2 of 30), P = 0.01], but the other methylation sites had no significant difference between the histopathological types. The p16 gene was methylated in one heteroplastic ACF from an FAP patient and in a mixed ACF from a patient with sporadic colon cancer. Methylation of the hMLH1 gene was present in two microsatellite-stable heteroplastic ACF from two patients with sporadic CRC (Figure 3) ▶ . The MGMT gene was methylated in six ACF (Figure 3) ▶ , including dysplastic ACF from FAP and sporadic patients and heteroplastic ACF from sporadic patients. Patient age, and site and size of ACF were not associated with the methylation status of the ACF.
K-ras mutation was more common in ACF from sporadic than FAP patients but was found in dysplastic ACF, whereas LOH of chromosome 1p was found only in heteroplastic ACF. K-ras mutation was present in 7% (2 of 27) of ACF from FAP patients but in 38% (13 of 34) of ACF from patients with sporadic carcinomas (P = 0.01). Mutation was present in 10% (3 of 30) of ACF with dysplasia, but 39% (12 of 31) of heteroplastic ACF (P = 0.02). Loss of 1p was present in 13% (3 of 23) of heteroplastic ACF, consisting of one ACF from an FAP patient and two ACF from two patients with sporadic CRC, but in none of 29 dysplastic or mixed ACF.
K-ras mutations and LOH of chromosome 1p were more common in ACF with CIM. K-ras mutation was present in 48% (10 of 21) of ACF with CIM, but only 13% (5 of 40) of ACF without methylation (P = 0.007). Two of three ACF with K-ras mutations and methylation of MGMT had G-to-A transition, as reported in a previous study of adenomas or carcinomas. 12 Similarly, chromosome 1p loss was present in 20% (3 of 18) of ACF with CIM, but in 0 of 34 ACF without methylation (P = 0.04).
When the clinicopathological and more frequent molecular markers were considered together, 92% (22 of 24) of dysplastic ACF from FAP patients lacked methylation or K-ras mutation (dysplasia +, methylation −, K-ras mutation −; Figure 3 ▶ ). In contrast, the heteroplastic, mixed, and dysplastic sporadic ACF and the heteroplastic ACF from FAP patients were much more heterogeneous than dysplastic FAP ACF: 35% (n = 13) were without methylation or K-ras mutation (methylation −, K-ras mutation −); 24% (n = 9) had methylation (methylation +, K-ras mutation −); 27% (n = 10) had both methylation and K-ras mutation (methylation +, K-ras mutation +); and 14% (n = 5) had K-ras mutation (methylation −, K-ras mutation +).
Discussion
The role of ACF in colorectal carcinogenesis has been contentious, and ACF could be precursors or innocent bystanders. 1-6 The dysplastic ACF-adenoma-carcinoma sequence is well-recognized because dysplastic ACF are frequent in FAP patients, and APC mutations are present in dysplastic ACF from FAP patients and a minority of sporadic dysplastic ACF. 5,6,22 The role of ACF as precursors of some CRCs is further corroborated by the presence of MSI in ACF from patients with hereditary nonpolyposis CRC in whom carcinoma with high levels of MSI is the hallmark, and in ACF from sporadic patients. 3,23 The role of heteroplastic ACF in colorectal carcinogenesis is more controversial because these ACF have frequent K-ras mutation but lack dysplasia or APC mutations, in contrast to most adenomas and carcinomas. 5,6,22 However, a role for heteroplastic ACF as a precursor to a subset of CRCs is supported by the following lines of evidence: heteroplastic ACF are clonal; 24 genetic alterations that are common in CRCs such as K-ras mutation, 1,2,4-6,22 chromosome 1p loss (the present study), and/or CIM (the present study) are present in heteroplastic ACF; ACF, adenomas, and carcinomas share similar incidence and anatomical distribution; 25-27 and ACF with mixed heteroplastic and dysplastic components exist. 28 A heteroplastic ACF-adenoma-carcinoma sequence has been proposed in which K-ras mutation precedes APC mutation in human and rodent studies. 6,29
An alternative pathway of colorectal carcinogenesis with a serrated polyp-dysplasia-adenocarcinoma sequence has also been proposed. 13-17 Jass 17 has reported in colorectal carcinogenesis the importance of serrated lesions, low levels of MSI (MSI-L), and ras mutation. In a previous study, we have reported concordant methylation of multiple hyperplastic polyps in patients with hyperplastic polyposis and colorectal neoplasia including serrated adenomas. 18 Heteroplastic ACF therefore may be precursors of serrated lesions that progress to CRCs.
