Abstract
Fluorescence in situ hybridization (FISH) represents an excellent method for profiling gene amplification in situ, but correlation with tissue morphology is difficult because of dark-field visualization. Validation of a bright-field assay for assessment of HER-2/neu gene amplification was investigated. Streptavidin-Nanogold was used to generate bright-field gene copy signals using GoldEnhance gold-based autometallography, catalyzed reported deposition, and a biotin-labeled probe. One hundred cases of invasive breast carcinoma were evaluated for which FISH gene copy results, and mRNA and oncoprotein gene expression, were known. Autometallographic signals were qualitatively evaluable without the use of oil immersion microscopy. Results correlated well with indirect and direct label FISH. Autometallographic gold-based in situ hybridization represents a promising bright-field assay for the assessment of HER-2/neu gene amplification.
Pharmacogenomics as a clinical and laboratory discipline has emerged as an important rational approach to the treatment of malignancy. Perhaps one of the best examples of pharmacogenomics is the relationship between oncogene genotype and responses to specific available therapy, such as the link between HER-2/neu oncogene genotyping (as reflected in overamplification of the gene) and response to the humanized monoclonal antibody Herceptin alone or in combination with adriamycin and other drugs. 1-6 HER-2/neu amplification/overexpression has been linked to progression of breast carcinoma and responsiveness to tamoxifen, taxanes, and adriamycin. 7-18 With some 180,000 new breast cancer cases diagnosed in women in the United States annually (http://www.cancer.org/statisticis/statistics.html), this new approach to therapy, which may be a paradigm for successful cancer treatment in the future, depends heavily on reliable clinical laboratory results assessing the suitability of a tumor for Herceptin therapy.
There are several ways to measure endogenous gene amplification, including extraction methods based on Southern, Northern, or Western blots, immunohistochemistry, and in situ hybridization. 10-26 Controversy persists as to which of these methods provides the most accurate, precise, and reproducible results as a basis for therapy. 20-36 Perhaps some of this angst reflects pathologists’ discomfort with “working in the dark,” ie, reliance on fluorescence in situ hybridization (FISH) in which morphology and gene amplification are primarily disconnected. Counting gene copy signals using oil immersion high-magnification microscopy, paying someone else to count signals, and/or purchasing special image analysis equipment is expensive and time consuming and not easily accommodated in the usual diagnostic surgical pathology practice. Other FISH limitations include fluorescent signal fading and bleaching limiting slide archiving.
We tested the hypothesis that a bright-field assay for HER-2/neu gene amplification, qualitatively evaluable by conventional light microscopy without the need for oil immersion microscopy, yields results for gene amplification equal to fluorescence-based methods (FISH). HER-2/neu was used as the endogenous gene of interest. This work was based on a series of cases of invasive ductal carcinomas of breast, using direct label FISH as the reference standard. Results were also correlated with digoxigenin-based indirect FISH, RNA:RNA in situ hybridization, and immunohistochemistry using both monoclonal and polyclonal antibodies.
Materials and Methods
Patient samples from 100 primary infiltrating mammary carcinomas were selected for study from a cohort recently published. 36 Each paraffin block from the 100 cases included sufficient redundant paraffin-embedded archived tissue for completion of all of the components of the study. All tissues were first evaluated with a presectioning hematoxylin- and eosin-stained section, as well as a section after preparation of the last unstained section, to confirm the presence of infiltrating carcinoma in material to be studied.
Direct FISH, indirect FISH (DigFISH), RNA:RNA autoradiographic in situ hybridization, and immunohistochemistry using CB11 monoclonal antibody (Ventana Medical Systems, Tucson, AZ) and HercepTest (Dako, Carpinteria, CA), were performed as previously described. 36-39 Statistical comparisons were made using Statview (SAS Institute, Cary, NC).
