Abstract
Pancreatic β cells respond to changes in blood glucose by secreting insulin and increasing insulin synthesis. To identify genes used in these responses, we have carried out expression profiling of β cells exposed to high (25 mM) or low (5.5 mM) glucose by using oligonucleotide microarrays. Functional clustering of genes that averaged a 2.2-fold or greater change revealed large groups of secretory pathway components, enzymes of intermediary metabolism, cell-signaling components, and transcription factors. Many secretory pathway genes were up-regulated in high glucose, including seven members of the endoplasmic reticulum (ER) translocon. In agreement with array analysis, protein levels of translocon components were increased by high glucose. Most dramatically, the α subunit of the signal recognition particle receptor was increased over 20-fold. These data indicate that the translocon and ribosome docking are major regulatory targets of glucose in the β cell. Analysis of genes encoding enzymes of intermediary metabolism indicated that low glucose brought about greater utilization of amino acids as an energy source. This conclusion was supported by observations of increased urea production under low-glucose conditions. The above results demonstrate genome-wide integration of β-cell functions at the level of transcript abundance and validate the efficacy of expression profiling in identifying genes involved in the β-cell glucose response.
The β cells of the pancreatic islets of Langerhans are a primary component in vertebrate glucose homeostasis. β cells sense glucose directly via its metabolism and respond by secretion of insulin from storage granules formed in the regulated secretory pathway. This stimulus-secretion coupling takes place on the order of seconds/minutes and entails ATP production from glucose metabolism (reviewed in ref. 1). β cells also respond to glucose longer-term (minutes/h) by increasing preproinsulin biosynthesis at the ER membrane (2–4). Derangement of this process leads to reduced insulin production and is a factor in the development of diabetes (5, 6) (reviewed in refs. 7, 8).
Under normal conditions, β cells maintain tight control over intracellular insulin stores (reviewed in ref. 9). Type 2 diabetes is characterized by reduced β-cell insulin stores, which suggests that the diabetic cell is unable to synthesize a sufficient quantity of insulin (10). Insulin production is regulated primarily by glucose at the level of preproinsulin mRNA translation (2, 11), whereas regulation of transcription of the preproinsulin gene can provide longer-term enhancement (12, 13). The mechanism by which glucose exerts translational control of preproinsulin mRNA is poorly understood.
The study of β-cell biology has been advanced greatly by the development of cultured β-cell lines (reviewed in ref. 14). Established cell lines offer several advantages over isolated islet material. Because the pancreatic islet is a complex tissue composed of varied cell types, cell lines offer a pure cell population with which to work. Moreover, β cells from freshly prepared islets exhibit considerable variability in their sensitivity to glucose stimulation and therefore represent a heterogeneous population (15, 16). MIN6 cells are among the best-studied clonal β-cell lines and have been shown in early passages to maintain accurate β-cell function, including robust insulin secretion in response to physiological changes in glucose levels (17, 18). Therefore, MIN6 cells provide a homogeneous β-cell population that responds synchronously and physiologically to changes in glucose concentration.
To identify glucose-responsive genes in β cells, we have carried out expression profiling of MIN6 cells exposed to either high- or low-glucose media by using Affymetrix oligonucleotide microarrays (Affymetrix, Santa Clara, CA). Our data demonstrate genome-wide integration of cellular processes in glucose-stimulated β cells.
Materials and Methods
Preparation of Labeled Targets for Hybridization.
Early-passage MIN6 murine pancreatic β cells (passages 17–19) were maintained as previously described (18). Cells were grown in high-glucose DMEM (GIBCO) supplemented with 10% heat-inactivated FBS (GIBCO) and 28.4 μM 2-mercaptoethanol (Sigma). Once cells reached 75–85% confluence, flasks were rinsed one time in either 25 mM glucose DMEM/10% FCS/28.4 μM 2-mercaptoethanol or 5.5 mM glucose DMEM/10% FCS/28.4 μM 2-mercaptoethanol and then incubated for 24 h in the same medium. Total RNA was prepared (Ambion, Austin, TX), and poly(A) + RNA was then isolated by using oligo-dT-conjugated latex beads (Qiagen, Chatsworth, CA).
Double-stranded cDNA was synthesized from 1 μg of poly(A)+ RNA by using the Superscript Choice System (GIBCO) with an HPLC-purified oligo-dT primer containing a T7 RNA polymerase promoter (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24-3′) (GENSET, La Jolla, CA). After second-strand synthesis, reactions were extracted with phenol/chloroform/isoamyl alcohol, cDNA precipitated with ethanol, and resuspended in 3 μl diethyl pyrocarbonate-treated water. In vitro transcription was carried out on 1.5 μl cDNA by using Bioarray High Yield RNA Transcript Labeling Reagents (Enzo Diagnostics, Farmingdale, NY) following the manufacturer's instructions and incorporating biotinylated CTP and UTP. In vitro transcription reactions yielded 50–70 μg biotin-labeled cRNA. Biotin-labeled cRNA was purified on a RNeasy affinity column (Qiagen) and fragmented at 94°C for 35 min in fragmentation buffer (40 mM/Tris-acetate, pH 8.1/100 mM KOAc/30 mM MgOAc).
Array Hybridization.