CIM of one or more loci was present in a third of ACF in our study, including both dysplastic and heteroplastic types in both FAP and sporadic patients. A previous study has reported methylation of the androgen receptor gene on the X chromosome in human ACF. 24 Because ACF are putative precursor lesions, these studies provide evidence that CIM is an early event in colorectal carcinogenesis.
Previous studies have identified genetic alterations in ACF, including APC gene alterations, K-ras mutations, and MSI. 1-6,22 We also found loss of chromosome 1p in 6% of ACF, and previous studies have identified chromosome 1p loss in colorectal hyperplastic polyps, adenomas, and carcinomas. 13,30,31 Our study therefore broadens the spectrum of molecular abnormalities that are identified in ACF as well as in tumors.
Methylation at multiple loci is present in 40 to 50% of sporadic colorectal carcinomas and adenomas, 7,10,11 and in hyperplastic polyps from patients with hyperplastic polyposis. 18 Methylation of two or more loci was present in 15% of ACF in our study, and methylation of genes known to be important in tumorigenesis was associated with methylation of multiple loci. In our study, four of six ACF with MGMT methylation, both ACF with hMLH1 methylation, and the one ACF with p16 methylation were methylated at more than one locus. CIM in ACF was associated with K-ras mutation, and G-to-A transition was present in three ACF with methylation of MGMT in our study. A previous study has shown an association between CIMP in cancer and K-ras mutation, 10 and another previous study reported an association between G-to-A transition in K-ras gene and methylation of MGMT in colorectal carcinomas and large adenomas. 12 These findings suggest that CIM of MGMT is an early event and precedes the G-to-A transition mutations of the K-ras gene. Similarly, methylation of the hMLH1 mismatch repair gene in ACF was not associated with the development of MSI, suggesting that hMLH1 methylation precedes MSI. These results emphasize the early occurrence of abnormal methylation as a pathogenic mechanism in colorectal carcinogenesis.
In our study, the frequency of CIM was related to the type of patient with ACF: methylation was present in 53% of ACF from patients with sporadic colorectal carcinomas but only 11% of ACF from FAP patients. Of note, methylation was more frequent in dysplastic sporadic ACF than in dysplastic FAP ACF (75% versus 8%). Previous studies have shown differences in frequencies of dysplasia and K-ras mutation in ACF from patients with sporadic colorectal carcinomas and FAP patients. 5,6,22 ACF from patients with sporadic carcinomas usually had CIM and/or K-ras mutation and lacked dysplasia. In contrast, ACF from FAP patients usually lacked CIM or K-ras mutation but were frequently dysplastic, reflecting the inherited predisposition to dysplasia resulting from germline APC gene mutation. Our data on methylation provide additional evidence that phenotypic and genetic progression in FAP differs from some sporadic colorectal carcinomas. Dysplastic ACF occur in sporadic patients, although at low frequency, and have characteristics such as those of dysplastic ACF in FAP patients but with less frequent APC mutation, 5,6 and more frequent methylation in our study. In addition, we found that MINT31, which is present 2-kb upstream of the CACNA1G, a T-type calcium channel gene, 21 was differentially methylated in heteroplastic but not dysplastic ACF with higher frequency in the heteroplastic type.
In conclusion, the epigenetic differences in methylation in ACF we found are analogous to the heterogeneity of phenotypic and genetic characteristics in colorectal carcinomas. Understanding of the events in ACF provides the opportunity for their prevention and suppression of their progression to malignancy.