In Situ Hybridization via Gold-Based Autometallography
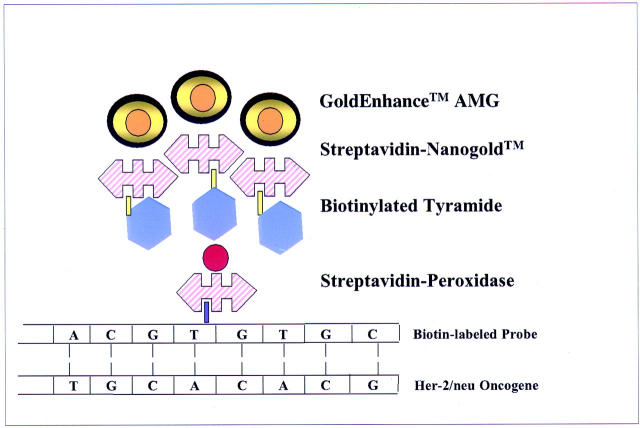
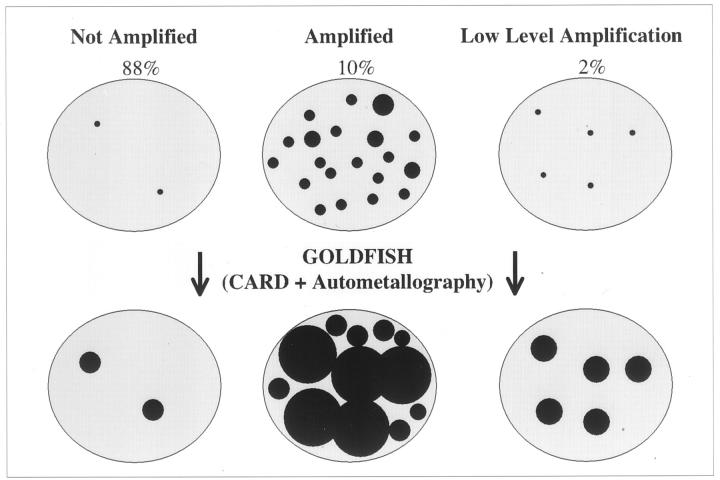
The method is summarized in Figure 1 ▶ . Unstained 4-μm paraffin sections on electrostatically charged slides (Superfrost; Fisher, Pittsburgh, PA) were baked overnight at 60°C, and deparaffinized and rehydrated in two changes of xylene, 5 minutes each, two 1-minute washes in absolute alcohol, two 1-minute washes in 95% alcohol, and then soaked in distilled water for 5 minutes. Sections were then cell conditioned using target retrieval solution (DAKO, Carpinteria, CA) for 40 minutes at 95°C, then allowed to cool for 20 minutes at ambient temperature. Multiple changes of distilled water throughout a 5-minute period were followed by enzyme digestion with Proteinase K (DAKO) at a 1:5000 dilution in 50 mm of Tris-HCl, pH 7.6, for 4 minutes at room temperature. After 5 minutes of washing and several changes of distilled water, endogenous peroxidase was blocked for 20 minutes using 3% hydrogen peroxide (H2O2) in absolute methanol at room temperature. After a 20-minute distilled water wash at room temperature, sections were dehydrated with graded alcohols (absolute, 95%, 70%), and air-dried. Biotinylated cDNA probe (Ventana Medical Systems International, Tucson, AZ) preincubated with COT 1 DNA (1 mg/ml; Roche, Indianapolis, IN), and salmon sperm DNA (Sigma Chemical Co., St. Louis, MO) was applied as 10 μl, and a coverslip applied and sealed with rubber cement. The probe and target were co-denatured for 6 minutes at 90°C, and allowed to hybridize at 37°C overnight in a humidified chamber. Coverslips were then removed by soaking in 2× standard saline citrate (Sigma Chemical Co.), and subjected to a stringency wash of 0.5× standard saline citrate at 72°C for 5 minutes. Sections were then washed with 1× phosphate-buffered saline (PBS) containing 0.1% Tween 20 for 3 minutes at room temperature. Streptavidin-horseradish peroxidase (DAKO SA/HRP from Gen Point Kit at 1:800) 100 μl total volume, was applied for 15 minutes at room temperature. Sections were washed in 1× PBS with 0.1% Tween 20 three times, 5 minutes each wash, at room temperature. Prediluted tyramide signal amplification (TSA) biotin (DAKO Gen Point Kit) was then applied to the sections for 5 minutes at room temperature. Sections were then washed with PBS/Tween 20 three times for 5 minutes each at room temperature, treated with Lugol’s iodine (Fisher) by immersion for 5 minutes at room temperature, followed by three rinses in distilled water, and cleared by immersion for ∼5 seconds in sodium thiosulfate (2.5%). The iodine solution was used to remove contaminating heavy metals in the sections that would interfere with autometallography. Sections were then washed in double-distilled water for a total of 7 minutes using five rinses. Slides were then immersed for 5 minutes in PBS, pH 7.6, containing 0.1% cold water fish gelatin (Sigma). Streptavidin-Nanogold (Nanoprobes, Inc., Yaphank, NY) prediluted to 1:400 with PBS, pH 7.6, containing 1% bovine serum albumin was applied and then incubated for 30 minutes at room temperature. Sections were then washed with PBS, pH 7.6, for two 5-minute washes, immersed in PBS, pH 7.6, with 0.1% cold water fish gelatin for 5 minutes, rinsed in double-distilled water for using multiple changes throughout 10 minutes, and the autometallographic signal developed with GoldEnhance LM/Blot (Nanoprobes, Inc.); the autometallographic signal was developed for 4 minutes at room temperature, and the reaction terminated by adding 500 μl of 2.5% sodium thiosulfate, and then double-distilled water for 3 minutes at room temperature (Figures 2 and 3) ▶ ▶ . Sections were counterstained with nuclear fast red (Newcomer, Middleton, WI) for 8 minutes, dehydrated in graded alcohols and xylene, and coverslipped and mounted with Cytoseal/60 (Stephens Scientific, Riverdale, NJ). Controls for each experiment included positive and negative controls, which consisted of tumors independently and previously evaluated in triplicate for the presence or absence of gene amplification by direct FISH and for which chromosome 17 enumeration had been established. An additional internal positive hybridization control was present on every slide, consisting of the identification of endogenous nonamplified HER-2/neu gene signals in nonneoplastic cells. Figures 2 and 3 ▶ ▶ illustrate the mechanism of signal development. Gold-facilitated fluorescence in situ hybridization (GOLDFISH) slides were interpreted as amplified, low-level amplified, or nonamplified using conventional microscopy at ×100 and ×400. GOLDFISH preparations were designated as “nonamplified” if only one or two small discreet black signals were identified within nuclei of the invasive carcinoma. Approximately 30 to 40% of tumor cell nuclei demonstrated one or two individual small discreet signals—a 4-μm section contains primarily truncated nuclei and the gene target may not be present in all of the cells in an individual 4-μm section. The designation “low-level amplification” was rarely made, and reserved for cases with four to eight (usually four or five) discreet black small nonconfluent signals within invasive tumor nuclei. The designation “HER-2/neu amplified” was reserved for invasive carcinomas demonstrating massive black confluent signals in invasive tumor cell nuclei that were usually located centrally in the nucleus and occupied most of the nuclear area.
