Abstract
Nasal natural killer (NK)/T-cell lymphoma (NL) frequently co-expresses Fas (Apo-1/CD95) and Fas ligand (FasL), but the tumor cells seldom undergo apoptosis. To determine the reason for failure of apoptosis, we examined Fas mRNA expression in 23 NL cases by reverse transcriptase-polymerase chain reaction and sequenced the entire coding region of the Fas gene in 15 of these cases for which the full-length Fas cDNA could be amplified. The reverse transcriptase-polymerase chain reaction analysis revealed that all of the 23 cases expressed Fas mRNA and the sequencing results showed that in addition to the commonly expressed wild-type Fas mRNA and four alternative splice variants detected in 7 cases, mutant Fas transcripts were present in 9 of the 15 (60%) cases sequenced. With confirmation of some Fas mutations at the gene level, 12 deletions in nine cases and one insertion in one case were eventually identified. To rule out any potential polymerase chain reaction artifacts, the same protocol was used to examine 10 reactive tonsils as a control. No aberrant transcripts associated with deletions were detected in these tonsils except for three alternative splice variants. All of the deletion variants detected in NL contained N-terminal preligand assembly domain but not C-terminal death domain and/or transmembrane domain. Co-detection of the wild-type allele and the mutated Fas alleles without the death domain suggested that a dominant-negative mechanism could block the apoptosis signaling. Moreover, loss of the transmembrane domain could protect the tumor cells from apo-ptosis by producing a soluble form of the Fas receptor. The actuarial 3-year survivals leveled off at 15% for patients carrying the Fas mutations and/or splice variants in the lesions and 49% for those carrying the wild type only, but the difference did not reach statistical significance on the univariate analysis (P = 0.396). Taken together, the findings in this study suggest that frequent Fas gene mutations in NL can result in resistance to apoptosis and may contribute to the pathogenesis of NL by adding to the tumor immune privilege.
Cell death by apoptosis is mediated via the coordinated interactions of many different gene products. Mutations in some of them, acting at different levels in the apoptotic cascade, have been identified as causes or contributing pathogenetic factors for many human diseases. 1 Fas (Apo-1/CD95) is an apoptosis-signaling cell surface receptor belonging to the tumor necrosis factor receptor superfamily 2 and was recently defined as a tumor suppressor gene. 3 Fas, together with its ligand (FasL), is a key element in maintaining the homeostasis of lymphocytes and many other types of cells. Autoimmune lymphoproliferative syndrome type 1a (ALPS-1a) provides the best model for the study of the human Fas gene mutations in vivo, because failure of immune clearance of autoimmune lymphocytes in individuals with ALPS-1a is consistently associated with inherited Fas gene defects. 4-10 In contrast, less frequent somatic mutations of the Fas gene have been described in some subtypes of lymphoid malignancies, such as B-cell lymphoma, 11-13 adult T-cell leukemia, 14,15 and Hodgkin’s disease, 16 whereas no somatic mutations of Fas gene were found in any of the 35 cases of peripheral T-cell lymphoma (unspecified) examined in a recent study. 11
In Hong Kong Chinese, nasal natural killer (NK)/T-cell lymphoma (NL) represents the second most frequent group of extra-nodal lymphomas 17 and consists of tumors of either NK-cell or T-cell lineage, with most being NK-cell and a smaller number being true T-cell lymphomas. 18-20 NL is closely associated with Epstein-Barr virus (EBV) infection, 21-23 and the tumor cells are able to provide target epitopes for cytotoxic T lymphocytes. 24 In vivo, however, the tumor cells seem to escape immune recognition. We have previously identified several mechanisms acquired by NL tumor cells to escape immune surveillance, including down-regulation of the immunogenic EBV nuclear antigens by alternative promoter usage, 25 preferential selection of the deletion genotype of latent membrane protein 1, 26 and expression of the immune suppressive human interleukin-10 (hIL-10). 24 It has been recently reported that NL cells frequently co-express Fas and FasL, but seldom undergo apoptosis, 27 suggesting that there may be defects in the Fas-triggered programmed cell death pathway.
In the current study, we investigated the expression and the somatic mutations of the Fas gene in NL biopsy samples to clarify whether Fas mutations play a role in limiting apoptosis, thereby providing the tumor with immune escape.