Hybridization solution (1 M NaCl/20 mM EDTA/100 mM 2-(N-morpholino)ethanesulfonic acid/0.01% Tween 20) was used to prehybridize Affymetrix Mu6500 oligonucleotide microarrays for 30 min at 40°C. The prehybridization solution was removed and replaced with 200 μl of hybridization solution containing 0.05 μg/μl of fragmented cRNA. The arrays were hybridized for 16 h at 40°C. After hybridization, arrays were washed on an Affymetrix fluidics station and stained with streptavidin-phycoerythrin (hybridization solution, 2 mg/ml acetylated BSA, and 5 μg/μl streptavidin R-phycoerythrin) (Molecular Probes). After staining, arrays were washed extensively in fresh hybridization buffer. Arrays were scanned on a Hewlett–Packard GeneArray Scanner, and the data obtained were analyzed by using Affymetrix genechip 3.2 software. Data are available as supplemental material at www.pnas.org.
Criteria for Selecting Induced/Suppressed Genes and Functional Assignment.
The following criteria were set for determining which genes are glucose responsive in MIN6 cells. Genes were considered up- or down-regulated if the fold change was at least 2.0 in individual experiments and the averaged fold change was 2.2 or greater in duplicate experiments. These limits are in general agreement with array experiments conducted in other mammalian systems (19). It was noted empirically that relaxing the criteria led to identification of large numbers of genes that were not functionally related and/or are not present in pancreatic β cells. Genes were assigned to functional clusters by database searches on Ovid, Pubmed, and the Kyoto Encyclopedia of Genes and Genomes (www.KEGG.com).
Northern Blots.
Northern blots were carried out as described previously (20). Poly(A)+ RNA (0.2 μg/lane) was separated on formaldehyde gels and transferred to Nytran neutral nylon membrane (Schleicher & Schuell). Probes were made by 32P nick translation (Amersham Pharmacia) of sequenced clones [Integrated Molecular Analysis of Genomes and Their Expression (I.M.A.G.E.) Consortium clones, Research Genetics, Huntsville, AL]. Blots were exposed to film and quantified by densitometry (Molecular Dynamics). Blots were reprobed for β actin and the resulting signal quantitated and used for normalization. I.M.A.G.E. clones used were (by GenBank accession no.): argininosuccinate synthetase (AA109145), ornithine decarboxylase (W45736), signal recognition particle receptor α (SRα) (W53731), SEC61α (W62742), translocon-associated protein γ (TRAPγ) (W83038), ornithine aminotransferase (W83543), and spermidine synthase (AA144166).
Taqman Assay.
Taqman 5′ nuclease fluorogenic quantitative PCR assay was carried out according to manufacturer's instructions by using poly(A)+ RNA from MIN6 cells grown for 24 h in either high- (25 mM) or low- (5.5 mM) glucose media. Reverse transcriptase reaction (PE Applied Biosystems) was carried out with 2 ng MIN6 poly(A)+ RNA by using an oligo d(T)16 primer. Taqman assays (PE Applied Biosystems) were carried out by using the following oligonucleotides (5′ to 3′): SRα forward primer TTGAAAAAGGCTATGGCAAGG, SRα reverse primer TACGTGCAAAGGCAATGGC, SRα probe 6FAM-CCATGGCAATGCCAGCAGCG-TAMRA, actin forward primer ACCCACACTGTGCCCATCTAC, actin reverse primer AGCCAAGTCCAGACGCAGG, actin probe VIC-AGGGCTATGCTCTCCCTCACGCCA-TAMRA.
Western Blots.
Western blots were carried out as previously described (21). Antibodies to Sec61α (22) and -β (23) were provided by T. Rapoport (Harvard Medical School, Boston, MA). Antibodies to SRα (24), and TRAPα and -β (25) were provided by C. Nicchitta (Duke University, Durham, NC). α-Tubulin antibody was purchased commercially (Calbiochem). Blot detection was carried out by enhanced chemiluminescence (Amersham Pharmacia) and quantified by densitometry (Molecular Dynamics). Fold induction was normalized by α tubulin.
Pulse Labeling and Immunoprecipitation.
MIN6 cells were incubated in high (25 mM) or low (5.5 mM) glucose for 24 h followed by pulse labeling with [35S]methionine for 45 min as described previously (26). Labeled proinsulin was immunoprecipitated with anti-insulin IgG/Affi-10 beads overnight at 4°C and was eluted into Laemmli buffer, separated on 16% Tricine-buffered polyacrylamide gels, processed for autoradiography, and exposed to film.
Assays.
Urea secretion was determined by assaying media from cells grown 24 h in either high- or low-glucose media. Media were removed and centrifuged at 430 × g for 5 min to remove cells. Urea content was measured by a urease/glutamate dehydrogenase assay on an ATAC 8000 Random Access Chemistry System (Bio-Chem Laboratory Systems, Lakewood, NJ).
Results
Glucose Modulates the Expression of Large Functionally Related Clusters of Genes in MIN6 Pancreatic β Cells.
To identify pancreatic β-cell genes that are glucose responsive, we carried out microarray analysis of murine β-cell line MIN6 cells exposed to either high (25 mM) or low (5.5 mM) glucose for 24 h. RNA, cDNA, and biotinylated cRNA were made as described in Materials and Methods. The resulting biotinylated cRNAs were hybridized to Affymetrix Mu6500 oligonucleotide arrays. Hybridization intensities were analyzed by using Affymetrix genechip analysis suite 3.2 software.