Figure 1.
Diagram of GOLDFISH procedure.
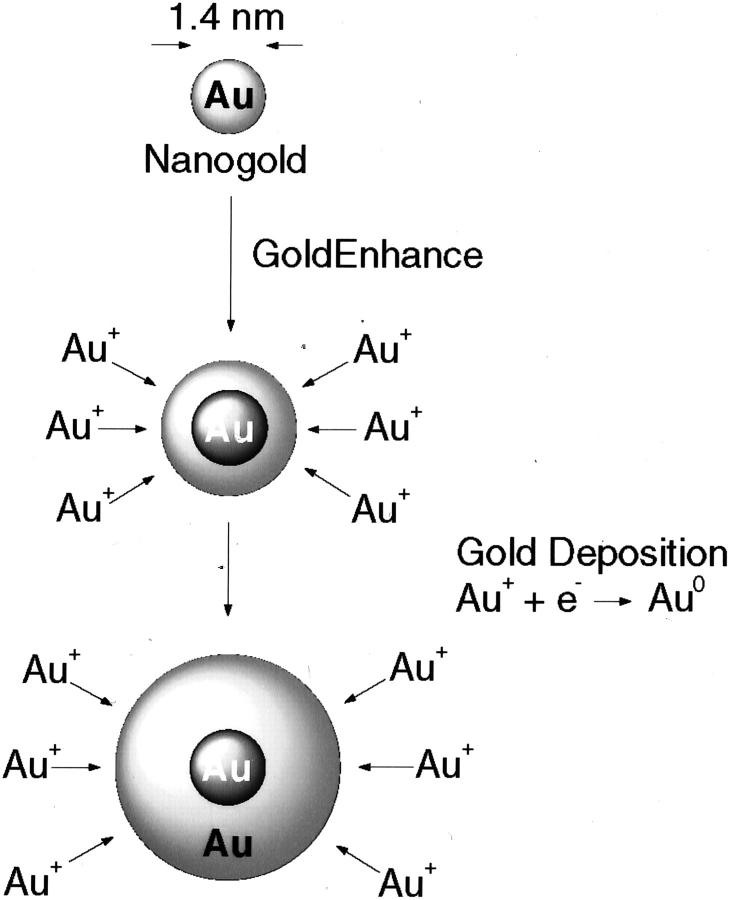
Figure 2.
Mechanism for metallic gold deposition via autometallography using GoldEnhance. Gold ions in solution are catalytically deposited onto the Nanogold particle as metallic gold (Au0). Particle grows in size with development time: shorter times are used for electron microscopy, longer times for light microscopy and blots.
Figure 3.
Mechanism of signal diameter and intensity of growth.
Results
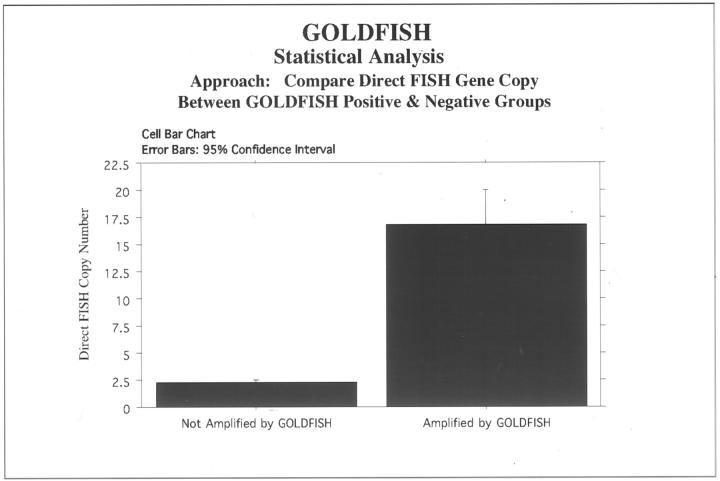
Results of the gold-based autometallographic assay were compared directly with two FISH assays (direct FISH and indirect DigFISH) using digoxigenin-labeled probe), mRNA autoradiographic in situ hybridization, and paraffin section immunohistology. Results are summarized in Table 1 ▶ . The correlation of the autometallographic assay with both DigFISH and the reference direct FISH standard was excellent and identical (Table 1 ▶ and Figures 4, 5, 6, and 7 ▶ ▶ ▶ ). When the assay results were compared to mRNA and protein expression, correlation with mRNA was reasonable but correlation with immunohistochemistry quite variable (Table 1) ▶ . Discrepancies between GOLDFISH and immunohistochemistry results were identified, consistent with the discordances well-documented in the literature when immunohistochemistry for HER-2/neu is compared with FISH. 20-36 Furthermore the discordances between GOLDFISH and immunohistochemistry were observed more frequency with theHercepTest assay than when the GOLDFISH results were compared with immunohistochemistry using the CB11 monoclonal antibody; this result also parallels previous studies contrasting FISH and immunohistochemistry for detection of HER-2/neu gene amplification/overexpression. 20-36
Table 1.