Materials and Methods
Case Description
Total RNA samples extracted from NL tumor biopsy specimens of 23 Chinese patients used in this study have been described previously. 25 The lymphomas were diagnosed according to the World Health Organization classification of lymphoid malignancies. 28 RNA extracted from 10 reactive tonsils was analyzed as a control. These tonsils were obtained from Chinese patients who underwent tonsillectomy for enlarged tonsils at the Queen Mary Hospital, Hong Kong.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) and Detection of Mutations
RT-PCR was performed on RNA extracted from 23 NL tumors and 10 reactive tonsils in a thermocycler (PTC-200; MJ Research, Waltham, MA) using the strategy as described, 7 with the GeneAmp RNA PCR kit (Perkin-Elmer-Cetus, Norwalk, CT) and three pairs of primers, forward primer 1 (F1) (5′-TCTTTCACTTCGGAGGATTGCT-3′)/reverse primer 1 (R1) (5′-GAACTTTCTGTTCTGCTGTGTCTTG-3′), F2 (5′-TGCCAAGAAGGGAAGGAGTA-3′)/R2 (5′-ACCAAGCAGTATTTACAGCCAG-3′) and F3 (5′-CCAAGACACAGCAGAACAGAAAG-3′)/R3 (5′-CACCAAGGCAAAAATGGAGAG-3′), to amplify the entire coding region corresponding to the Fas mRNA sequences nucleotides 165 to 1020, nucleotides 447 to 1287, and nucleotides 995 to 1972 (GenBank accession no. M67454), respectively (Figure 1a) ▶ . For enrichment of the specific PCR products for Fas mRNA, nested PCR was performed on the diluted first-round PCR products using primers FI (5′-GGAGGATTGCTCAACAACCAT-3′)/RI (5′-CATTGTCATTCTTGATCTCATCTATTT-3′) for F1/R1 and FII (5′-GGAAGGAGTACACAGACAAAG-3′)/RII (5′-ACAGCCAGCTATTAAGAATC-3′) for F2/R2, which will give 818-bp and 817-bp PCR products, respectively. To verify the quality of RNA samples used in this study, a pair of primers (5′-ACCAGGGCTGCTTTTAACTCT-3′ and 5′-GGGTCTCTCTCTTCCTCTTGTG-3′) were used in the RT-PCR to amplify a 1005-bp cDNA fragment of glyceraldehyde-3-phosphate dehydrogenase as the template quality control. Total RNA from the phytohemagglutinin-activated Jurkat cell line served as the positive control and distilled water served as the negative control.
Figure 1.
Schematic representation of the normal and the defective Fas genes in NL.a: Structure of the Fas cDNA is illustrated, consisting of nine coding exons and the flanking untranslated regions (UTR). The locations of the primers used in the study for RT-PCR are marked in the figure. The structure of Fas protein is illustrated, which is composed of the PLAD, the transmembrane domain (TM), and the death domain (DD). b: A variety of Fas mRNA sequences detected in NL are summarized at the left, including variants of alternative splicing, deletion, and insertion, which are represented respectively by dashed line, continuous line, and arrow labeled with the number of position involved. Each variant is coded in the form of case no.-sequence no. The regions affected by frame-shift effects are drawn in shaded boxes, and the predicted positions of stop codons are marked with asterisks. Fas domains affected by these nucleotide alterations are summarized at the right. +, retained; ±, partially retained; and −, lost.
RT-PCR products were analyzed in a 2% agarose gel, followed by purification of the DNA fragments from gel slices using the QIAEX II gel extraction kit (Qiagen GmbH, Hilden, Germany). The purified nested PCR products of unexpected sizes were directly sequenced from both directions using the DYEnamic ET terminator cycle sequencing kit (Amersham Pharmacia Biotech, Piscataway, NJ) by the ABI PRISM 377 DNA Sequencer (Applied Biosystems, Foster City, CA). The first-round normal-sized PCR products of F1/R1 and F2/R2 were directly sequenced from both directions; they were also cloned into the pGEM-T Easy vector (Promega, Madison, WI) and at least 10 clones for each PCR fragment were sequenced. To rule out PCR artifacts, RT-PCR analysis was repeated using the Expand High Fidelity PCR System (Boehringer Mannheim, Mannheim, Germany), and the PCR products were sequenced again as described above. When the same mutation was found in both sequence analyses, the data were regarded as confirmed.
Confirmation of Mutations at the DNA Level
In the NL cases with apparent alternative splicing (sequences represented by dashed lines in Figure 1 ▶ ), genomic DNA was analyzed to detect any potential Fas gene mutations at the gene level mimicking alternative splicing by deletions of exons or aberrant splicing because of mutations at the splice sites. The genomic sequences covering exons 3, 4, 6, and 7 were examined by PCR using the Expand Long Template PCR system (Boehringer Mannheim) and primers Fa (5′-CAGAAATCAATAAAATTCTCTTCATG-3′)/Ra (5′-GGGAAAGGAGGATATAACCG-3′), Fb (5′-GGAATCATCAAGGAATGCACACTCACC-3′)/Rb (5′-GAGCAAGACTCCATCTCAAA-CAAAATGAAA-3′) and Fc (5′-ATAATATGCCAATGTTC-CAACC-3′)/Rc (5′-TTTACTCTGAAATTGGCCTATTAC-3′), which will give PCR products corresponding to the Fas DNA sequences from EMBL DNA database 29 (nucleotide 189, entry no. X82280; nucleotide 391, entry no. X82283; nucleotide 305, entry no. X82283; nucleotide 292, entry no. X82284; nucleotide 482, entry no. X82283; nucleotide 266, entry no. X82285, respectively). The PCR products were analyzed by electrophoresis in a 1% agarose gel to detect the abnormal-sized fragments with deletions of exons. Purified PCR products were directly sequenced from both directions; they were also cloned into the pGEM-T Easy vector (Promega) and at least 10 clones for each PCR fragment were sequenced for detection of exon deletions or mutations at the splice sites within exon/intron boundaries.