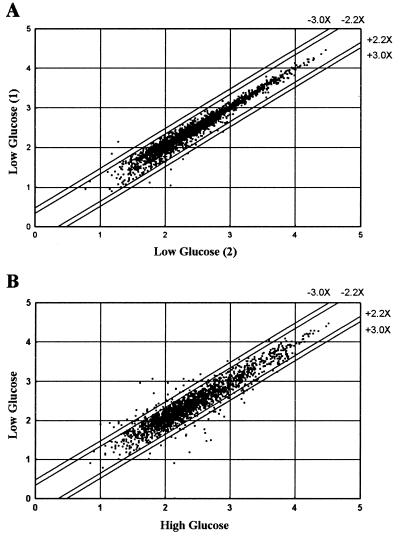
Scatter plot analysis comparing expression levels (log Avg Diff) in two independent experiments where cells were exposed to low-glucose medium show a closely linear distribution of expression levels along the diagonal, demonstrating reproducibility of expression levels in cells treated similarly (Fig. 1A). Scatter plot analysis comparing expression levels in high-glucose vs. low-glucose media demonstrated that many β-cells genes are glucose responsive (Fig. 1B), with 78 demonstrating a 2.2-fold or greater change in transcript levels. The resulting list of β-cell glucose-responsive genes is shown in Table 1 along with GenBank accession nos.
Figure 1.
Scatter plot analysis of expression levels. The expression level for each probe set was plotted to determine the reproducibility of the array-based hybridization signals and to compare gene expression levels of MIN6 cells grown in high- vs. low-glucose media. Transcript expression levels [logΣ (PM-MM)/pairs in average, log Avg Diff] of two separate flasks of MIN6 cells grown in low-glucose medium for 24 h were compared (A). Transcript expression levels of MIN6 cells grown in high- vs. low-glucose media for 24 h were compared (B). The parallel lines flanking the diagonal indicate 2.2- and 3.0-fold changes in gene expression.
Table 1.
Transcripts differentially regulated in high vs. low glucose
| Cluster/GenBank database no. | Gene name | Exp. 1 | Exp. 2 | Avg. |
|---|---|---|---|---|
| Secretion | ||||
| Translocation | ||||
| W83038 | TRAPγ | +9.7 | +13.0 | +11.4 |
| W62742 | SEC61 α | +6.9 | +9.2 | +8.1 |
| W70905 | SRP receptor β | +9.1 | +6.2 | +7.7 |
| AA036265 | TRAPα | +3.3 | +4.8 | +4.1 |
| W53731 | SRP receptor α | +3.1 | +3.7 | +3.4 |
| AA118729 | TRAM protein | +2.5 | +2.8 | +2.7 |
| W08293 | TRAPδ | +2.3 | +2.3 | +2.3 |
| Glycosylation | ||||
| X61172 | α-mannosidase II | +2.6 | +3.1 | +2.9 |
| X65603 | GlcNAc-1-P transferase | +2.7 | +2.2 | +2.5 |
| D87990 | UDP-galactose transporter | +2.5 | +2.4 | +2.5 |
| Vesicle Trans. | ||||
| AA028877 | KDEL receptor 2 | +3.2 | +3.7 | +3.5 |
| AA119959 | SEC23 | +3.0 | +2.5 | +2.8 |
| AA108956 | SEC13 | +2.8 | +2.5 | +2.7 |
| Fold/Process | ||||
| AA048301 | Peptidyl-prolyl cis-trans isomerase | +5.3 | +6.0 | +5.7 |
| M58589 | Prohormone convertase 3 | +4.3 | +3.5 | +3.9 |
| U79523 | Peptidyl α-amidating monooxy | +2.6 | +3.5 | +3.1 |
| Protein Synth. | ||||
| M98035 | GEF, δ for eIF-2B | +2.5 | +2.4 | +2.5 |
| AA104459 | eIF4G | +2.2 | +2.5 | +2.4 |
| U39473 | Histidyl-tRNA synthetase | +2.2 | +2.1 | +2.2 |
| Unassigned | ||||
| Z46845 | Preproglucagon | +5.5 | +4.3 | +4.9 |
| W62646 | NEFA protein | +3.4 | +2.3 | +2.9 |
| Metabolism | ||||
| Carbohydrate | ||||
| Y00309 | Lactate dehydrogenase | +16.3 | +10.6 | +13.5 |
| W65920 | α enolase (1) | +3.3 | +2.8 | +3.1 |
| W33721 | α enolase (2) | +2.4 | +2.3 | +2.4 |
| U27014 | Sorbitol dehydrogenase | −4.2 | −4.5 | −4.4 |
| AA044561 | PEPCK | −3.9 | −3.3 | −3.6 |
| Nitrogen | ||||
| X64837 | Ornithine aminotransferase | +5.0 | +3.1 | +4.1 |
| J03733 | Ornithine decarboxylase | +5.2 | +2.1 | +3.7 |
| L19311 | Spermidine synthetase | +3.8 | +2.8 | +3.3 |
| M31690 | Argininosuccinate synthetase | −6.5 | −7.6 | −7.1 |
| Mitochondria | ||||
| M63445 | Methylene-dehydrogenase | −18.5 | −8.6 | −13.6 |
| AA061310 | Mitochondrial lon protease | −12.7 | −6.5 | −9.6 |
| X57349 | Transferin receptor | −4.5 | −3.3 | −3.9 |
| Unassigned | ||||
| X97755 | Δ8-Δ7 sterol isom. | +3.2 | +4.3 | +3.8 |
| AA03494 | Glutathione peroxidase related | +2.2 | +2.8 | +2.5 |
| J04758 | Tryptophan hydrolase | +2.8 | +2.2 | +2.4 |