HER-2/neu Gene Amplification Concordance of GOLDFISH Results with Other Methods
| GOLDFISH | Amplified by direct FISH | Amplified by Dig FISH | HercepTest IMH | CB11 IMH | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No | Yes | No | Yes | 0 or 1+ | 2+ or 3+ | 0 or 1+ | 2+ or 3+ | |||
| Not amplified | 81 | 0 | 81 | 0 | 55 | 18 | 8 | 75 | 6 | 0 |
| Low-level amplification | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 0 | 1 |
| Amplified | 1 | 15 | 1 | 15 | 1 | 2 | 13 | 4 | 1 | 11 |
Figure 4.
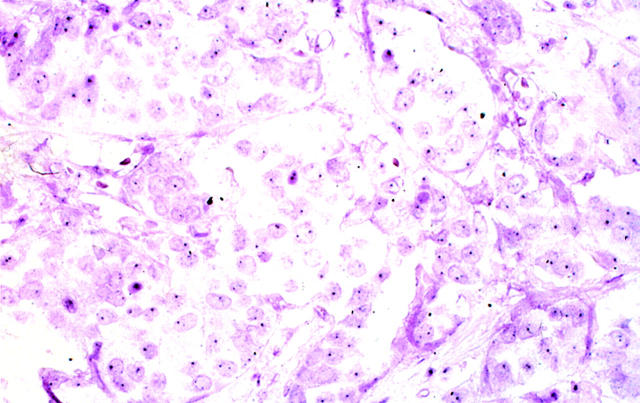
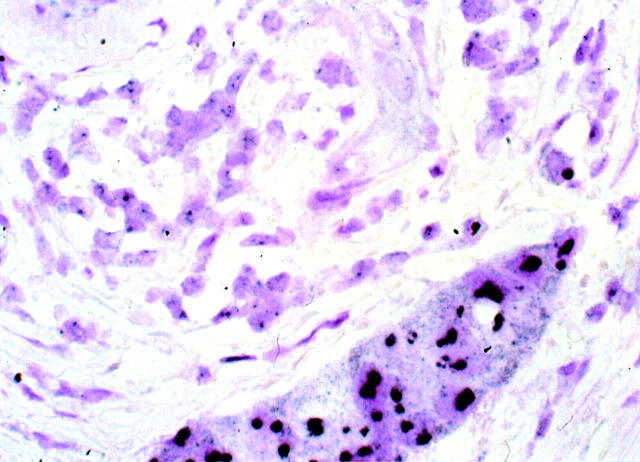
Photomicrograph showing infiltrating ductal carcinoma having a normal (nonamplified) HER-2/neu gene copy and an absence of increased mRNA and oncoprotein expression. One or two HER-2/neu gene copies are identified (black granules, arrows). Paraffin sections contain truncated and partially sectioned rather than whole nuclei; some nuclei therefore have no or only one signal because the target was not present in the section. GOLDFISH with nuclear fast red counterstains; original magnification, ×400.
Figure 5.
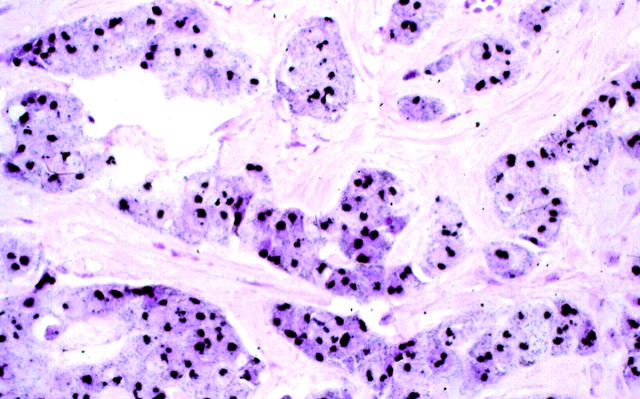
Photomicrograph of a breast carcinoma displaying HER-2/neu gene amplification by FISH, mRNA enhanced expression, and overexpression of the oncoprotein by immunohistochemistry. Most of the nuclear area of the tumor cells contains large confluent black metallic gold signal generated through autometallography. GOLDFISH with nuclear fast red counterstain; original magnification, ×400.
Figure 6.
A nonneoplastic duct displaying normal endogenous (nonamplified) HER-2/neu gene copy (left) is surrounded by infiltrating duct carcinoma demonstrating HER-2/neu gene amplification (right). GOLDFISH with nuclear fast red counterstains; original magnification, ×400.
Statistical analysis using StatView comparing the GOLDFISH results to the direct FISH reference standard reflected excellent concordance of the two methods. The mean difference was −15.042, with the 95% lower confidence at −18.211 and 95% upper confidence limit at −11.873, with a T value of −9.973 and a P value of< 0.0001 (paired means comparison with hypothesized difference = 0).