To confirm the Fas gene deletion in variant 9-2 (25-bp deletion within exon 3), the sequence encompassing the deleted region was amplified by PCR on the genomic DNA from case 9 using primers Fd (5′-TGCCAAGAAGGGAAGGAGTA-3′)/Rd (5′-TGCATTTTAAGACTCTTACCATGTC-3′) (nucleotides 281 to 381, EMBL DNA database entry no. X82281). The PCR products were examined in an 8% polyacrylamide gel electrophoresis and the purified DNA fragments were analyzed by direct sequencing. Moreover, different cycle numbers of PCR were performed to get a better evaluation of the ratio between the wild-type (101 bp) and the deletion (76 bp) alleles at the DNA level. For confirmation of 1-bp insertion at nucleotide 1095 in exon 9 sequence of a cDNA variant in case 21, the DNA from the same case was examined by PCR using primers Fe (5′-CAATGTCCAAGACACAGCAGAACAGAAAGT-3′)/Re (5′-CCAAGCAGTATTTACAG-CCAGCTATTAAGAATCT-3′) (nucleotides 989 to 1286, GenBank accession no. M67454). The PCR products were cloned into pGEM-T Easy vector (Promega) and 90 clones were sequenced.
Immunostaining of Fas and FasL
Only 16 of 23 cases had paraffin blocks available for the immunohistochemical detection of Fas and FasL proteins. The primary antibodies were rabbit polyclonal FAS (C-20) (1:100; Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit polyclonal FAS-L (N-20) (1:100; Santa Cruz Biotechnology). The EnVision staining system (DAKO, Carpinteria, CA) was used in the subsequent steps for the visualization of the antigens.
Statistical Analysis
The significance of Fas mutations on the overall survival rates was estimated using the standard Kaplan-Meier survival analysis.
Results
Expression of the human Fas gene was examined in RNA samples from 23 cases of NL by RT-PCR using three pairs of primers: F1/R1, F2/R2, and F3/R3 (Figure 1a) ▶ . The F3/R3 fragment, which is at the 3′ end of Fas mRNA, could be easily amplified in all of the cases (data not shown), demonstrating consistent Fas mRNA expression in NL; whereas the RT-PCR products of F1/R1 and F2/R2, which in combination cover the entire coding region, could be detected in only 15 of 23 cases, possibly because of partial degradation of RNA in some samples. Some abnormal-sized fragments were clearly observed among the first-round PCR products in many cases (data not shown); nevertheless, a nested PCR was performed to rule out the nonspecific PCR products for Fas mRNA. Besides the expected bands of normal size, various small-sized fragments ranging from 200 bp to 800 bp could be easily observed during electrophoresis in most cases, raising the possibility that partial Fas gene deletion or alternative pre-mRNA splicing might have occurred in these cases of NL (Figure 2a) ▶ . Sequencing of the RT-PCR products of the entire coding region of the Fas gene and further confirmation of some mutations at the DNA level identified four alternative splice variants in seven cases, 12 deletions in nine cases and one insertion in one case (Figure 1b) ▶ . Several mutant alleles with different deletions were also detected in single tumor specimens from cases 10, 15, and 17 (Figures 1b ▶ and 2b ▶ ).
Figure 2.
Somatic mutations of the Fas mRNA in NL. a: Various RT-PCR products of unexpected sizes were detected by electrophoresis (open arrowheads), suggestive of Fas deletion or splicing. M, molecular marker of 100-bp ladder; number, case number; −, negative control of PCR using distilled water; +, positive control of PCR using cDNA from phytohemagglutinin-activated Jurkat cell line. b: Sequence analysis shows several distinct mutant forms of deletions in case 10 (three deletion variants) and cases 15 and 17 (two deletion variants each), in addition to the normal transcripts and alternative splice variants. The sequences of the left and the right break points of a deletion combined with an insertion of a 205-bp Fas fragment in 15-2 are presented.
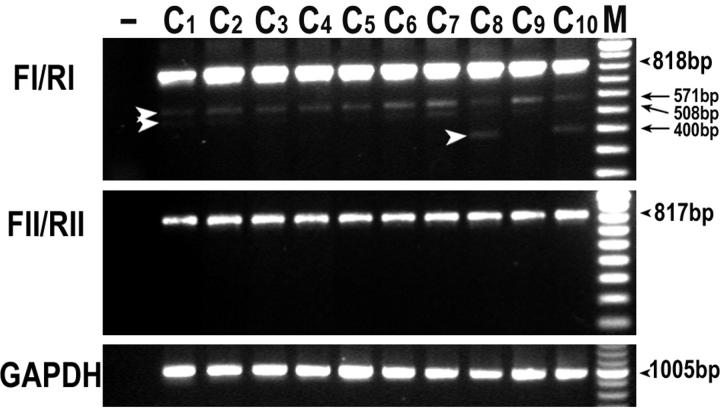
PCR artifacts caused by nucleotide mismatching were ruled out by the repetition of RT-PCR. To exclude any rare deletion artifact during PCR, the identical protocol for the NL analysis was used in parallel to examine 10 cases of reactive tonsils by RT-PCR as an additional control. As shown in Figure 3 ▶ , no aberrant transcripts associated with deletions were detected in these unselected cases of tonsils, except for three alternative splice variants in which the whole exons 3 + 4 (571 bp), exons 3 + 4 + 6 (508 bp), and exons 3 + 4 + 6 + 7 + 8 (400 bp) were skipped, respectively, confirming the reliability of our results.