| W85270 | Inorganic pyrophosphatase | +2.2 | +2.2 | +2.2 |
| U06670 | VLDL receptor | −10.3 | −6.1 | −8.2 |
| X07888 | HMG-CoA reductase | −2.9 | −4.1 | −3.5 |
| U12961 | Menadione oxidoreductase | −4.1 | −2.0 | −3.1 |
| Signaling | ||||
| W09590 | Protein-tyrosine phosphatase ζ | +15.0 | +15.0 | +15.0 |
| U28423 | Protein kinase inhibitor p58 | +16.8 | +5.9 | +11.4 |
| AA048650 | Estradiol dehydrogenase | +4.5 | +2.5 | +3.5 |
| U50413 | PI 3-kinase reg. subunit | +3.0 | +4.0 | +3.5 |
| L13593 | Pseudo-prolactin receptor | +3.8 | +2.4 | +3.1 |
| W64628 | Guanine nucleotide-binding protein γ7 | +3.1 | +2.7 | +2.9 |
| AA106492 | cAMP-dep. protein kinase, reg. | +2.0 | +2.6 | +2.3 |
| X15373 | P400 protein | −5.3 | −3.9 | −4.6 |
| D10939 | ERK2 | −3.5 | −5.2 | −4.4 |
| W47946 | Gephyrin | −3.7 | −3.5 | −3.6 |
| L02241 | Protein kinase inhibitor | −3.1 | −2.6 | −2.9 |
| L34214 | Glucocorticoid regulatory protein | −2.7 | −2.8 | −2.8 |
| Transcription | ||||
| M31885 | HLH DNA-binding protein regulator | +3.3 | +3.4 | +3.4 |
| X67083 | CHOP-10 protein | −19.2 | −14.5 | −16.9 |
| U29762 | Albumin gene D-box binding protein | −6.0 | −5.5 | −5.8 |
| L20450 | DNA-binding protein | −3.1 | −2.2 | −2.7 |
| U64828 | Steroid receptor coactivator 1a | −2.6 | −2.7 | −2.7 |
| Z67747 | Zinc finger protein | −2.7 | −2.2 | −2.5 |
| AA041651 | Transcription factor BTF3 | −2.4 | −2.2 | −2.2 |
| Apoptosis | ||||
| L22472 | Bax α | −2.3 | −2.0 | −2.2 |
| Cell Cycle | ||||
| L76150 | CDK4/CDK6 inhibitor protein | −2.9 | −10.9 | −6.9 |
| RNA Splicing | ||||
| AA035984 | Polypyrimidine tract-binding protein | +3.3 | +6.8 | +5.1 |
| AA003990 | Splicing factor SRP20 | +2.1 | +2.3 | +2.2 |
| AA146248 | U2-snRNPb (pRNP11) | −2.4 | −2.3 | −2.4 |
| Miscellaneous | ||||
| M10114 | κ casein | +13.1 | +3.8 | +8.5 |
| U38981 | Uterine mRNA | +6.0 | +5.7 | +5.9 |
| M36120 | Keratin 19 | +5.0 | +4.6 | +4.8 |
| X58438 | Proline-rich protein | +4.1 | +4.4 | +4.3 |
| AA051446 | CDR 2 | +3.7 | +3.8 | +3.8 |
| X85994 | Semaphorin E | +3.0 | +3.7 | +3.4 |
| X59520 | Cholecystokinin | +3.4 | +2.6 | +3.0 |
| AA051500 | Unknown | +2.6 | +3.0 | +2.8 |
| X16151 | Early T-lymphocyte activated protein | +3.2 | +2.2 | +2.7 |
| M55413 | Vitamin D-binding protein | +3.2 | +2.0 | +2.6 |
| AA059763 | β-2 tubulin | +2.2 | +2.9 | +2.6 |
| AA060064 | I3 protein | −4.3 | −3.1 | −3.7 |
| M33212 | Nucleolar protein NO38 | −3.5 | −2.6 | −3.1 |
| AA049662 | Plasma retinol-binding protein Prec. | −3.0 | −2.9 | −3.0 |
List of mRNAs that are increased or decreased an average of ≥2.2-fold in high-glucose media. Identity of columns, from left to right: functional cluster assignment and representative clone GenBank accession no, name of gene, fold change observed in experiment 1, fold change observed in experiment 2, and averaged fold change. Complete data sets are available at www.pnas.org. TRAP, translocon-associated protein; TRAM, translocating-chain associating membrane protein; Exp., experiment; Avg., average.
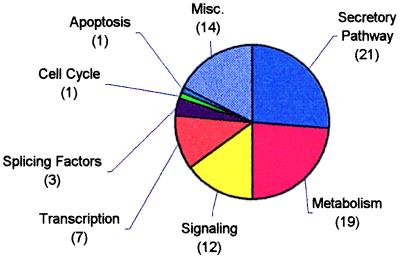
These glucose-responsive genes and expressed sequence tags (ESTs) were clustered according to known cellular functions assigned to the genes or by sequence similarity to genes of known function. Significant numbers of these genes were found to function in the secretory pathway, metabolism, signaling, and transcriptional regulation. These clusters account for over 75% of the identified genes and ESTs (Fig. 2).
Figure 2.
Pie chart demonstrating the relative sizes of functional clusters of genes up- or down-regulated by incubation of MIN6 cells for 24 h in high (25 mM) or low (5.5 mM) glucose.