We pursued the autometallographically enhanced gold 40 and tyramide amplification approach after initial disappointing results using an enzyme immunohistochemistry approach. The peroxidase-based immunohistochemical assay often required oil immersion to view the signals in HER 2/neu nonamplified invasive breast carcinoma and even considering the truncated nature of nuclei and paraffin sections, there was uncertainty regarding the ability to detect nonamplified endogenous gene copy in a consistent manner. The GOLDFISH approach relies on both tyramide signal amplification and autometallography to expand the diameter of individual signals, (Figures 2 and 3) ▶ ▶ . The result for 16 amplified tumors was a massive confluent black nuclear deposit that obliterated most of the nuclear area (Figures 5 and 6) ▶ ▶ . Nonamplified cases (n = 81) demonstrated one or two single discreet small black signals (Figure 4) ▶ in approximately a third to one-half of the nuclei (a 4-μm section consists of a mixture of whole intact nuclei, and partially sectioned truncated nuclei only some of which contain the target gene). The designation “low-level amplification” was used for three cases fulfilling the criteria outlined above.
Discussion
Interphase in situ hybridization for detection of endogenous human genes has almost exclusively been based on fluorescence microscopy (FISH), and its clinical applications have been principally exercised under the purview of cytogenetics laboratories. As more applications for interphase FISH are developed and validated, more molecular pathology and other pathology laboratories, and the pathologists that staff them, will recognize the value of direct visualization of signals derived from endogenous and exogenous DNA and RNA. Pathologists are, at the root of their practice, morphologists. Evaluating DNA and RNA “in the dark” using FISH is for many pathologists counterintuitive; a DNA or RNA assay that is sensitive enough to detect single copies of an endogenous or exogenous gene in the context of morphological changes would have great potential practical value.
Herein we report such a system for bright-field detection of amplification of the oncogene HER-2/neu. Detection is accomplished by using the small 1.44-nm Nanogold gold label, combined with gold-based autometallographic signal enhancement. 40-42 During the course of development of these assays, it became clear that recognition of overamplification was not the challenge for this molecular morphology system. Rather, the development and consistent performance of an assay that reliably detected nonamplified genes represented the greater challenge. Conventional chromogenic enzyme-based in situ hybridization systems 43 were suboptimal in this important regard in our preliminary experience. Of course, one cannot simply assume the absence of gene amplification if normal endogenous gene signals are not detectable, and the confirmation of nonamplification is just as important as the recognition of overamplification of the gene.
We have evaluated gold-enhanced autometallographic in situ hybridization results in comparison with direct FISH and digoxigenin-based indirect FISH assays and found them to be highly comparable. The signals obtained from the use of Streptavidin-Nanogold combined with gold-based autometallography are optically well-defined, sharp, crisp, and dense black signals. 40-42 By design, signal confluency precluded precise enumeration of individual gene copy—our specific intent was to develop a system that did not require counting signals.
The rarity of low-level amplification should be emphasized—only 3 of 100 cases (3%) in our series were classified in this manner (Table 1) ▶ . This unusual category demonstrates FISH gene copy between 4.1 and 6.0 (usually <5.0), and is nearly always nonproductive of transcribed mRNA, and is associated with variable immunohistochemistry (IMH) results (Table 1) ▶ . Copy numbers in this 4.1 to 5.0 range are often pseudoamplifications, that is, more than four copies of chromosome 17 are present, each with a nonamplified copy of the HER-2/neu gene at 17q11.2-q12. The clinical significance of these rare borderline cases will not be known with certainty until results from large clinical outcome studies are reported.
Apparent discrepancies between GOLDFISH and direct FISH results initially occurred in six cases (6%). The investigation of these discrepancies is detailed in Table 2 ▶ . Two cases not amplified by GOLDFISH were shown to actually represent FISH pseudoamplifications because of chromosome 17 polysomy. One low-level GOLDFISH-amplified case was not amplified by FISH; on review, most of the tumor tissue on the GOLDFISH slide was not amplified, but rare focal areas demonstrated background staining and low-level positive GOLDFISH staining. In one instance initial low-level amplification was reviewed and restained and interpreted as focally GOLDFISH-positive (concordant); FISH in this case demonstrated one focal area of amplification. Two cases remained unresolved.
Table 2.
Discrepancies between FISH and GOLDFISH
| GOLDFISH (Preview) | GOLDFISH (Rereview) | Initial direct FISH | HER/ 17 ratio | Dig FISH | Repeat direct FISH | Repeat HER/17 ratio | mRNA | HercepTest | CB11 | Resolution |
|---|---|---|---|---|---|---|---|---|---|---|
| Not amplified | Not amplified | 4.9 | 1.3 | 2.9 | 2.0 | 0.6 | Negative | 3+ | 0 | FISH pseudoamplification* |
| Not amplified | Not amplified | 4.3 | 1.7 | 5.7 | 3.6 | 1.3 | Negative | 3+ | 2+ | FISH pseudoamplification* |
| Low-level amplified | Local low-level amplification | 2.1 | 1.1 | 1.7 | 2.2 | 1.0 | Negative | 0 | 0 | Most of tissue on GOLDFISH not amplified. Rare focal areas demonstrated background and low level positive GOLDFISH staining. |
| Low-level amplified | Amplified | 16.7 | 14.0 | 13.2 | See Resolution | See Resolution | Negative | 3+ | 3+ | GOLDFISH interpreted as focally positive on rereview and again on repeat staining. Repeat direct FISH demonstrated one small area with HER-2/neu amplification and only one chromosome 17 signal per nucleus. |
| Not amplified | Not amplified | 4.8 | 3.3 | 3.9 | 9.3 | 4.2 | Negative | 2+ | 2+ | Unresolved |
| Low-level amplified | Low-level amplified | 3.5 | 1.6 | 4.0 | 3.4 | 1.2 | Negative | 2+ | 0 | Unresolved |
*FISH pseudoamplification was defined as the presence of apparent low-level HER-2/neu gene amplification attributable to a polysomic state of chromosome 17, based on the results of the reference direct dual-color FISH reference standard. In such cases, the HER-2/neu gene copy number was >4, but <8 (usually between 4.1 and 5.0) and multiple copies of chromosome 17 were present, each with a nonamplified HER-2/neu gene at locus 17q11.2-q12, resulting in a HER-2/chromosome 17 ratio < 2.0.