Figure 3.
RT-PCR analysis of the Fas mRNA in tonsillar controls. The identical protocol used for analysis of the NL tumors was applied on 10 cases of reactive tonsils (C1 to C10). No aberrant transcripts associated with deletion were detected except for three alternative splice variants as indicated by open arrowheads (571 bp, 508 bp, and 400 bp). −, negative control using distilled water; M, molecular marker of 100-bp ladder.
In the NL cases with alternative splice variants (sequences represented by dashed lines in Figure 1 ▶ ), PCR amplification and sequencing of the chromosomal DNA covering the skipped exons were performed to rule out potential Fas gene mutations mimicking alternative splicing by deletions of exons or aberrant splicing because of mutations at the splice sites. The PCR results on chromosomal DNA showed that no aberrant fragments associated with deletions were detectable; moreover, no mutations were found by sequencing the splice sites at exon/intron boundaries in these cases (data not shown). Therefore, these aberrant-sized transcripts with skipped exons should be regarded as alternative splice variants.
Further confirmation of Fas gene mutations was performed at the chromosomal DNA level in cases in which small sequence changes were detected at the mRNA level. In case 9, an aberrant transcript with a 25-bp deletion in the exon 3 sequence was detected (variant 9-2 in Figure 1b ▶ ). As shown in Figure 4 ▶ , PCR on the genomic DNA of the sequence encompassing the deleted region of 9-2 confirmed the presence of this short deletion at the DNA level, and also showed that the ratio of the wild-type versus the mutant allele in case 9 was ∼2.5:1. We next examined a 1-bp insertion at nucleotide 1095 in the exon 9 sequence of a transcript from case 21 (variant not included in Figure 1b ▶ ). Insertion of one adenosine at nucleotide 1095 of Fas cDNA from case 21 could not be detected in the chromosomal DNA from the same case, even though 90 clones of the amplified fragment encompassing nucleotide 1095 were sequenced from both directions.
Figure 4.
Examination of chromosomal DNA for confirmation of the 25-bp deletion in the exon 3 sequence of case 9 (variant 9-2). Different cycle numbers of PCR were performed to obtain the semiquantitative measure of DNA ratio between the wild type (101 bp) and the deletion (76 bp) in the sample. −, negative control using distilled water; M, pBSK/MspI.
Immunostaining of Fas and FasL was performed on the paraffin sections of 16 cases. Fas and FasL proteins were expressed in the lymphoma cells and in the surrounding reactive lymphoid cells. Fifteen (94%) cases showed positive staining for Fas and 11 (69%) cases were positive for FasL (Figure 5) ▶ .
Figure 5.

Expression of Fas and FasL proteins in NL. The positive immunostaining of Fas (a) and FasL (b) was detected in case 17. Immunoperoxidase staining, original magnifications, ×200.
Clinical and prognostic data of the NL patients included in the study are presented in Table 1 ▶ and the patients were divided into two groups with respect to whether they are carrying Fas gene mutations and/or alternative splice variants in the lesions or carrying the wild-type Fas only. There were two NL patients who are still alive with relatively long overall survival in each group; by contrast, most others died of the disease in a very short period. Disseminated NL was incompatible with prolonged survival, and it was only found in patients with Fas mutations. However, the number of cases was too small to give statistical significance (P = 0.52, chi-square test). The actuarial 3-year survivals leveled off at 15% for patients carrying the Fas mutations and/or splice variants in the lesions and 49% for those carrying the wild type only, but the difference did not reach statistical significance on the univariate analysis (P = 0.396) (Figure 6) ▶ .
Table 1.