Glucose Regulation of the Secretory Pathway.
Secretory pathway genes accounted for 21 of the 78 genes found to be glucose responsive. Subgroupings showed transcriptional regulation of members of the multiprotein ER translocon, enzymes involved in glycosylation, coat proteins for vesicle-mediated transport and sorting, and protein folding/processing. Because secretory pathway transcripts comprised the largest functional cluster identified, we chose to investigate further the regulation of their biosynthesis.
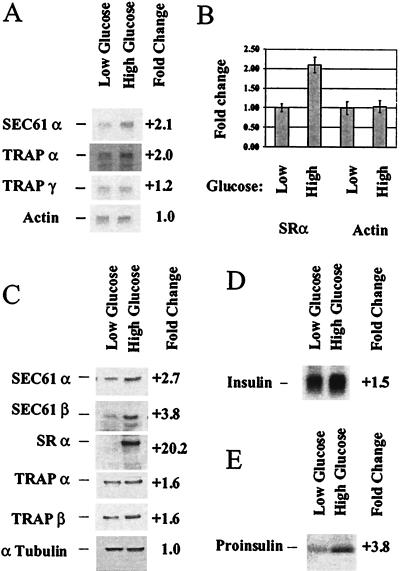
Translocon components represented the largest subgrouping of the secretory pathway genes and included SRα and β, the translocon pore-forming component (SEC61α), less characterized TRAPα, -γ, and -δ, and translocating-chain-associating membrane protein. We carried out Northern blots on several members to verify these results. Northern blotting confirmed that SEC61α and TRAPα are up-regulated in high glucose, whereas TRAPγ was up-regulated to a lesser degree (Fig. 3A). These data confirm the results found with the microarrays. Northern blotting, however, showed less dramatic changes in transcript levels than seen by microarray analysis. Northern blotting for the SRα was not successful, suggesting that very little of this transcript was present in either high- or low-glucose conditions. We therefore carried out a Taqman assay to quantify changes in SRα gene expression. These results show that the SRα transcript is also up-regulated by glucose over 2-fold (Fig. 3B).
Figure 3.
Analysis of representative translocon component transcripts and proteins confirms changes in mRNA levels and demonstrates changes in encoded protein levels. Northern blotting was carried out on poly(A)+ RNA (0.2 μg/lane) from MIN6 cells incubated for 24 h in high (25 mM) or low (5.5 mM) glucose to determine changes in expression level in translocon components (A). Each blot was reprobed for β actin to normalize quantitation. Taqman assay was carried out on 0.2 ng poly(A)+ RNA from MIN6 cells treated as in A to determine changes in expression level of SRα (B). Protein was extracted from MIN6 cells treated as in A and immunoblots of translocon components performed (C). Northern blotting was carried out on poly(A)+ RNA (0.05 μg/lane) from MIN6 cells treated as in A and probed for insulin (D). MIN6 cells treated as in A were pulse labeled and insulin immunoprecipitated (E).
We next carried out Western blots on translocon components to determine whether the effects seen at the transcript level were reflected in the abundance of translocon proteins. For this analysis, antibodies raised to canine translocon proteins were used (provided by T. Rapoport and C. Nicchitta). In all cases, the antibodies recognized predominately a single protein of the appropriate molecular weight. This analysis demonstrated modest up-regulation in high glucose in the amount of TRAPα and β (1.6-fold), whereas significantly more stimulation of translocon pore proteins SEC61α and β was observed (2.7- and 3.8-fold, respectively). By contrast, a very dramatic up-regulation of SRα protein was observed that constituted an over 20-fold change in the level of receptor in high-glucose conditions. These data indicate that SRα is a major target of glucose regulation and may be a limiting component in glucose-responsive insulin synthesis in pancreatic β cells.
Proinsulin biosynthesis under the above experimental conditions was also investigated. We found with our experimental design that, whereas there is a modest effect on the amount of preproinsulin transcript, 1.5-fold difference between high- and low-glucose conditions (Fig. 3D), there is a greater effect, 3.8-fold, on the biosynthetic rate of proinsulin at the ER membrane (Fig. 3E). These results are consistent with the regulation of the translocon and SRα that we see.
Glucose Regulation of Intermediary Metabolism.
The second largest cluster of glucose-responsive genes identified were enzymes involved in intermediary metabolism. These included up-regulation of enzymes for forward movement through the glycolytic pathway and down-regulation of enzymes for gluconeogenesis, nitrogen disposal, and mitochondrial protein import and synthesis. Although the effects of glycolytic flux and mitochondrial function on β cells are well characterized, we were intrigued by differential expression of genes that function in and around the disposal of nitrogen through the urea cycle.
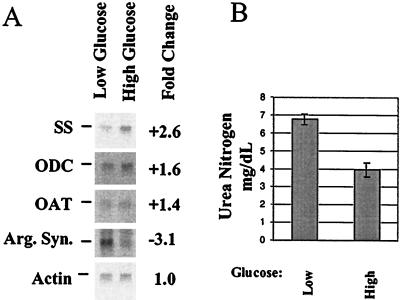
The urea cycle requires free ornithine as the acceptor for ammonia derived from amino acid catabolism. Northern blots confirmed that under low-glucose conditions, transcripts for enzymes that catabolize ornithine (ornithine aminotransferase) or polymerize ornithine (ornithine decarboxylase and spermidine synthase) were down-regulated (Fig. 4A). These data suggested β cells scavenge free ornithine under low-glucose conditions. Under the same conditions, the transcript for argininosuccinate synthetase was up-regulated (Fig. 4A). This enzyme processes citrulline derived from the condensation of ornithine, ammonia, and carbon dioxide through the second step of the urea cycle. These data suggested that under low-glucose conditions, cells used the scavenged free ornithine in the urea cycle. Thus cells in low glucose are predicted to produce more urea than cells in high glucose. We assayed the amount of urea secreted by cells exposed to high or low glucose and determined that cells exposed to low glucose produce almost twice as much urea as cells grown in high glucose (Fig. 4B). We infer that the observed increase in urea production indicates increased utilization of amino acids as an energy supply.
