Abstract
We have recently reported that overrepresentation of 8q24 (c-myc) is associated with clinical progression in prostate cancer. In this study, we map the boundaries of the overrepresented region within 8q23-q24 using interphase fluorescent in situ hybridization analysis of paraffin-embedded prostate cancer specimens. One hundred primary prostate cancers and three prostate cancer cell lines were evaluated, and the minimally overrepresented region could be narrowed to the ∼8.2-Mb region between D8S514 and H47317. This region includes c-myc and is wholly within 8q24. Eukaryotic translation initiation factor 3 subunit 3 does not seem to be overrepresented independent of c-myc in prostate cancer. The cell lines PC3 and DU145 have and do not have 8q24 overrepresentation, respectively. We then selected 39 expressed sequence tags (ESTs) within and surrounding the minimally overrepresented region and performed expression analysis using Northern blot hybridization. Five ESTs/genes including c-myc were overexpressed in both the PC3 cell line and DU145, but the PC3 to DU145 expression ratios were <2. Seven ESTs were overexpressed twofold or more in PC3 compared to DU145. This group included hyaluronan synthase 2, nephroblastoma-overexpressed gene, eukaryotic translation initiation factor 3 subunit 3, and an EST (R69368) encoding a hypothetical protein (BM009). These seven genes as well as c-myc are candidate target genes within the overrepresented 8q24 region and their overexpression may be associated with prostate cancer progression.
Human prostate cancers commonly have alterations of chromosome arms 1q, 7q, 8p, 8q, 10q, 13q, 16q, 17p, 17q, and 18q, as well as the X chromosome. 1,2 We have recently shown that overrepresentation of 8q24 is associated with cancer progression and patient prognosis in both clinically localized and metastatic prostate cancer. 3,4 It has been known that the oncogene c-myc is located within 8q24, 5 and the gene is involved in several neoplastic disorders. 6,7 In breast and small cell lung cancer, the minimally overrepresented 8q24 region has been reported to be ∼16 cM and 3 Mb, respectively. 8,9 However, the overrepresented region in prostate cancer has not been reported. It is possible that other genes besides c-myc may also be targets within the commonly overrepresented region. For example, the overrepresentation of 20q is commonly observed in breast and ovarian cancer, and several candidate genes have been found in the region. 10 Even if c-myc is the target of the overrepresentation of 8q24, then at least several of these genes may be of diagnostic importance or may be unique and potentially specific therapeutic targets.
In this study, we mapped the overrepresented region of 8q24 in prostate cancer using interphase fluorescent in situ hybridization (FISH) analysis of archival paraffin-embedded tumor specimens. Additionally, we screened the expression level of 39 known genes or expressed sequence tags (ESTs) within and surrounding the minimally overrepresented 8q24 region to discover candidates of target genes associated with prostate cancer.
Materials and Methods
Samples and Slide Preparation
A total of 100 cases of prostate cancer, selected newly or from previously reported studies, and three cell lines (ie, LNCaP, DU145, and PC3) were investigated. Forty of the 100 cases were previously reported to have c-myc overrepresentation relative to the centromere of chromosome 8, 36 cases had gain of c-myc and the centromere of the chromosome, and 24 cases had no apparent c-myc anomaly. 3,4 Of the 100 cases, 13, 74, and 13 cases were stage pT2N0M0, pT3N0M0, and pT2–3N1–2M0, respectively. For FISH analysis, 15 serial 5-μm sections were sliced from each paraffin-embedded tumor block and mounted on glass slides. The first and the last sections were stained by hematoxylin and eosin to determine the region of interest. The regions that contained tumor were precisely recorded. The locations of these regions were transferred to unstained sections by etching the opposite side of the glass slides. The same regions were then analyzed by each of the FISH probe sets. The LNCaP, DU145, and PC3 cell lines were obtained from the American Type Culture Collection (Rockville, MD) and were cultured in RPMI 1640 medium (Sigma, St. Louis, MO) supplemented with 5% serum at 37°C in a 5% CO2 humidified incubator. The cells were grown to ∼80% confluence and were harvested for mRNA isolation. For FISH analysis of these cell lines, slides were prepared as described previously. 11
Bacterial Artificial Chromosome (BAC) Probe Preparation
Microsatellite markers and EST markers surrounding c-myc were selected using the databases of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nih.gov) and the Whitehead Institute for Biomedical Research/MIT Center for Genome Research (http://www-genome.wi.mit.edu/). These markers were used to screen arrays of a BAC library (Research Genetics, Huntsville, AL) using the protocols supplied by the company. Probes mapped in 8q23-q24 were first selected at 10-cM intervals, and then at ∼1-cM intervals to narrow the overrepresented region. These selected probes were labeled with digoxigenin and detected with a rhodamine-labeled anti-digoxigenin antibody (Intergen, Purchase, NY).
FISH Analysis
Metaphase FISH analyses of normal lymphocyte metaphase chromosomes were performed to confirm the chromosomal location of the isolated BAC clone as previously described. 10 Twelve of 20 BAC probes mapped to the correct region. Dual-probe interphase FISH on the paraffin-embedded section was then performed as described previously to analyze the copy number of each respective locus-specific probe (LSP) with reference to the centromere of chromosome 8. 3 The probe for c-myc was provided from Vysis, Inc. (Downers Grove, IL).