Clinical and Prognostic Features of 15 Cases of NL Carrying the Wild-Type Fas Only or Carrying the Mutated Fas and/or Alternative Splice Variants in the Lesion
| Case no. | Age/sex | Biopsy site | Initial stage | Treatment | Survival | Cause of death§ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group 1: NL carrying the wild-type Fas only | ||||||||||||
| 7 | 61 /M | Nasal cavity | I | Chemotherapy | 1 month | DOD | ||||||
| 12 | 60 /F | Oropharynx | I | Chemotherapy | 4 months | DOD | ||||||
| 14 | 18 /F | Nasal cavity | I | Chemotherapy | >23 years† | NA (alive) | ||||||
| 23 | 72 /M | Nasal cavity | I | Radiotherapy | >11 years | NA (alive) | ||||||
| Group 2: NL carrying the mutated Fas and/or alternative splice variants | ||||||||||||
| 8 | 55 /M | Nasal cavity | I | Chemotherapy | 2 months | DOR | ||||||
| 9 | 32 /M | Nasopharynx | I | Chemotherapy | 12 months | DOR | ||||||
| 10 | 57 /M | Nasopharynx | IV | Nil | 1 month | DOD | ||||||
| 11 | 84 /F | Nasal cavity | I | Chemotherapy | <1 month | DOD | ||||||
| 13 | 73 /M | Nasal cavity | I | Chemotherapy | 21 months | DOR | ||||||
| 15 | 34 /M | Nasal cavity | I | Chemotherapy | 8 months | DOR | ||||||
| 17 | 32 /M | Nasal cavity | IV | Chemotherapy | 7 months | DOD | ||||||
| 18 | 30 /F | Nasal cavity | I | Chemotherapy | 20 months | DOR | ||||||
| 21 | 42 /M | Testis* | I | Chemotherapy | 12 months | DOR | ||||||
| 16 | 17 /F | Nasal cavity | I | Radiotherapy | >7.5 years | NA (alive) | ||||||
| 19 | 64 /F | Nasal cavity | I | Chemotherapy+ radiotherapy | >12 years‡ | NA (alive) | ||||||
*Metastatic lesions from the primary nasal site.
†Relapse after 17 years of disease-free survival.
‡Relapse locally after 9 years of disease-free survival.
§DOD, died of disease; DOR, died of relapse; NA, data not available.
Figure 6.
Cumulative survival rates of NL patients. Actuarial comparison of survival between patients carrying the Fas mutations and/or the splice variants in the lesions and those carrying the wild-type Fas only showed no statistical significance (P = 0.396).
Discussion
Until now, most of the studies on the detection of somatic mutations of the Fas gene in human cancers have reported that point mutations occur in the death domain of the Fas gene and that these mutations occur at relatively low frequencies. 11,12,16,30-36 Insertions or deletions of short sequences in the Fas gene have been reported only occasionally. 13-15 The frequency of Fas mutations detected in NL in the present study is significantly higher than that reported in any other lymphoid neoplasm. Furthermore, most of the mutations contain deletions of very large DNA fragments in the coding region, which have not been described in any other malignancy. These results in NL are also in contrast to the findings in peripheral T-cell lymphomas (unspecified) where no mutations of Fas were found in any of 35 cases studied. 11
Among the deletion variants detected in NL, eight break points (9-2, 10-1, 10-2, 11-1, 17-2, 18-1, 19-1, and 15-2) were localized within a 23-bp region at nucleotides 496 to 518, identifying this location as a potential hotspot region of break points for Fas gene deletion in NL (Figure 1b) ▶ .
As illustrated in Figure 2 ▶ , in many of the cases, various small-sized PCR products were detected along with the normal-sized fragments. This observation suggests that the mutant Fas alleles were co-amplified with the wild-type allele and that a dominant-negative mechanism might be operative in NL, which could block the apoptosis signaling. However, it is also possible that the normal transcripts were derived from normal tissues because many nontumor cells with wild-type alleles might be present in the cellular extractions from the tumor biopsy samples. Sequence analysis showed several distinct mutant forms of deletions in case 10 (three deletion variants) and cases 15 and 17 (two deletion variants each), in addition to the normal transcripts and alternative splice variants (Figures 1b ▶ and 2b ▶ ). The deleted regions overlap with each other, so the deletions cannot co-exist in a single mRNA from one allele, suggesting subclonal generation of Fas mutations at least in case 10. In cases 15 and 17, it is also feasible that the two deletion variants were derived from two mutated alleles within the same lymphoma clones and the normal transcripts might come from the nonneoplastic population. Single cell studies in the future can be useful in addressing the above issues. Taken together, the detection of several different deletion variants in the same tumor specimen from these cases points to a process in which somatic mutations in the Fas gene accumulate during tumor progression and suggests that the Fas gene mutation is a late event in the progression of NL.
Because deletions were originally detected in PCR products of cDNA, confirmation of some of these mutations in the chromosomal DNA from a few cases can reassure us of the reliability of the sequence data derived from RT-PCR. In those cases with small sequence changes, such as the 25-bp deletion in sequence 9-2 or the 1-bp insertion at nucleotide 1095 in case 21, wild-type and mutant alleles from genomic DNA should be amplified with similar efficiencies. As a result, we can obtain a semiquantitative measure of allele ratios in the samples. The PCR analysis on the genomic DNA of the sequence encompassing the 25-bp deleted region of case 9 confirmed the presence of this short deletion at the chromosomal DNA level (Figure 4) ▶ , and also showed that the ratio of the wild-type versus the mutant allele in case 9 was ∼2.5:1. This result indicates that the short deletion sequence of 9-2 was present in a significant proportion of the NL tumor cells in the sample.