This bright-field assay using gold-based autometallography compares favorably with the reference standard, direct label FISH, for the detection of the amplified endogenous oncogene, HER-2/neu. Pathologists may use this assay to simultaneously evaluate HER-2/neu gene amplification and morphological changes by conventional bright-field microscopy. Signal enumeration and oil immersion microscopy are unnecessary. Further studies are underway to evaluate the interobserver reproducibility of the assay, its portability to automated instrumentation, its relevance to clinical therapeutic outcome, and the utility of GOLDFISH for profiling other gene amplification applications.
Figure 7.
Correlation of GOLDFISH with direct FISH.
Footnotes
Address reprint requests to Raymond R. Tubbs, D.O., Cleveland Clinic Foundation, Department of Clinical Pathology (L11), 9500 Euclid Ave., Cleveland, OH 44195-5131. E-mail: tubbsr@ccf.org.
Supported by STTR grant 1R11 CA83618-01 from the National Cancer Institute to Nanoprobes, Inc. and the Cleveland Clinic Foundation.
References
- 1.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Pharm D, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L: Use of chemotherapy plus a monoclonal antibody against Her-2 for metastatic breast cancer that overexpresses Her-2. N Engl J Med 2001, 344:783-792 [DOI] [PubMed] [Google Scholar]
- 2.Seidman AD, Fornier MN, Esteva FJ, Tan L, Kaptain S, Bach A, Panageas KS, Arroyo C, Valero V, Currie V, Gilewski T, Theodoulou M, Moynahan ME, Moasser M, Sklarin N, Dickler M, D’Andrea G, Cristofanilli M, Rivera E, Hortobagyi GN, Norton L, Hudis C: Weekly Trastuzumab and Paclitaxel therapy for metastatic breast cancer with analysis of efficacy by Her-2 immunophenotype and gene amplification. J Clin Oncol 2001, 19:2587-2595 [DOI] [PubMed] [Google Scholar]
- 3.Baselga J, Tripathy D, Mendelsohn J, Baughman S, Benz CC, Dantis L, Sklarin NT, Seidman AD, Hudis CA, Moore J, Rosen PP, Twaddell T, Henderson IC, Norton L: Phase II study of weekly intravenous recombinant humanized anti-p185HER2 monoclonal antibody in patients with HER2/neu-overexpressing metastatic breast cancer. J Clin Oncol 1996, 14:697-699 [DOI] [PubMed] [Google Scholar]
- 4.Baselga J, Norton L, Albanell J, Kim YM, Mendelsohn J: Recombinant humanized anti-HER2 antibody (Herceptin) enhances the antitumor activity of Paclitaxel and doxorubicin against HER2/neu overexpressing human breast cancer xenografts. Cancer Res 1998, 58:2825-2831 [PubMed] [Google Scholar]
- 5.Cobleigh MA, Vogle CL, Tripath D, Robert NJ, Scholl S, Fehrenbacher L, Paton V, Shak S, Lieberman G, Slamon D: Efficacy and safety of Herceptin (humanized anti-HER2 antibody) as a single agent in 222 women with HER2 overexpression who relapsed following chemotherapy for metastatic breast cancer. Proc Am Soc Clin Oncol 1998, 17:97a. [DOI] [PubMed] [Google Scholar]
- 6.Slamon D, Leyland-Jones B, Shak S: Addition of Herceptin (humanized anti-HER2 antibody) to first line chemotherapy for HER2 overexpressing metastatic breast cancer (HER2+/MBC) markedly increases anticancer activity: a randomized, multinational controlled phase III trial. Proc Am Soc Clin Oncol 1998, 17:98a [Google Scholar]
- 7.Press MF, Bernstein L, Thomas PA, Meisner LF, Zhou JY, Ma Y, Hung G, Robinson RA, Harris C, El-Naggar A, Slamon DJ, Phillips RN, Ross JS, Wolman SR, Flom DJ: HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol 1997, 15:2894-2904 [DOI] [PubMed] [Google Scholar]
- 8.Ro J, El-Naggar A, Ro J: C-erb B-2 amplification in node-negative human breast cancer. Cancer Res 1989, 49:6941-6944 [PubMed] [Google Scholar]
- 9.Seshadri R, Firgaria FA, Horsfall DJ, McCaul K, Setlur V, Kitchen P: Clinical significance of Her-2/neu oncogene amplification in primary breast cancer. J Clin Oncol 1993, 11:1936-1942 [DOI] [PubMed] [Google Scholar]
- 10.Slamon DJ, Godolphin W, Jones L, Holt J, Wong S, Keith D, Levin W, Stuart S, Udove J, Ulrich A, Press M: Studies of the Her-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989, 244:707-712 [DOI] [PubMed] [Google Scholar]