The chromosome enumeration probe 8 (CEP8; Vysis, Inc.) and LSP signals in each dual-probe hybridization were enumerated in each total of 300 nuclei. The normal and abnormal criteria were defined by a previously reported normal control study. 4 Briefly, in histologically benign prostate, the ranges for percentage of epithelial nuclei with 0 or 1, 2, and ≥3 c-myc signals were 12.7% to 37.5%, 67.5% to 84.6%, and 0% to 5.1%, respectively. The ranges for percentage of epithelial nuclei with 0 or 1, 2, and ≥3 CEP8 signals were 14.0% to 33.0%, 63.6% to 85.1%, and 0% to 6.0%, respectively. The range for the mean c-myc/CEP8 ratio was 0.93 to 1.04. The normal value ranges for other LSP probes were similar to those of c-myc and CEP8 (data not shown). On the basis of the normal control study, the threshold values for the following categories were determined. An LSP or CEP8 was classified as normal if <10% of the nuclei had three or more signals and if <55% of the nuclei had 0 or 1 signal. An LSP or CEP8 was classified as loss if 55% or more nuclei had 0 or 1 signal. An LSP or CEP8 was classified as gain if 10% or more nuclei had three or more signals. For the additional increase (AI) category, in addition to the gain criteria, it was necessary that the overall mean LSP/CEP8 ratio be >1.3. The boundary of the overrepresented region was defined by a change in the LSP copy number status.
Northern Blot Hybridization Analysis
Thirty-nine known genes and ESTs within and surrounding the minimally overrepresented region of 8q23-q24 and the surrounding region were selected using the NCBI and the University of California Santa Cruz databases (http://genome.ucsc.edu/) as shown in Table 1 ▶ . Prostate stem cell antigen (PSCA), which is located in 8q24.3, was included with this study because it has been previously reported to be frequently co-overrepresented with c-myc. 12 These clones were purchased from Research Genetics and were incubated overnight in Luria Broth (LB) agar containing an adequate antibiotic. Plasmid DNAs were isolated using a Plasmid Mini kit (Qiagen, Valencia, CA) and were labeled with [32P]dCTP (RadPrime DNA labeling System; Life Technologies, Inc., Rockville, MD) after hybridization. Normal prostate tissues were obtained from surgical specimens of radical prostatectomy and were kept frozen at −70°C until use. These tissues were pathologically confirmed to have neither benign prostate hyperplasia or cancer lesions. The tissues were shaved with scalpels on dry ice and were homogenized using Tissue-Tearor (BioSpec Products, Inc., Bartlesville, OK) at 15,000 rpm for 15 seconds three times in the presence of lysis buffer. Total RNA was extracted from normal prostate tissues, PC3, and DU145 using an RNeasy mini kit (Qiagen). Ten μl of total RNA was electrophoretically fractionated in 1% agarose gels containing formaldehyde and morpholinopropanesulfonic acid and transformed to nylon membranes (Hybond N+; Amersham Life Science, Inc., Cleveland, OH). Blots were prehybridized at 42°C in a hybridization buffer and then hybridized at 42°C overnight with a cDNA probe for each locus. Hybridized blots were washed with 2× standard saline citrate/0.1% sodium dodecyl sulfate for 30 minutes at room temperature and then 0.1× standard saline citrate/0.05% sodium dodecyl sulfate for 15 minutes at 50°C. The hybridization signals were detected by phosphorimaging (PhosphorImager; Molecular Dynamics, Sunnyvale, CA) and were semiquantified by ImageQuant (Molecular Dynamics). For each probe, the Northern blots were repeated at least three times using independent normal tissue and cell line RNAs. β-actin was used as a loading control.
Table 1.
Northern Blot Analysis of 8q24 Genes/ESTs in Normal Prostate Tissue, PC3, and DU145 Cell Lines*
| NCBI accession no. of EST/known gene | T/N Ratio† | PC3/DU145 | ||
|---|---|---|---|---|
| PC3 | DU145 | |||
| Group I (21 genes/ESTs): not detected in either PC3 or DU145 | AA195113/TNFRSF11B | –‡ | – | – |