The phenomenon of 1-bp insertion at nucleotide 1095 was recently described in Fas cDNA from the thyroid lymphomas of B cell lineage. 13 The same mutation was also detected at the RNA level in the transcripts from case 21 in this study, yet this insertion could not be found at the chromosomal DNA level. Because there is a poly-(A) run from nucleotides 1089 to 1094 of the Fas mRNA sequence, it is likely that the single adenosine insertion at the poly-(A) run occurs as a posttranscriptional RNA modification resulting from the nontemplated form of RNA editing 37-40 or it might be simply because of polymerase slippage during transcription or reverse transcription.
As illustrated in Figure 1 ▶ , the deletions and the alternative splice variants of Fas detected in NL result in loss of the death domain, which is in the cytoplasmic region of the Fas receptor and is essential for the apoptosis signaling. The exon 9 sequence, encoding the death domain, was deleted partially or completely in the mutant alleles of six NL cases (10-2, 11-1, 13-1, 15-2, 16-1, and 17-2), causing a shift of the reading frame. In contrast, an intact exon 9 sequence was preserved in the other deletions and splice variants (8-1, 9-1, 9-2, 10-3, 15-1, 15-3, 15-5, 17-1, and 19-1), but the death domain was likely to be lost because the frame shift effect causes premature termination before the death domain. Overall, only three of the detected variants of deletion and splicing would generate Fas receptors with intact death domains.
Another effect of deletion and alternative splice variants can be loss of the transmembrane domain, which may result in a soluble form of the Fas receptor. Except for two variants (9-1 and 16-1), most of the deletions and the alternative splice variants detected in NL are predicted to generate a soluble form of Fas. It has been reported in peripheral lymphocytes that alternative splicing can provide protection from Fas-mediated apoptosis by generating a soluble form of the Fas receptor. 41-43 Similar alternative splicing was also found in the reactive tonsils in this study. In NL, the same splice variant skipping exons 3 and 4 sequence was detectable as in the reactive tonsils. The other three splice variants detected in NL associated with exons 6 and 7 sequence skipping have been previously described in the peripheral lymphocytes. 41-43 Thus, the alternative splicing mechanism on the control of cellular susceptibility to the FasL signaling might be common to both normal lymphocytes and lymphoma cells. In contrast to the regulatory role of the soluble forms of Fas receptor by alternative splicing among the normal lymphocytes, the NL tumor cells are prone to generate soluble forms of Fas receptor by deletion and alternative splicing to limit apoptosis induced by FasL.
Interestingly, as shown in Figure 1 ▶ , all of these abnormal Fas proteins derived from deletions and alternative splicing possess the common characteristic of preserving the first cysteine-rich domain while losing the death domain and/or the transmembrane domain. Both effects may render the tumor cells insensitive to autocrine FasL that is generated by NL tumor cells on immune attack. 27,44,45
Based on an in vitro study, 46 the presence of the first cysteine-rich domain in the mutated Fas protein will cause dominant-negative interference with wild-type Fas within a functional receptor trimer. The preligand assembly domain (PLAD), localized within the first extracellular cysteine-rich domain, is an essential element in the formation of Fas trimer before FasL cross-linking, and dominant-negative interference is observed to be based on the ligand-independent interaction of mutant and wild-type Fas receptors at this domain. To cause the interference, mutant proteins must physically interact with wild-type proteins in a functional complex on the cell surface. Removal of the PLAD from aberrant Fas proteins lacking the death domain reverses the dominant blockage of apoptotic signal transduction in vitro. In a large number of ALPS case reports, 4-10 heterozygosity and dominant-negative interference have been observed in almost any size of prematurely terminated Fas proteins. A common characteristic among these mutants is preservation of the PLAD and absence of the death domain. The deletion mutants found in the present study on NL are predicted to generate truncated Fas proteins encoding the first 76 to 177 amino acids, sharing the characteristics of the mutant Fas proteins with dominant-negative interference reported in ALPS.
It has been reported that NL cells co-express Fas and FasL, whereas the percentage of apoptotic tumor cells is low. 27 O’Connell and colleagues 47 have recently suggested that FasL can play a double role of pro- and anti-inflammatory mediator subject to its context and level of expression, with the anti-inflammatory role prominent in numerous studies demonstrating FasL expression in many types of human cancer. In the context of cancer, FasL is thought to contribute to tumor immune privilege or immune escape. Reactive cytotoxic T lymphocytes are usually observed in the midst of NL tumor cells that are associated with EBV infection. 24 After activation in response to the virus antigens, Fas-sensitive EBV-specific cytotoxic T lymphocytes could be vulnerable to killing by FasL-expressing NL cells. Therefore, a selection pressure would operate in favor of both FasL expression and disruption of the Fas-triggered apoptotic pathway among the tumor cells. Consequently, various Fas mutants would accumulate in the population of tumor cells.