Figure 4.
Analysis of nitrogen metabolism transcripts confirms changes in mRNA levels, and analysis of urea production confirms the functional consequence of this regulation. Northern blotting was carried out on poly(A)+ RNA (0.2 μg/lane) from MIN6 cells grown for 24 h in high (25 mM) or low (5.5 mM) glucose to determine changes in expression level in nitrogen metabolism components (A). Each blot was reprobed for β actin to normalize quantitation. Media from cells grown as above were analyzed to determine the concentration of secreted urea (B).
Discussion
Pancreatic β cells sense glucose by changes in metabolic flux and respond by synthesis and secretion of insulin. The data presented here confirm and extend this model by presenting an overview of glucose-responsive gene expression in the mouse β-cell line MIN6. By using Affymetrix Mu6500 microarrays and cRNA produced from cells maintained 24 h in high glucose (25 mM) or low glucose (5.5 mM), we have demonstrated the regulation of transcript abundance of several large functionally related gene clusters. These include multiple genes that function in the secretory pathway, intermediary metabolism, cell signaling, and transcriptional regulation. Our analysis focused on the two largest clusters, the secretory pathway and intermediary metabolism.
The single largest cluster encompassed 21 transcripts that encode components of the early secretory pathway and regulated secretory pathway. These included seven members of the ER translocon, transcripts encoding genes necessary for glycosylation, protein folding, and vesicle-mediated transport, sorting, and processing. The majority of these genes is involved in ER function and indicates that regulation of ER function is important in β-cell glucose response. The regulated secretory pathway components included the processing enzyme prohormone convertase 3 and peptidylglycine α-amidating monooxygenase. Previous work on PC3 has demonstrated its response to glucose (11). Together, these results highlight the core β-cell function in the synthesis and secretion of insulin and other hormonal peptides through the early steps of the secretory pathway and ultimately into the regulated secretory granule.
The predominance of ER translocon components suggests that regulation of synthesis at the ER membrane may be a major mechanism in regulating insulin production in β cells. Our data demonstrate that not only does the β cell regulate the transcript abundance of multiple translocon components, but also this leads to changes in the amounts of the encoded proteins. These changes in protein levels were most dramatically demonstrated for the SRα subunit. The level of SRα protein was over 20-fold up-regulated in high-glucose medium. Because the change in observed protein levels is much greater than the observed change in transcript levels, it is likely that glucose-responsive translational control of SRα synthesis contributes significantly to this increase.
The SRα protein serves to dock the signal recognition particle-arrested ribosome/nascent polypeptide complex onto the ER membrane (27). On binding the SRα, SRP is released from the ribosome/nascent polypeptide complex, and the synthesis of the protein resumes as it passes through the translocon and into the ER lumen. It has been demonstrated that a primary means of regulating insulin biosynthesis is at the level of mRNA translation (2–4, 11). Under the conditions of our experimental design, the rate of proinsulin biosynthesis was 3.8-fold greater in high glucose compared with low glucose. Under the same conditions, the amount of preproinsulin transcript changed only 1.5-fold. Therefore glucose regulation of translation contributes significantly to glucose regulation of insulin biosynthesis. We propose that a part of this translational control includes the amount of SRα present to derepress SRP-arrested protein synthesis. Indeed, studies by Welsh and colleagues demonstrated that SRα was limiting in movement of SRP/ribosome/nascent preproinsulin complex onto the ER (3). Our present results demonstrate that β cells actively regulate the abundance of the SRα transcript and protein in response to glucose and thereby may constitute at least one component in translational regulation.
In addition to glucose regulation of the ER translocon, we have also observed the up-regulation of components of translation initiation (eIF-2B guanine nucleotide exchange factor and eIF-4G). Translation initiation factors have been hypothesized to participate in the regulation of insulin biosynthesis (28). Our data support this interpretation and therefore indicate that glucose regulation is exerted at multiple levels in insulin biosynthesis.
Metabolism is a key component of β-cell function. In addition to enzymes involved in glucose metabolism, we identified differential expression of multiple genes involved in ammonia nitrogen disposal. Although glucose metabolism has been demonstrated to be uniquely important in β-cell function, our data indicate that metabolism of amino acids and the resulting need for nitrogen disposal may play a greater role in β-cell function than previously appreciated. Our results showed that under low-glucose conditions, transcripts for multiple enzymes that remove free ornithine from the cell were depressed. Concomitantly, the transcript for an enzyme that initiates the synthesis of urea from free ornithine and amino acid-derived ammonia is up-regulated. Functional data demonstrated that cells incubated in low-glucose medium produced significantly more urea than cells incubated in high-glucose medium. These data suggest that adaptation of β cells to low glucose includes greater utilization of amino acids as an energy source.