| AI033054/SNTB1 | – | – | – | |
| AA041285/COL14A1 | – | – | – | |
| AA115496/PFDN2 | – | – | – | |
| AI027678/KIAA0429§ | – | – | – | |
| N35082 | – | – | – | |
| AI373141 | – | – | – | |
| AA652226 | – | – | – | |
| AA131866 | – | – | – | |
| AA662447 | – | – | – | |
| H99761 | – | – | – | |
| H43165 | – | – | – | |
| R06440 | – | – | – | |
| T83344 | – | – | – | |
| AA234000 | – | – | – | |
| N54807 | – | – | – | |
| AA134929 | – | – | – | |
| H47317 | – | – | – | |
| AA236935 | – | – | – | |
| H81291 | – | – | – | |
| AI093325 | – | – | – | |
| Group II (6 genes/ESTs): not overexpressed either in PC3 or DU145 compared to normal prostate | N33223 | 2.1 ± 0.1 | 1.2 ± 0.2 | 1.8 ± 0.2 |
| AA505468 | 1.7 ± 0.2 | 0.7 ± 0.1 | 2.4 ± 0.5 | |
| AI014960 | 1.7 ± 0.0 | 0.8 ± 0.1 | 2.0 ± 0.3 | |
| AA012865 | 1.7 ± 0.2 | 0.7 ± 0.0 | 2.4 ± 0.4 | |
| AA039790 | 0.8 ± 0.2 | 0.9 ± 0.1 | 0.9 ± 0.3 | |
| AI139599/PSCA¶ | 0.08 ± 0.01 | 0.02 ± 0.00 | 4.5 ± 0.9 | |
| Group III (5 genes/ESTs): overexpressed both in PC3 and DU145 compared to normal prostate | BG503734/RAD21 | 4.3 ± 1.7 | 2.6 ± 0.3 | 1.6 ± 0.6 |
| AA253165/EXT1 | 3.5 ± 2.5 | 2.2 ± 1.0 | 1.5 ± 0.4 | |
| AA400196/SQLE | 5.3 ± 1.8 | 5.5 ± 0.1 | 1.0 ± 0.3 | |
| AA975567/C-MYC | 5.5 ± 0.4 | 3.2 ± 0.3 | 1.7 ± 0.0 | |
| AI168842/KIAA1249§ | 3.6 ± 0.8 | 2.8 ± 0.9 | 1.8 ± 0.5 | |
| Group IV (7 genes/ESTs): overexpressed greater than 2 fold in PC3 relative to DU145 | AA604355/NOV | 2.4 ± 0.5 | 0.2 ± 0.1 | 15.0 ± 7.0 |
| AA024720/EIF3S3 | 4.2 ± 0.3 | 1.9 ± 0.8 | 2.4 ± 0.6 | |
| AA279171/PRO2577§ | 3.6 ± 0.3 | 1.2 ± 0.0 | 2.9 ± 0.4 | |
| AA031811/KIAA0196§ | 3.1 ± 0.6 | 1.5 ± 0.9 | 2.3 ± 1.0 | |
| AA166821/TRC8 | 2.8 ± 0.1 | 0.8 ± 0.2 | 3.5 ± 0.9 | |
| AI142961/HAS2 | +∥ | − | ∞ | |
| R69368/BM009§ | 11.6 ± 2.0 | 3.7 ± 0.5 | 3.1 ± 0.2 | |
*Data are represented as mean ± SD.
†Tumor/normal ratio adjusted for b-actin expression.
‡−, No detectable expression either in PC3 or DU145.
§Hypothetical proteins.
¶Located in 8q24.3.
∥No detectable expression in normal tissue.
Results
FISH Analysis
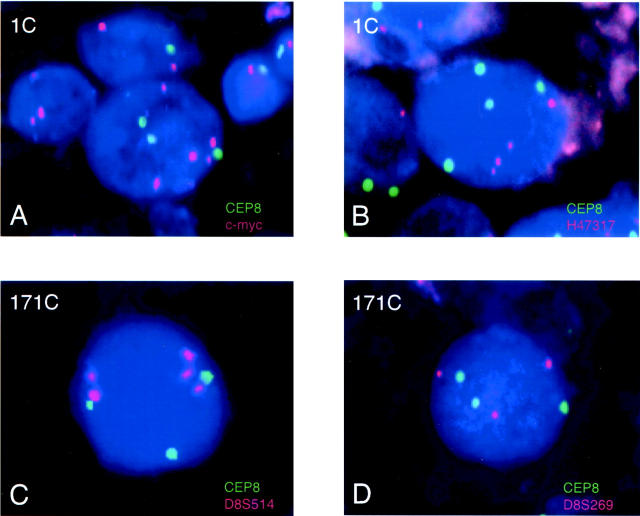
A total of 12 LSPs including a c-myc probe were suitable for FISH analysis; (from centromere to telomere) D8S207, EBAG9, EIF3S3, D8S592, D8S269, D8S514, c-myc, H47317, D8S1720, D8S557, D8S554, and PSCA. Representative FISH results are shown in Figure 1 ▶ . Figure 1A ▶ demonstrates a nucleus from case 1C with three CEP8 signals (green) and five c-myc signals (orange) suggesting gain of CEP8 and AI of c-myc. Figure 1B ▶ demonstrates a nucleus from case 1C with four signals for CEP8 and four signals for H47317, suggesting that whereas both probes were gained, H47317 does not exhibit AI. Statistical analysis of the population of 300 nuclei examined for both LSPs supports these qualitative conclusions (data not shown). Similarly, Figure 1C ▶ demonstrates a nucleus from case 171C showing gain of CEP8 and AI of D8S514, whereas Figure 1D ▶ demonstrates a nucleus from 171C that does not have AI of D8S269 but simple gain of both CEP8 and D8S269.
Figure 1.
Dual-probe FISH with a centromere probe for chromosome 8 (CEP8, green) and a LSP (orange) in critical cases for determination of the minimally overrepresented region of 8q24. The cases examined are indicated in the top left of each panel. Case 1C showed an AI in c-myc signals with a gain of CEP8 signals (A), but no AI of H47317 (B). For case 1C the ratio of LSP to CEP8 was 1.32 for c-myc and 1.17 for H47317. The percentage of nuclei with three or more CEP8 signals was 23.7% for c-myc and 25.6% for H47317. The mean number of CEP8 signals per nucleus was 2.15 for c-myc and 2.19 for H47317. Case 171C showed an AI of D8S514 with a gain of CEP8 (C), but no AI of D8S269 (D). For case 171C the ratio of LSP to CEP8 was 1.51 for D8S514 and 1.06 for D8S269. The percentage of nuclei with three or more CEP8 signals was 21.8% for D8S514 and 25.57% for D8S269. The mean number of CEP8 signals per nucleus was 1.96 for D8S514 and 2.10 for D8S269.