In this study, we have demonstrated frequent somatic mutations of the human Fas gene in NK/T-cell lymphoid malignancies and widespread expression of FasL. All of the Fas mutants preserved the sequences encoding the N-terminal PLAD domain but not the C-terminal death and/or transmembrane domains. Although Fas gene mutations seemed to be associated with more disseminated disease and inferior survival, the difference did not reach statistical significance because of the limited number of cases in this study. We have previously shown that NL tumors also express hIL-10, 24 an inhibitor of T-cell growth. We propose here that these two mechanisms might interact in NL to improve tumor immune privilege. We speculate that with a background of Fas mutation, hIL-10 and FasL combine to protect NL cells from the immune destruction by activated EBV-specific cytotoxic T lymphocytes. Successful immunotherapy against NL will need to take these defenses into account.
Footnotes
Address reprint requests to Gopesh Srivastava, Department of Pathology, The University of Hong Kong, Queen Mary Hospital Compound, 102 Pokfulam Rd., Hong Kong. E-mail: gopesh@pathology.hku.hk.
Supported by grants from the Research Grants Council of Hong Kong Special Administrative Region, P. R. China (HKU 7299/98M and 7348/02M to G. S.).
This paper was presented in abstract form at the 7th Congress of the European Hematology Association, Florence, Italy, June 2002.
References
- 1.Mullauer L, Gruber P, Sebinger D, Buch J, Wohlfart S, Chott A: Mutations in apoptosis genes: a pathogenetic factor for human disease. Mutat Res 2001, 488:211-231 [DOI] [PubMed] [Google Scholar]
- 2.Nagata S: Apoptosis by death factor. Cell 1997, 88:355-365 [DOI] [PubMed] [Google Scholar]
- 3.Muschen M, Warskulat U, Beckmann MW: Defining CD95 as a tumor suppressor gene. J Mol Med 2000, 78:312-325 [DOI] [PubMed] [Google Scholar]
- 4.Drappa J, Vaishnaw AK, Sullivan KE, Chu JL, Elkon KB: Fas gene mutations in the Canale-Smith syndrome, an inherited lymphoproliferative disorder associated with autoimmunity. N Engl J Med 1996, 335:1643-1649 [DOI] [PubMed] [Google Scholar]
- 5.Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, Strober W, Lenardo MJ, Puck JM: Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell 1995, 81:935-946 [DOI] [PubMed] [Google Scholar]
- 6.Martin DA, Zheng L, Siegel RM, Huang B, Fisher GH, Wang J, Jackson CE, Puck JM, Dale J, Straus SE, Peter ME, Krammer PH, Fesik S, Lenardo MJ: Defective CD95/APO-1/Fas signal complex formation in the human autoimmune lymphoproliferative syndrome, type Ia. Proc Natl Acad Sci USA 1999, 96:4552-4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rieux-Laucat F, Le Deist F, Hivroz C, Roberts IA, Debatin K, Fischer A, De Villartay JP: Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science 1995, 268:1347-1349 [DOI] [PubMed] [Google Scholar]
- 8.Rieux-Laucat F, Blachere S, Danielan S, De Villartay JP, Oleastro M, Solary E, Bader-Meunier B, Arkwright P, Pondare C, Bernaudin F, Chapel H, Nielsen S, Berrah M, Fischer A, Le Deist F: Lymphoproliferative syndrome with autoimmunity: a possible genetic basis for dominant expression of the clinical manifestations. Blood 1999, 94:2575-2582 [PubMed] [Google Scholar]
- 9.Sneller MC, Wang J, Dale JK, Strober W, Middelton LA, Choi Y, Fleisher TA, Lim MS, Jaffe ES, Puck JM, Lenardo MJ, Straus SE: Clinical, immunologic, and genetic features of an autoimmune lymphoproliferative syndrome associated with abnormal lymphocyte apo-ptosis. Blood 1997, 89:1341-1348 [PubMed] [Google Scholar]
- 10.Vaishnaw AK, Orlinick JR, Chu JL, Krammer PH, Chao MV, Elkon KB: The molecular basis for apoptotic defects in patients with CD95 (Fas/Apo-1) mutations. J Clin Invest 1999, 103:355-363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gronbaek K, Straten PT, Ralfkiaer E, Ahrenkiel V, Andersen MK, Hansen NE, Zeuthen J, Hou-Jensen K, Guldberg P: Somatic Fas mutations in non-Hodgkin’s lymphoma: association with extranodal disease and autoimmunity. Blood 1998, 92:3018-3024 [PubMed] [Google Scholar]
- 12.Landowski TH, Qu N, Buyuksal I, Painter JS, Dalton WS: Mutations in the Fas antigen in patients with multiple myeloma. Blood 1997, 90:4266-4270 [PubMed] [Google Scholar]