- 11.Tsuda H, Hirohashi S, Shimosato Y, Hirota T, Tsugane S, Yamamoto H, Miyajima N, Toyoshima K, Yamamoto T, Yokota J, Yoshda T, Skamoto H, Terada M, Sugimura T: Correlation between long-term survival in breast cancer patients and amplification of two putative oncogene coamplification units: hst-1 1/int-2 and c-erb B-2/er-1. Cancer Res 1989, 49:3104-3108 [PubMed] [Google Scholar]
- 12.Kallioniemi OP, Holli K, Visakorpi T, Koivula T, Helin HH, Isola JJ: Association of c-erbB-2 protein over-expression with high rate of cell proliferation, increased risk of visceral metastasis and poor long-term survival in breast cancer. Int J Cancer 1991, 49:650-655 [DOI] [PubMed] [Google Scholar]
- 13.Babiak J, Hugh J, Poppema S: Significance of c-erbB-2 amplification and DNA aneuploidy. Analysis in 78 patients with node-negative breast cancer. Cancer 1992, 70:770-776 [DOI] [PubMed] [Google Scholar]
- 14.Toikkanen S, Helin H, Isola J, Joensuu H: Prognostic significance of HER-2 oncoprotein expression in breast cancer: a 30-year follow-up. J Clin Oncol 1992, 10:1044-1048 [DOI] [PubMed] [Google Scholar]
- 15.Thor AD, Berry DA, Budman DR, Muss HB, Kute T, Henderson IC, Barcos M, Cirrincione C, Edgerton S, Allred C, Norton L, Liu ET: erbB2, p53, and efficacy of adjuvant therapy in lymph node-positive breast cancer. J Natl Cancer Inst 1998, 90:1346-1360 [DOI] [PubMed] [Google Scholar]
- 16.Paik S, Bryant J, Park C, Fisher B, Tan-Chiu E, Hyams D, Fisher ER, Lippman ME, Wickerham DL, Wolmark N: erbB-2 and response to doxorubicin in patients with axillary lymph node-positive, hormone receptor-negative breast cancer. J Natl Cancer Inst 1998, 90:1361-1370 [DOI] [PubMed] [Google Scholar]
- 17.Bianco AR, De Laurentiis M, Carlomagno C: 20 year update of the Naples GUN trial of adjuvant breast cancer therapy: evidence of interaction between c-erb-B2 expression and tamoxifen efficacy. Proc Am Soc Clin Oncol 1998, 17:97a [Google Scholar]
- 18.Gusterson BA, Gelbert RD, Goldhirsch A, Price KN, Save-Soderborgh J, Anbazhagan R, Styles J, Rudenstam CM, Golouh R, Reed R: Prognostic importance of c-erbB-2 expression in breast cancer. J Clin Oncol 1992, 10:1049-1056 [DOI] [PubMed] [Google Scholar]
- 19.Pauletti G, Godolpin W, Press MF, Slamon DJ: Detection and quantitation of HER-2/neu gene amplification in human breast cancer archival material using fluorescence in situ hybridization. Oncogene 1996, 13:63-72 [PubMed] [Google Scholar]
- 20.Jimenez RE, Wallis T, Tabasczka P, Visscher DW: Determination of Her-2/neu status in breast carcinoma: comparative analysis of immunohistochemistry and fluorescent in situ hybridization (FISH). Mod Pathol 2000, 13:37-45 [DOI] [PubMed] [Google Scholar]
- 21.Press MF, Hung G, Godolphin W, Slamon DJ: Sensitivity of HER-2/neu antibodies in archival tissue samples: potential source of error in immunohistochemical studies of oncogene expression. Cancer Res 1994, 54:2771-2777 [PubMed] [Google Scholar]
- 22.Roche PC, Ingle JN: Increased Her-2 with US Food and Drug Administration approved antibody. J Clin Oncol 1999, 17:434-435 [DOI] [PubMed] [Google Scholar]
- 23.Roche PC: Letter to editor. CAP Today, June1999, :16 [Google Scholar]
- 24.Tubbs RR: Breast cancer tests: her-2/neu versus FISH. Lab Med 2001, 32:518 [Google Scholar]
- 25.Persons DL, Bui MM, Lowery MC, Mark HFL, Yung JF, Birkmeier JM, Wong EY, Yang SJ, Masood S: Fluorescence in situ hybridization (FISH) for detection of Her-2/neu amplification in breast cancer: a multicenter portability study. Ann Clin Lab Sci 2000, 30:41-48 [PubMed] [Google Scholar]
- 26.Bankfalvi A, Simon, Brandt B, Burger H, Vollmer I, Dockhorn-Dworniczak B, Lelle R-J, Boecker W: Comparative methodological analysis of erbB-2/Her-2 gene dosage, chromosomal copy number and protein over-expression in breast cancer tissues for diagnostic use. Histopathology 2000, 37:411-419 [DOI] [PubMed] [Google Scholar]
- 27.Gancberg D, Lespagnard L, Rouzs G, Paesmans M, Piccart M, DiLeo A, Nogaret JM, Hertens D, Verhest A, Larsimont D: Sensitivity of Her-2/neu antibodies in archival tissue samples of invasive breast carcinomas. Am J Clin Pathol 2000, 113:675-682 [DOI] [PubMed] [Google Scholar]