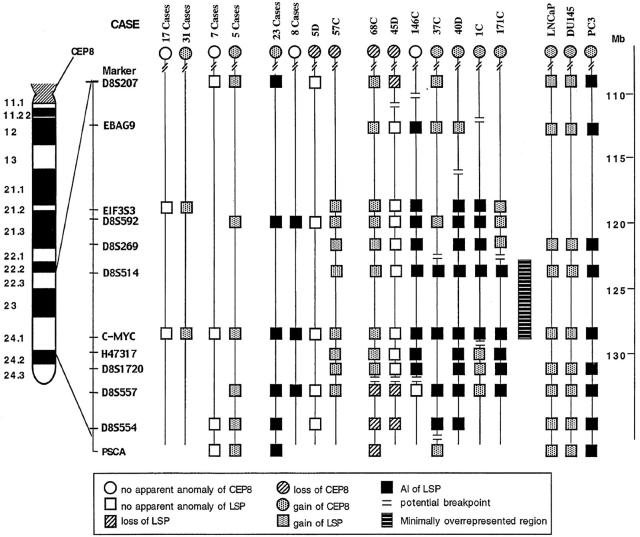
Figure 2 ▶ summarizes the copy number status of all of the probes evaluated in our cohort of 100 cases and three cell lines. Twenty-four cases showed no apparent anomaly of any LSP or CEP8. Thirty-six cases and two cell lines (LNCaP and DU145) had similar gains of all of the LSPs tested and of CEP8, suggesting a simple gain of all of chromosome 8 or of 8q. Twenty-three cases and one cell line (PC3) demonstrated AI of all 8q23-q24 LSPs as well as gain of CEP8, suggesting that they had gain of the centromere of chromosome 8 as well as additional overrepresentation of all of 8q23-q24. Eight cases exhibited AI of LSPs with a normal CEP8 copy number, suggesting they had a normal centromere 8 copy number with additional overrepresentation of the 8q23-q24. Case 5D and case 57C showed normal or gained copy numbers of all of the LSPs examined with loss of CEP8, suggesting that the centromere of chromosome 8 was lost with retention or duplication of 8q23-q24.
Figure 2.
Summary of the CEP8 and LSP8 status in 100 prostate cancer cases and three prostate cell lines. Six common patterns are summarized at the left of the figure and the number of cases is listed above each pattern. Seven cases have unique patterns. The genetic distance along chromosome 8 (in Mb using the NCBI database) is indicated in the scale on the right. A bold line indicates the minimally overrepresented region.
Seven cases had evidence of overrepresentation of a delimited region within 8q23-q24 (Figure 2) ▶ . For example, case 68C had loss of CEP8 while exhibiting gain of the proximal 8q23-q24 LSPs D8S207 through D8S1720 and losing distal 8q24 LSPs D8S557 through PSCA. Case 45D lost CEP8 and three LSPs while retaining LSPs EBAG9 through D8S1720. Case 1C defined the telomeric boundary of the minimally overrepresented 8q24 region. Case 1C showed gain of distal 8q24 LSPs H47317 through D8S557 while having AI of proximal LSPs EIF3S3 through c-myc. Thus, the distal boundary lies between c-myc and H47317. Case 171C defines the centromeric boundary of the minimally overrepresented region. This case showed gain of CEP8 and proximal LSPs EIF3S3 through D8S269, while exhibiting AI of distal LSPs D85S514 through D8S557. Thus, the proximal boundary lies between D8S514 and D8S269. EIF3S3 is centromeric to this boundary (Figure 2) ▶ .
Overrepresentation of EIF3S3 independent of c-myc has been reported in breast cancer. 13 We selected 17 cases with no alteration of chromosome 8 and 31 cases with simple gain of chromosome 8 to determine whether EIF3S3 is overrepresented independently of c-myc. None of the 48 cases showed overrepresentation of EIF3S3 (Figure 2) ▶ .