Footnotes
Address reprint requests to Asif Rashid M.D., Ph.D., Department of Pathology, Box 85, M. D. Anderson Cancer Center, 1515 Holcombe Blvd., Houston, TX, 77030-4095. E-mail: arashid@mdacc.tmc.edu.
Supported in part by Public Health Service grant DK56338 (to R.R.B.) which funds the Texas Gulf Coast Digestive Disease Center.
References
- 1.Nucci MR, Robinson CR, Longo P, Campbell P, Hamilton SR: Phenotypic and genotypic characteristics of aberrant crypt foci in human colorectal mucosa. Hum Pathol 1997, 28:1396-1407 [DOI] [PubMed] [Google Scholar]
- 2.Pretlow TP, Brasitus TA, Fulton NC, Cheyer C, Kaplan EL: K-ras mutations in putative preneoplastic lesions in human colon. J Natl Cancer Inst 1993, 85:2004-2007 [DOI] [PubMed] [Google Scholar]
- 3.Heinen CD, Shivapurkar N, Tang Z, Groden J, Alabaster O: Microsatellite instability in aberrant crypt foci from human colons. Cancer Res 1996, 56:5339-5341 [PubMed] [Google Scholar]
- 4.Yamashita N, Minamoto T, Ochiai A, Onda M, Esumi H: Frequent and characteristic K-ras activation and absence of p53 protein accumulation in aberrant crypt foci of the colon. Gastroenterology 1995, 108:434-440 [DOI] [PubMed] [Google Scholar]
- 5.Otori K, Konishi M, Sugiyama K, Hasebe T, Shimoda T, Kikuchi-Yanoshita R, Mukai K, Fukushima S, Miyaki M, Esumi H: Infrequent somatic mutation of the adenomatous polyposis coli gene in aberrant crypt foci of human colon tissue. Cancer 1998, 83:896-900 [DOI] [PubMed] [Google Scholar]
- 6.Takayama T, Ohi M, Hayashi T, Miyanishi K, Nobuoka A, Nakajima T, Satoh T, Takimoto R, Kato J, Sakamaki S, Niitsu Y: Analysis of K-ras, APC, and β-catenin in aberrant crypt foci in sporadic adenoma, cancer, and familial adenomatous polyposis. Gastroenterology 2001, 121:599-611 [DOI] [PubMed] [Google Scholar]
- 7.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa J-PJ: CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999, 96:8681-8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bird AP: CpG-rich islands and the function of DNA methylation. Nature 1986, 321:209-213 [DOI] [PubMed] [Google Scholar]
- 9.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa J-PJ: Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 1998, 72:141-196 [PubMed] [Google Scholar]
- 10.Toyota M, Ohe-Toyota M, Ahuja N, Issa J-PJ: Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA 2000, 97:710-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rashid A, Shen L, Morris JS, Issa JPJ, Hamilton SR: CpG island methylation in colorectal adenomas. Am J Pathol 2001, 159:1129-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, Issa J-PJ, Sidransky D, Baylin SB, Herman JG: Inactivation of the DNA repair gene O6methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res 2000, 60:2368-2371 [PubMed] [Google Scholar]
- 13.Rashid A, Houlihan PS, Booker S, Peterson GM, Giardiello FM, Hamilton SR: Phenotypic and molecular characteristics of hyperplastic polyposis. Gastroenterology 2000, 119:323-332 [DOI] [PubMed] [Google Scholar]
- 14.Jass JR, Cottier DS, Pokos V, Parry S, Winship IM: Mixed epithelial polyps in association with hereditary non-polyposis colorectal cancer providing an alternative pathway of cancer histogenesis. Pathology 1997, 29:28-33 [DOI] [PubMed] [Google Scholar]