Identification of genes whose encoded proteins function in the mitochondria highlights the role this organelle plays in β-cell function. Genes encoding components necessary for protein import/turnover (lon protease) as well as mitochondrial protein synthesis (methylenetetrahydrofolate dehydrogenase) were up-regulated in low-glucose conditions. These data suggest that the mitochondria adapt to low-glucose conditions by modifying protein components, possibly to adapt to a greater utilization of amino acids, as discussed above. Alternatively, these changes may relate to the production of coupling factors in the mitochondria that relate glucose concentrations to insulin secretion (29, 30).
Through our analysis, we also identified many genes that operate in cell-signaling pathways. Many of the metabolic signals and ion fluxes that bring about glucose-stimulated insulin secretion are known (1); however, the longer-term effects of glucose on β-cell function are poorly understood. The signaling components uncovered by the described experiments indicate that longer-term changes in β-cell function, from increased insulin synthesis to proliferative responses, may be initiated by signaling pathways that are themselves glucose responsive. Recent efforts toward treatment of insulin-dependent diabetes mellitus (IDDM) via β-cell transplants of harvested β cells or in vitro differentiation of stem cells should benefit from an understanding of which signaling pathways operate in β cells.
The last large cluster of genes identified was multiple glucose-responsive transcription factors. This group included the transcript encoding Chop10/GADD153, which demonstrated the single largest change in transcript level found in our experiments. This transcription factor is a nutrient-responsive gene that is up-regulated when nutrients are reduced (31, 32). Our results reconfirm this finding and indicate that Chop10 may play a role in adaptation of β cells to low-glucose conditions as well.
There are important β-cell genes that we did not see regulated in our experiments. One of these is, of course, insulin. Northern blot data indicated that glucose led to only a 1.5-fold change in the abundance of the preproinsulin transcript (Fig. 3D). This agrees with the microarray data that indicated 1.7-fold more preproinsulin transcript in cells incubated in high-glucose medium (data not shown). As discussed above, this suggests that under our experimental conditions, glucose regulation of insulin biosynthesis takes place predominately at the level of protein translation. It should be noted, however, that preproinsulin transcripts demonstrate half-lives of greater than 24 h, and therefore transcript abundance may not accurately reflect transcription rates (33). A second protein important to β-cell function is the glycolytic enzyme glucokinase. The function of this enzyme as a sensor of glucose levels in well established (reviewed in ref. 34). Previous findings indicate that glucose has little effect on the β-cell-specific glucokinase mRNA (35, 36). Like insulin, glucose modulation of β-cell glucokinase activity is likely to be at the level of mRNA translation and effector proteins.
The Affymetrix Mu6500 oligonucleotide array allows interrogation of 6,500 murine genes and ESTs found in the GenBank database. This collection of probes corresponds largely to genes of known cellular function, which has greatly facilitated analysis of the presented data by permitting functional cluster of β-cell glucose-responsive transcripts. The experimental design followed has both confirmed and extended previous analysis of pancreatic β cells. Current efforts in the lab are under way both to expand this analysis to larger numbers of transcripts and to observe the time course of β-cell glucose stimulation. This will not only identify novel candidate genes and functional pathways but also will demonstrate the temporal profile of β-cell glucose stimulation.
In summary, our current experiments on MIN6 cells identify glucose-responsive expression of many large functional clusters of genes. The two largest clusters encode secretory pathway components and enzymes of intermediary metabolism. In the secretory pathway, SRα is a major glucose-responsive protein and suggests a mechanism by which β cells regulate insulin biosynthetic levels. Data from our study also indicate that amino acid metabolism and the subsequent need for ammonia disposal are responsive to glucose levels in the β cell. Overall, the described results are the first direct demonstration of widespread integration of these important processes in β cells and illustrate the complex dynamics of whole-cell adaptation to changes in glucose levels. Our findings also demonstrate that microarray-based technology provides a powerful tool for identifying genes involved in the function and regulation of pancreatic β cells.
Supplementary Material
Acknowledgments
We thank Margaret Milewski, Paul Gardner, and Jeff Stein for technical assistance. We also thank Clive Slaughter and Steve Madden at Howard Hughes Medical Institute/University of Texas Southwestern Medical Center (Dallas) for expert assistance with oligonucleotide array analysis. Our thanks also to Graeme Bell, Kenneth Polonsky, Louis Philipson, and Joseph Bass for encouragement, discussions, and helpful criticism of this work. This work was supported by the Howard Hughes Medical Institute and by grants from the National Institutes of Health (DK13914 and DK20595).