Northern Blot Hybridization Analysis
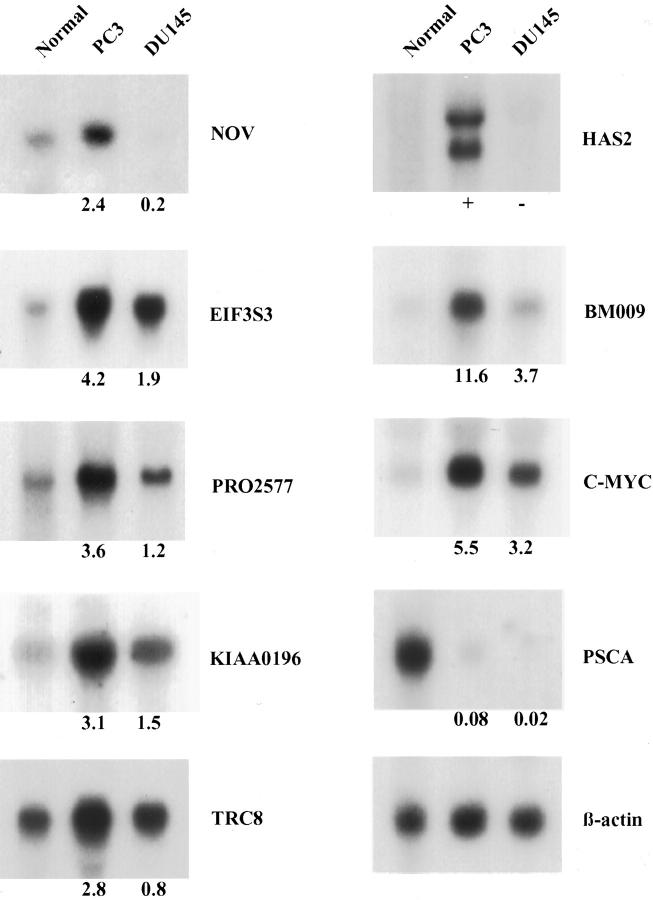
Table 1 ▶ summarizes the PC3 and DU145 expression results for known genes or ESTs in 8q23-q24. These 39 genes were categorized into four different expression pattern groups. Four known genes [tumor necrosis factor receptor superfamily 11B (TNFRSF11B), syntrophin, β1 (SNTB1), collagen type XIV (COL14A1), and prefoldin 2 (PFDN2)], one hypothetical protein (KIAA0429), and 16 ESTs were not detected in either normal prostate tissue or the two cell lines (group I). One known gene (PSCA) and five ESTs were not overexpressed in either of the cell lines compared to normal prostate tissue (group II). For example, PSCA was expressed in normal prostate tissue but was down-regulated in both PC3 and DU145 (Figure 3) ▶ . The remaining four ESTs were similarly expressed in normal prostate tissue and the cell lines. Four known genes and one EST were overexpressed to similar extents in both PC3 and DU145 (the expression ratio of PC3 to DU145 was <2) (group III). This group included RAD21 (Schizosaccharomyces pombe) homologue, multiple exostoses 1 (EXT1), squalene epoxidase (SQLE), c-myc, and a hypothetical protein (KIAA1249) encoded by the EST. Four known genes and three ESTs were overexpressed twofold or more in PC3 compared to DU145 (group IV). Group IV included the nephroblastoma-overexpressed gene (NOV), EIF3S3, patched-related protein translocated in renal cancer (TRC8), and hyaluronan synthase 2 (HAS2). Three of the ESTs encoded hypothetical proteins (PRO2577, KIAA0196, and BM009).
Figure 3.
Differential expression of selected known genes and ESTs for 8q23-24 in normal prostate tissue and in the PC3 and DU145 prostate cancer cell lines. Each lane contained 10 μg of total RNA from the indicated tissue or cell line. The blots were hybridized with the probes to the indicated genes/ESTs. The expression level was semiquantitated by phosphorimage analysis (PhosphorImager). Differences in loading were normalized using β-actin as a control. The number below each blot indicates the cell line/normal tissue expression ratio of each gene/EST for PC3 and DU145.
Discussion
Overrepresentation of 8q24 and c-myc is frequent in prostate cancer. 3,4,14 We recently demonstrated that the overrepresentation of c-myc is observed in 12.8%, 19.4%, and 44% of pT2N0M0, pT3N0M0, and pT2–3N1–2M0 prostate cancers, respectively (N Tsuchiya, JM Slezak, MM Lieber, EJ Bergstralh, RB Jenkins, unpublished data). 3,4 Overrepresentation was correlated with cancer progression and patient prognosis. 3,4 Despite the frequency of 8q24 overrepresentation in prostate cancer, especially hormone-refractory or metastatic prostate cancer, 4,14 the overrepresented region has not been mapped. In the present study, we mapped the minimally overrepresented 8q24 region in prostate cancers. The centromeric boundary of the region, D8S269, is ∼8 Mb proximal to c-myc, whereas the telomeric boundary, H47317, is <0.2 Mb distal to the c-myc locus and is wholly within 8q24 according to the NCBI database.
Nupponen and colleagues 13 recently demonstrated that several hormone-refractory prostate cancers, primary breast cancers, breast cancer cell lines, and prostate cancer cell lines exhibit overrepresentation and high expression levels of EIF3S3, which is located in 8q23. They also mapped the minimally overrepresented region surrounding EIF3S3 using primary breast cancers and breast cancer cell lines. 15 The region was determined to be ∼2.5 cM in size between markers D8S166 and WI-7959, and it did not apparently include c-myc. When we began these experiments, the locations of EIF3S3, D8S592, and D8S269 were unclear. Molecular cytogenetic mapping studies of several of the markers in the region (including EIF3S3, D8S269, c-myc, D8S514) demonstrated that EIF3S3 was located centromeric to both D8S592 and D8S269 (data not shown). The genomic data in the most recent releases of the NCBI, Whitehead Institute, and University of California Santa Cruz databases now agree with this locus order. Thus, the minimally overrepresented region that we determined in this study does not encompass EIF3S3. Saramaki and colleagues 16 recently reported that 1 of 79 prostate cancers with 8q23-q24 overrepresentation had overrepresentation of EIF3S3 without overrepresentation of c-myc. Among 48 cases without overrepresentation of c-myc that we examined, no case showed overrepresentation of EIF3S3. Together these data suggest that EIFS3S overrepresentation independent of c-myc is rare in prostate cancer. However, EIF3S3 is commonly co-overrepresented with c-myc. In this study 34 of 36 cases and in the study of Saramaki and colleague 16 68 of 68 cases co-overrepresented c-myc and EIF3S3 (Figure 2) ▶ . EIF3S3 is a component of EIF3, which is involved in the initiation of protein synthesis. 17 Although the function of EIF3S3 is still unclear, some members of the EIF family are suggested to be associated with apoptosis, cell growth, and proliferation. 13,18,19
C-myc has already been mapped within the 8q24 region and has been assumed to be the primary target of the overrepresentation. The oncoprotein c-myc exerts its effects by transcriptionally activating genes, such as cyclin D and cyclin-dependent kinases (Cdks), which promote cell-cycle progression. Conversely, c-myc expression inactivates several Cdk-inhibitors, such as p27Kip1 and p21Cip1, which also results in cell proliferation. 20 Overexpression of c-myc in model systems increases cellular proliferation rates, induces apoptosis, and increases tumorigenicity. 20 Interestingly, c-myc was not overexpressed in PC3 compared to DU145, although it was highly overexpressed in PC3 relative to normal prostate tissue. The DU145 results demonstrate that the overexpression of c-myc is thus induced both by genomic overrepresentation and by other genetic mechanisms.