- 15.Iino H, Jass JR, Simms LA, Young J, Leggett B, Ajioka Y, Watanabe H: DNA microsatellite instability in hyperplastic polyps, serrated adenomas, and mixed polyps: a mild mutator pathway for colorectal cancer? J Clin Pathol 1999, 52:5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jass JR, Iino H, Ruszkiewicz A, Painter D, Solomon MJ, Koorey DJ, Cohn D, Furlong KL, Walsh MD, Palazzo J, Edmonston TB, Fishel R, Young J, Leggett BA: Neoplastic progression occurs through mutator pathways in hyperplastic polyposis of the colorectum. Gut 2000, 47:43-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jass JR: Serrated route to colorectal cancer: back street or super highway? J Pathol 2001, 193:283-285 [DOI] [PubMed] [Google Scholar]
- 18.Chan AO, Issa J-PJ, Morris JS, Hamilton SR, Rashid A: Concordant CpG island methylation in hyperplastic polyposis. Am J Pathol 2002, 160:529-536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moskaluk CA, Kern SE: Microdissection and polymerase chain reaction amplification of genomic DNA from histological tissue sections. Am J Pathol 1997, 150:1547-1552 [PMC free article] [PubMed] [Google Scholar]
- 20.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996, 93:9821-9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toyota M, Ho C, Ohe-Toyota M, Baylin SB, Issa J-PI: Inactivation of CAGNA1G, a T-type calcium channel gene, by aberrant methylation of its 5′ CpG island in human tumors. Cancer Res 1999, 59:4535-4541 [PubMed] [Google Scholar]
- 22.Jen J, Powell SM, Papadopoulos N, Smith KJ, Hamilton SR, Vogelstein B, Kinzler KW: Molecular determinants of dysplasia in colorectal lesions. Cancer Res 1994, 54:5523-5526 [PubMed] [Google Scholar]
- 23.Augenlicht LH, Richards C, Corner G, Pretlow TP: Evidence for genomic instability in human colonic aberrant crypt foci. Oncogene 1996, 12:1767-1772 [PubMed] [Google Scholar]
- 24.Sakurazawa N, Tanaka N, Onda M, Esumi H: Instability of X chromosome methylation in aberrant crypt foci of the human colon. Cancer Res 2000, 60:3165-3169 [PubMed] [Google Scholar]
- 25.Bouzourene H, Chaubert P, Seelentag W, Bosman FT, Saraga E: Aberrant crypt foci in patients with neoplastic and nonneoplastic colonic disease. Hum Pathol 1999, 30:66-71 [DOI] [PubMed] [Google Scholar]
- 26.Shpitz B, Bomstein Y, Mekori Y, Cohen R, Kaufman Z, Neufeld D, Galkin M, Bernheim J: Aberrant crypt foci in human colons: distribution and histomorphological characteristics. Hum Pathol 1998, 29:469-475 [DOI] [PubMed] [Google Scholar]
- 27.Roncucci L, Modica S, Pedroni M, Tamassia MG, Ghidoni M, Losi L, Fante R, Di Gregoria C, Manenti A, Gafa L, Ponz de Leon M: Aberrant crypt foci in patients with colorectal cancer. Br J Cancer 1998, 77:2343-2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otori K, Sugiyama K, Haseve T, Fukushima S, Esumi H: Emergence of adenomatous aberrant crypt foci (ACF) from hyperplastic ACF with concomitant increase in cell proliferation. Cancer Res 1995, 55:4743-4746 [PubMed] [Google Scholar]
- 29.De Filippo C, Caderni G, Bazzicalupo M, Briani C, Giannini A, Fazi M, Dolara P: Mutations of the Apc gene in experimental colorectal carcinogenesis induced by azomymethane in F344 rats. Br J Cancer 1998, 77:2148-2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bardi G, Pandis N, Fenger C, Kronborg O, Bomme L, Heim S: Deletion of 1p36 as a primary chromosomal aberration in intestinal tumorigenesis. Cancer Res 1993, 53:1895-1898 [PubMed] [Google Scholar]
- 31.Lothe RA, Andersen SN, Hofstad B, Meling GI, Peltomaki P, Heim S, Brogger A, Vatn M, Rognum TO, Borresen A-L: Deletion of 1p loci and microsatellite instability in colorectal polyps. Genes Chromosom Cancer 1995, 14:182-188 [DOI] [PubMed] [Google Scholar]