Abbreviations
- ER
endoplasmic reticulum
- SRα and -β
signal recognition particle receptor α and β
- TRAPα
-γ, and -δ, translocon-associated proteins α, γ, and δ
- EST
expressed sequence tag
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100126597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100126597
References
- 1.Lang J. Eur J Biochem. 1999;259:3–17. doi: 10.1046/j.1432-1327.1999.00043.x. [DOI] [PubMed] [Google Scholar]
- 2.Itoh N, Okamoto H. Nature (London) 1980;283:100–102. doi: 10.1038/283100a0. [DOI] [PubMed] [Google Scholar]
- 3.Welsh M, Scherberg N, Gilmore R, Steiner D F. Biochem J. 1986;235:459–467. doi: 10.1042/bj2350459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollheimer L C, Skelly R H, Chester M W, McGarry J D, Rhodes C J. J Clin Invest. 1998;101:1094–1101. doi: 10.1172/JCI420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tasaka Y, Marumo K, Inoue Y, Hirata Y. Acta Endocrinol. 1986;113:355–362. doi: 10.1530/acta.0.1130355. [DOI] [PubMed] [Google Scholar]
- 6.Shima K, Zhu M, Noma Y, Mizuno A, Murakami T, Sano T, Kuwajima M. Diabetes Res Clin Pract. 1997;35:11–19. doi: 10.1016/s0168-8227(96)01357-5. [DOI] [PubMed] [Google Scholar]
- 7.Kahn C R, Vicent D, Doria A. Annu Rev Med. 1996;47:509–531. doi: 10.1146/annurev.med.47.1.509. [DOI] [PubMed] [Google Scholar]
- 8.Kahn B B. Cell. 1998;92:593–596. doi: 10.1016/s0092-8674(00)81125-3. [DOI] [PubMed] [Google Scholar]
- 9.Halban P A. Diabetologia. 1991;34:767–778. doi: 10.1007/BF00408349. [DOI] [PubMed] [Google Scholar]
- 10.Hosokawa Y A, Leahy J L. Diabetes. 1997;46:808–813. doi: 10.2337/diab.46.5.808. [DOI] [PubMed] [Google Scholar]
- 11.Alarcon C, Lincoln B, Rhodes C J. J Biol Chem. 1993;268:4276–4280. [PubMed] [Google Scholar]
- 12.Brunstedt J, Chan S J. Biochem Biophys Res Commun. 1982;106:1383–1389. doi: 10.1016/0006-291x(82)91267-0. [DOI] [PubMed] [Google Scholar]
- 13.Schuppin G T, Rhodes C J. Biochem J. 1996;313:259–268. doi: 10.1042/bj3130259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poitout V, Olson L K, Robertson R P. Diabetes Metab. 1996;22:7–14. [PubMed] [Google Scholar]
- 15.Van Schravendijk C F, Kiekens R, Pipeleers D G. J Biol Chem. 1992;267:21344–21348. [PubMed] [Google Scholar]
- 16.Kiekens R, In ‘t Veld P, Mahler T, Schuit F, Van De Winkel M, Pipeleers D. J Clin Invest. 1992;89:117–125. doi: 10.1172/JCI115551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skelly R H, Schuppin G T, Ishihara H, Oka Y, Rhodes C J. Diabetes. 1996;45:37–43. doi: 10.2337/diab.45.1.37. [DOI] [PubMed] [Google Scholar]
- 18.Miyazaki J, Araki K, Yamato E, Ikegami H, Asano T, Shibasaki Y, Oka Y, Yamamura K. Endocrinology. 1990;127:126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 19.Fambrough D, McClure K, Kazlauskas A, Lander E S. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 20.Virca G D, Northemann W, Shiels B R, Widera G, Broome S. BioTechniques. 1990;8:370–371. [PubMed] [Google Scholar]
- 21.Bass J, Chiu G, Argon Y, Steiner D F. J Cell Biol. 1998;141:637–646. doi: 10.1083/jcb.141.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorlich D, Prehn S, Hartmann E, Kalies K U, Rapoport T A. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- 23.Gorlich D, Rapoport T A. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 24.Nicchitta C V, Migliaccio G, Blobel G. Cell. 1991;65:587–598. doi: 10.1016/0092-8674(91)90091-c. [DOI] [PubMed] [Google Scholar]
- 25.Migliaccio G, Nicchitta C V, Blobel G. J Cell Biol. 1992;117:15–25. doi: 10.1083/jcb.117.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furukawa H, Carroll R J, Swift H H, Steiner D F. Diabetes. 1999;48:1395–1401. doi: 10.2337/diabetes.48.7.1395. [DOI] [PubMed] [Google Scholar]
- 27.Gilmore R, Walter P, Blobel G. J Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu G, Marshall C A, Lin T A, Kwon G, Munivenkatappa R B, Hill J R, Lawrence J C, Jr, McDaniel M L. J Biol Chem. 1998;273:4485–4491. doi: 10.1074/jbc.273.8.4485. [DOI] [PubMed] [Google Scholar]
- 29.Eto K, Tsubamoto Y, Terauchi Y, Sugiyama T, Kishimoto T, Takahashi N, Yamauchi N, Kubota N, Murayama S, Aizawa T, et al. Science. 1999;283:981–985. doi: 10.1126/science.283.5404.981. [DOI] [PubMed] [Google Scholar]
- 30.Maechler P, Wollheim C B. Nature (London) 1999;402:685–689. doi: 10.1038/45280. [DOI] [PubMed] [Google Scholar]
- 31.Carlson S G, Fawcett T W, Bartlett J D, Bernier M, Holbrook N J. Mol Cell Biol. 1993;13:4736–4744. doi: 10.1128/mcb.13.8.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Q, Lau S S, Monks T J. Biochem J. 1999;341:225–231. [PMC free article] [PubMed] [Google Scholar]
- 33.Welsh M, Nielsen D A, MacKrell A J, Steiner D F. J Biol Chem. 1985;260:13590–13594. [PubMed] [Google Scholar]
- 34.Bell G I, Pilkis S J, Weber I T, Polonsky K S. Annu Rev Physiol. 1996;58:171–186. doi: 10.1146/annurev.ph.58.030196.001131. [DOI] [PubMed] [Google Scholar]
- 35.Tiedge M, Lenzen S. Biochem J. 1991;279:899–901. doi: 10.1042/bj2790899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magnuson M A. J Cell Biochem. 1992;48:115–121. doi: 10.1002/jcb.240480202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.