However, it is possible that other genes besides c-myc may also be targets within the commonly overrepresented region. Even if c-myc is the target of the overrepresentation of 8q24, these other genes may be used for a diagnostic or therapeutic purpose. We therefore examined the expression of 39 genes/ESTs within and surrounding the minimally overrepresented 8q23-q24 region by Northern blot hybridization. As shown in Figure 2 ▶ , the PC3 and DU145 are prostate cancer cell lines, which have and do not have overrepresentation of 8q23-q24, respectively. Thus, these cell lines can be used to determine which genes in 8q23-q24 are overrepresented and overexpressed.
We found seven ESTs in 8q23-q24 that were overexpressed specifically in PC3 relative to normal prostate and DU145. The most interesting gene found in this study was hyaluronan synthase 2 (HAS2) because HAS2 was expressed only in PC3 and was undetectable in both normal prostate tissue and DU145. HAS is considered to regulate hyaluronan production, 21 which is a ubiquitous polysaccharide expressed in skin, soft connective tissues, and some epithelia. 22,23 HAS has been also known to regulate cell proliferation and migration, 24,25 and overexpression of hyaluronan is associated with cancer cell invasion, metastatic potential, and poor prognosis. 23,26,27 Recently, Simpson and colleagues 28 reported that HAS2 and HAS3 were overexpressed in PC3 and PC3M-LN4, both of which had highly metastatic potential compared to DU145 and LNCaP. It is possible that overrepresentation of 8q24 is a mechanism for HAS2 overexpression and that this genetic alteration may partly explain the invasive and metastatic behavior of prostate cancer.
Other interesting ESTs/genes overexpressed in PC3 are R69368 and NOV. EST clone R69368 encodes the hypothetical protein BM009 (NCBI accession no. AAF64265). This protein shows 85% homology to a mouse protein (NCBI accession no. BAB27204), although the function of this mouse protein is still unknown. NOV was originally found in myeloblastosis-associated virus type 1 (MAV1)-induced nephroblastoma. 29 This gene is a member of the CCN (connective tissue growth factor, CYR61, and NOV) family that stimulates the growth of fibroblast and endothelial cells. 30 NOV is overexpressed in some human cancers, and it may also be involved in prostate cancer. 31
PSCA is often co-overrepresented with c-myc and is overexpressed specifically in high-grade, advanced, and metastatic prostate cancer. 12,32 In this study, we observed five cases that had AI of c-myc without AI of PSCA (cases 68C, 45D, 146C, 37C, and 1C in Figure 2 ▶ ). Thus, our data suggest that PSCA is not the primary target in the overrepresented 8q24 region. In addition, we observed that PSCA was not overexpressed in PC3 or DU145 compared to normal prostate tissue. As the expression level of PSCA may be affected by confluency status in vitro or by castration and testosterone administration in vivo, it is likely that PSCA may be regulated by other factors such as the cell-cell contact or androgen. 33,34 It should be noted that to optimize our PSCA expression studies, we used RNA from 80% confluent cultures in our Northern blots.
The most frequent anomaly in prostate cancer is loss of 8p. The rate of 8p22 loss has been reported to be 29 to 50% in prostatic intraepithelial neoplasia, 32 to 69% in primary prostate cancer, and 65 to 100% in metastatic cancer. 3,35 Several studies using comparative genomic hybridization analysis have revealed that most cases that had 8p22 loss exhibited deletions of the distal two-thirds or all of the 8 p-arm. 14,36,37 In a previous report, we hypothesized that the alteration of 8p is an early event in the development of prostate cancer and is followed by 8q gain. 3 One chromosomal mechanism for this alteration is isochromosome 8q formation [I(8q)]. I(8q) has been suggested to contribute to both 8p loss and 8q gain in a prostate cancer cell line. 38 Although our data suggest that some cases have a delimited region of 8q24 gain, most of the cases we analyzed gained all of 8q23-q24 and probably all of 8q. Thus, isochromosome 8q formation is likely to be the explanation for most, but not all, of the cases with 8q24 overrepresentation. We recentlydemonstrated that AI of 8q is associated with a poor patient prognosis, particularly when it is accompanied with loss of 8p in prostate cancer. 3 The finding suggests an importance of I(8q) formation to acquire the further aggressiveness in cancer progression.