- 28.Lebeau A, Deimling D, Kaltz C, Sendelhofert A, Iff A, Luthardt B, Untch M, Löhrs U: Her-2/neu analysis in archival tissue samples of human breast cancer: comparison of immunohistochemistry and fluorescence in situ hybridization. J Clin Oncol 2001, 19:354-363 [DOI] [PubMed] [Google Scholar]
- 29.Couturier J, Vincent-Salomon A, Nicolas A, Beuzeboc P, Mouret E, Zafrani B, Sastre-Garau X: Strong correlation between results of fluorescent in situ hybridization and immunohistochemistry for the assessment of the ERBB2 (Her-2/neu) gene status in breast carcinoma. Mod Pathol 2000, 13:1238-1243 [DOI] [PubMed] [Google Scholar]
- 30.Tubbs RR, Stoler MH: The quality of Her-2/neu predictive immunohistochemistry: something FISHy? Mod Pathol 2000, 12:1-3 [DOI] [PubMed] [Google Scholar]
- 31.Hoang MP, Sahin AA, Ordòñez NG, Sneige N: Her-2/neu gene amplification compared with Her-2/neu protein overexpression and interobserver reproducibility in invasive breast carcinoma. Am J Clin Pathol 2000, 113:852-859 [DOI] [PubMed] [Google Scholar]
- 32.Ridolfi RL, Jamehdor MR, Arber JM: Her-2/neu testing in breast carcinoma: a combined immunohistochemical and fluorescence in situ hybridization approach. Mod Pathol 2000, 13:866-873 [DOI] [PubMed] [Google Scholar]
- 33.O’Malley FP, Parkes R, Latta E, Tjan S, Zadro T, Mueller R, Arneson N, Blackstein M, Andrulis I: Comparison of Her-2/neu status assessed by quantitative polymerase chain reaction and immunohistochemistry. Am J Clin Pathol 2001, 115:504-511 [DOI] [PubMed] [Google Scholar]
- 34.Kakar S, Puangsuvan N, Stevens JM, Serenas R, Mangan G, Sahai S, Mihalov ML: HER-2/neu assessment in breast cancer by immunohistochemistry and fluorescence in situ hybridization: comparison of results and correlation with survival. Mol Diagn 2000, 5:199-207 [DOI] [PubMed] [Google Scholar]
- 35.Kakar S, Puangsuvan N, Stevens JM, Serenas R, Mangan G, Sahai S, Mihalov M: Comparison of PathVysion and INFORM fluorescence in situ hybridization kits for assessment of Her-2/neu status in breast carcinoma. Mol Diagn 2000, 5:193-197 [DOI] [PubMed] [Google Scholar]
- 36.Tubbs RR, Pettay JD, Roche PC, Stoler MH, Jenkins RB, Grogan TM: Discrepancies in clinical laboratory testing of eligibility for Trastuzumab therapy: apparent immunohistochemical false-positives do not get the message. J Clin Oncol 2001, 19:2714-2721 [DOI] [PubMed] [Google Scholar]
- 37.Tubbs RR, Pettay JD, Roche PC, Stoler MH, Jenkins RB, Myles J, Grogan TM: Concomitant oncoprotein detection with fluorescence in situ hybridization (CODFISH): a fluorescence-based assay enabling simultaneous visualization of gene amplification and encoded protein expression. J Mol Diagn 2000, 2:78-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angerer LM, Stoler MH, Angerer RC: In situ hybridization with RNAprobes—an annotated recipe. Valentino K Eberwine J Barchus J eds. In-Situ Hybridization Applications to Neurobiology. 1987, :pp 42-70 Oxford University Press, New York [Google Scholar]
- 39.Stoler MH: Tissue in situ hybridization. Henry JB eds. Clinical Diagnosis and Management by Laboratory Methods. 1996, :pp 1400-1412 W. B. Saunders New York [Google Scholar]
- 40.Hainfeld JF, Powell RD, Stein JK, Hacker GW, Hauser-Kronberger C, Cheung ALM, Schofer C: Gold-based autometallography. Bailey GW Jerome WG McKernan S Mansfield JF Price RL eds. Microscopic Microanalysis 5, Suppl. 2, Proceedings. 1999, :pp 486-487 Springer-Verlag, New York [Google Scholar]
- 41.Cheung AL, Graf AH, Hauser-Kronberger C, Dietze O, Tubbs RR, Hacker GW: Detection of human papillomavirus in cervical carcinoma: comparison or peroxidase, Nanogold, and catalyzed reporter deposition (CARD)-Nanogold in situ hybridization. Mod Pathol 1999, 12:689-696 [PubMed] [Google Scholar]
- 42.Graf AH, Cheung AL, Hauser-Kronberger C, Dandachi N, Tubbs RR, Dietze O, Hacker GW: Clinical relevance of HPV 16/18 testing methods in cervical squamous cell carcinoma. Appl Immunohistochem Mol Morphol 2000, 8:300-309 [PubMed] [Google Scholar]
- 43.Tanner M, Gancberg D, DiLeo A, Larsimont D, Rouas G, Piccart MJ, Isola J: A practical alternative for fluorescence in situ hybridization to detect Her-2/neu oncogene amplification in archival breast cancer samples. Am J Pathol 2000, 157:1467-1472 [DOI] [PMC free article] [PubMed] [Google Scholar]