Prostate cancer is now the second cause of cancer-related death of men in the United States. Despite the recent progress in treatments for prostatic cancer, we have no efficient treatments for those patients who suffer from metastases or refractory disease. Therefore, the development of novel therapeutic interventions is critical for such patients. In this study, we found several candidate genes lying on the overrepresented 8q24 region. It is likely that these genes may be targets for such novel therapies for prostate cancer.
Footnotes
Address reprint requests to Robert B. Jenkins, Department of Laboratory Medicine and Pathology, Mayo Clinic, 200 First St. SW, Rochester, MN 55905. E-mail: rjenkins@mayo.edu.
Supported in part by Public Health Service grant CA15083 from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services (to R. B. J.).
References
- 1.Xu J, Meyers D, Freije D, Isaacs S, Wiley K, Nusskern D, Ewing C, Wilkens E, Bujnovszky P, Bova GS, Walsh P, Isaacs W, Schleutker J, Matikainen M, Tammela T, Visakorpi T, Kallioniemi OP, Berry R, Schaid D, French A, McDonnell S, Schroeder J, Blute M, Thibodeau S, Trent J, et al: Evidence for a prostate cancer susceptibility locus on the X chromosome. Nat Genet 1998, 20:175-179 [DOI] [PubMed] [Google Scholar]
- 2.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman JG, Jen J, Isaacs WB, Bova GS, Sidransky D: Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res 1997, 57:4997-5000 [PubMed] [Google Scholar]
- 3.Sato K, Qian J, Slezak JM, Lieber MM, Bostwick DG, Bergstralh EJ, Jenkins RB: Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst 1999, 91:1574-1580 [DOI] [PubMed] [Google Scholar]
- 4.Jenkins RB, Qian J, Lieber MM, Bostwick DG: Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res 1997, 57:524-531 [PubMed] [Google Scholar]
- 5.Neel BG, Jhanwar SC, Chaganti RS, Hayward WS: Two human c-onc genes are located on the long arm of chromosome 8. Proc Natl Acad Sci USA 1982, 79:7842-7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong AJ, Ruppert JM, Eggleston J, Hamilton SR, Baylin SB, Vogelstein B: Gene amplification of c-myc and N-myc in small cell carcinoma of the lung. Science 1986, 233:461-464 [DOI] [PubMed] [Google Scholar]
- 7.Masramon L, Arribas R, Tortola S, Perucho M, Peinado MA: Moderate amplifications of the c-myc gene correlate with molecular and clinicopathological parameters in colorectal cancer. Br J Cancer 1998, 77:2349-2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokota T, Yoshimoto M, Akiyama F, Sakamoto G, Kasumi F, Nakamura Y, Emi M: Frequent multiplication of chromosomal region 8q24.1 associated with aggressive histologic types of breast cancers. Cancer Lett 1999, 139:7-13 [DOI] [PubMed] [Google Scholar]
- 9.Okazaki T, Takita J, Kohno T, Handa H, Yokota J: Detection of amplified genomic sequences in human small-cell lung carcinoma cells by arbitrarily primed-PCR genomic fingerprinting. Hum Genet 1996, 98:253-258 [DOI] [PubMed] [Google Scholar]
- 10.Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang SI, Polikoff D, Nonet G, Cochran J, Myambo K, Jay KE, Froula J, Cloutier T, Kuo WL, Yaswen P, Dairkee S, Giovanola J, Hutchinson GB, Isola J, Kallioniemi OP, Palazzolo M, Martin C, Ericsson C, Pinkel D, Albertson D, Li W-B, Gray JW: Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc Natl Acad Sci USA 1998, 95:8703-8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law M, Jalal SM: M-FISH technique: how to set up and analyze. J Assoc Genet Technol 2000, 26:51-53 [Google Scholar]
- 12.Reiter RE, Sato I, Thomas G, Qian J, Gu Z, Watabe T, Loda M, Jenkins RB: Coamplification of prostate stem cell antigen (PSCA) and MYC in locally advanced prostate cancer. Genes Chromosom Cancer 2000, 27:95-103 [DOI] [PubMed] [Google Scholar]
- 13.Nupponen NN, Porkka K, Kakkola L, Tanner M, Persson K, Borg A, Isola J, Visakorpi T: Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am J Pathol 1999, 154:1777-1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nupponen NN, Kakkola L, Koivisto P, Visakorpi T: Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am J Pathol 1998, 153:141-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nupponen NN, Isola J, Visakorpi T: Mapping the amplification of EIF3S3 in breast and prostate cancer. Genes Chromosom Cancer 2000, 28:203-210 [PubMed] [Google Scholar]
- 16.Saramaki O, Willi N, Bratt O, Gasser TC, Koivisto P, Nupponen NN, Bubendorf L, Visakorpi T: Amplification of EIF3S3 gene is associated with advanced stage in prostate cancer. Am J Pathol 2001, 159:2089-2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asano K, Vornlocher HP, Richter-Cook NJ, Merrick WC, Hinnebusch AG, Hershey JW: Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J Biol Chem 1997, 272:27042-27052 [DOI] [PubMed] [Google Scholar]
- 18.Clemens MJ, Bushell M, Jeffrey IW, Pain VM, Morley SJ: Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ 2000, 7:603-615 [DOI] [PubMed] [Google Scholar]
- 19.Li BD, Liu L, Dawson M, De Benedetti A: Overexpression of eukaryotic initiation factor 4E (eIF4E) in breast carcinoma. Cancer 1997, 79:2385-2390 [PubMed] [Google Scholar]
- 20.Amati B, Alevizopoulos K, Vlach J: Myc and the cell cycle. Front Biosci 1998, 3:D250-D268 [DOI] [PubMed] [Google Scholar]
- 21.Spicer AP, McDonald JA: Characterization and molecular evolution of a vertebrate hyaluronan synthase gene family. J Biol Chem 1998, 273:1923-1932 [DOI] [PubMed] [Google Scholar]
- 22.Laurent TC, Fraser JR: Hyaluronan. FASEB J 1992, 6:2397-2404 [PubMed] [Google Scholar]
- 23.Ropponen K, Tammi M, Parkkinen J, Eskelinen M, Tammi R, Lipponen P, Agren U, Alhava E, Kosma VM: Tumor cell-associated hyaluronan as an unfavorable prognostic factor in colorectal cancer. Cancer Res 1998, 58:342-347 [PubMed] [Google Scholar]
- 24.West DC, Kumar S: The effect of hyaluronate and its oligosaccharides on endothelial cell proliferation and monolayer integrity. Exp Cell Res 1989, 183:179-196 [DOI] [PubMed] [Google Scholar]
- 25.Savani RC, Wang C, Yang B, Zhang S, Kinsella MG, Wight TN, Stern R, Nance DM, Turley EA: Migration of bovine aortic smooth muscle cells after wounding injury. The role of hyaluronan and RHAMM. J Clin Invest 1995, 95:1158-1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertrand P, Girard N, Delpech B, Duval C, d’Anjou J, Dauce JP: Hyaluronan (hyaluronic acid) and hyaluronectin in the extracellular matrix of human breast carcinomas: comparison between invasive and non-invasive areas. Int J Cancer 1992, 52:1-6 [DOI] [PubMed] [Google Scholar]
- 27.Itano N, Sawai T, Miyaishi O, Kimata K: Relationship between hyaluronan production and metastatic potential of mouse mammary carcinoma cells. Cancer Res 1999, 59:2499-2504 [PubMed] [Google Scholar]
- 28.Simpson MA, Reiland J, Burger SR, Furcht LT, Spicer AP, Oegema J, McCarthy JB: Hyaluronan synthase elevation in metastatic prostate carcinoma cells correlates with hyaluronan surface retention, a prerequisite for rapid adhesion to bone marrow endothelial cells. J Biol Chem 2001, 276:17949-17957 [DOI] [PubMed] [Google Scholar]
- 29.Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B: Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1-induced nephroblastomas. Mol Cell Biol 1992, 12:10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kireeva ML, Mo FE, Yang GP, Lau LF: Cyr61, a product of a growth factor-inducible immediate-early gene, promotes cell proliferation, migration, and adhesion. Mol Cell Biol 1996, 16:1326-1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirasaki S, Koide N, Ujiie K, Shinji T, Tsuji T: Expression of Nov, CYR61 and CTGF genes in human hepatocellular carcinoma. Hepatol Res 2001, 19:294-305 [DOI] [PubMed] [Google Scholar]
- 32.Gu Z, Thomas G, Yamashiro J, Shintaku IP, Dorey F, Raitano A, Witte ON, Said JW, Loda M, Reiter RE: Prostate stem cell antigen (PSCA) expression increases with high Gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene 2000, 19:1288-1296 [DOI] [PubMed] [Google Scholar]
- 33.Bahrenberg G, Brauers A, Joost HG, Jakse G: Reduced expression of PSCA, a member of the LY-6 family of cell surface antigens, inbladder, esophagus, and stomach tumors. Biochem Biophys Res Commun 2000, 275:783-788 [DOI] [PubMed] [Google Scholar]
- 34.Dubey P, Wu H, Reiter RE, Witte ON: Alternative pathways to prostate carcinoma activate prostate stem cell antigen expression. Cancer Res 2001, 61:3256-3261 [PubMed] [Google Scholar]
- 35.Qian J, Jenkins RB, Bostwick DG: Genetic and chromosomal alterations in prostatic intraepithelial neoplasia and carcinoma detected by fluorescence in situ hybridization. Eur Urol 1999, 35:479-483 [DOI] [PubMed] [Google Scholar]
- 36.Cher ML, MacGrogan D, Bookstein R, Brown JA, Jenkins RB, Jensen RH: Comparative genomic hybridization, allelic imbalance, and fluorescence in situ hybridization on chromosome 8 in prostate cancer. Genes Chromosom Cancer 1994, 11:153-162 [DOI] [PubMed] [Google Scholar]
- 37.Visakorpi T, Kallioniemi AH, Syvanen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP: Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res 1995, 55:342-347 [PubMed] [Google Scholar]
- 38.Virgin JB, Hurley PM, Nahhas FA, Bebchuk KG, Mohamed AN, Sakr WA, Bright RK, Cher ML: Isochromosome 8q formation is associated with 8p loss of heterozygosity in a prostate cancer cell line. Prostate 1999, 41:49-57 [DOI] [PubMed] [Google Scholar]