- 13.Takakuwa T, Dong Z, Takayama H, Matsuzuka F, Nagata S, Aozasa K: Frequent mutations of Fas gene in thyroid lymphoma. Cancer Res 2001, 61:1382-1385 [PubMed] [Google Scholar]
- 14.Maeda T, Yamada Y, Moriuchi R, Sugahara K, Tsuruda K, Joh T, Atogami S, Tsukasaki K, Tomonaga M, Kamihira S: Fas gene mutation in the progression of adult T cell leukemia. J Exp Med 1999, 189:1063-1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamiya S, Etoh K, Suzushima H, Takatsuki K, Matsuoka M: Mutation of CD95 (Fas/Apo-1) gene in adult T-cell leukemia cells. Blood 1998, 91:3935-3942 [PubMed] [Google Scholar]
- 16.Muschen M, Re D, Brauninger A, Wolf J, Hansmann ML, Diehl V, Kuppers R, Rajewsky K: Somatic mutations of the CD95 gene in Hodgkin and Reed-Sternberg cells. Cancer Res 2000, 60:5640-5643 [PubMed] [Google Scholar]
- 17.Ho FC, Todd D, Loke SL, Ng RP, Khoo RK: Clinico-pathological features of malignant lymphomas in 294 Hong Kong Chinese patients, retrospective study covering an eight-year period. Int J Cancer 1984, 34:143-148 [DOI] [PubMed] [Google Scholar]
- 18.Chiang AK, Srivastava G, Lau PW, Ho FC: Differences in T-cell-receptor gene rearrangement and transcription in nasal lymphomas of natural killer and T-cell types: implications on cellular origin. Hum Pathol 1996, 27:701-707 [DOI] [PubMed] [Google Scholar]
- 19.Jaffe ES, Chan JK, Su IJ, Frizzera G, Mori S, Feller AC, Ho FC: Report of the workshop on nasal and related extranodal angiocentric T/natural killer cell lymphomas definitions, differential diagnosis, and epidemiology. Am J Surg Pathol 1996, 20:103-111 [DOI] [PubMed] [Google Scholar]
- 20.Tao Q, Chiang AK, Srivastava G, Ho FC: TCR-CD56+CD2+ nasal lymphomas with membrane-localized CD3 positivity: are the CD3+ cells neoplastic or reactive? Blood 1995, 85:2993-2996 [PubMed] [Google Scholar]
- 21.Harabuchi Y, Yamanaka N, Kataura A, Imai S, Kinoshita T, Mizuno F, Osato T: Epstein-Barr virus in nasal T-cell lymphomas in patients with lethal midline granuloma. Lancet 1990, 335:128-130 [DOI] [PubMed] [Google Scholar]
- 22.Ho FC, Srivastava G, Loke SL, Fu KH, Leung BP, Liang R, Choy D: Presence of Epstein-Barr virus DNA in nasal lymphomas of B and ‘T’ cell type. Hematol Oncol 1990, 8:271-281 [DOI] [PubMed] [Google Scholar]
- 23.Tao Q, Ho FC, Loke SL, Srivastava G: Epstein-Barr virus is localized in the tumour cells of nasal lymphomas of NK, T or B cell type. Int J Cancer 1995, 60:315-320 [DOI] [PubMed] [Google Scholar]
- 24.Shen L, Chiang AK, Liu WP, Li GD, Liang RH, Srivastava G: Expression of HLA class I, beta(2)-microglobulin, TAP1 and IL-10 in Epstein-Barr virus-associated nasal NK/T-cell lymphoma: implications for tumor immune escape mechanism. Int J Cancer 2001, 92:692-696 [DOI] [PubMed] [Google Scholar]
- 25.Chiang AK, Tao Q, Srivastava G, Ho FC: Nasal NK- and T-cell lymphomas share the same type of Epstein-Barr virus latency as nasopharyngeal carcinoma and Hodgkin’s disease. Int J Cancer 1996, 68:285-290 [DOI] [PubMed] [Google Scholar]
- 26.Chiang AK, Wong KY, Liang AC, Srivastava G: Comparative analysis of Epstein-Barr virus gene polymorphisms in nasal T/NK-cell lymphomas and normal nasal tissues: implications on virus strain selection in malignancy. Int J Cancer 1999, 80:356-364 [DOI] [PubMed] [Google Scholar]
- 27.Ng CS, Lo ST, Chan JK: Peripheral T and putative natural killer cell lymphomas commonly coexpress CD95 and CD95 ligand. Hum Pathol 1999, 30:48-53 [DOI] [PubMed] [Google Scholar]
- 28.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD: World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J Clin Oncol 1999, 17:3835-3849 [DOI] [PubMed] [Google Scholar]
- 29.Cheng J, Liu C, Koopman WJ, Mountz JD: Characterization of human Fas gene. Exon/intron organization and promoter region. J Immunol 1995, 154:1239-1245 [PubMed] [Google Scholar]
- 30.Bertoni F, Conconi A, Luminari S, Realini C, Roggero E, Baldini L, Carobbio S, Cavalli F, Neri A, Zucca E: Lack of CD95/FAS gene somatic mutations in extranodal, nodal and splenic marginal zone B cell lymphomas. Leukemia 2000, 14:446-448 [DOI] [PubMed] [Google Scholar]
- 31.Bertoni F, Conconi A, Carobbio S, Realini C, Codegoni AM, Zucca E, Cavalli F: Analysis of Fas/CD95 gene somatic mutations in ovarian cancer cell lines. Int J Cancer 2000, 86:450. [DOI] [PubMed] [Google Scholar]
- 32.Lee SH, Shin MS, Park WS, Kim SY, Dong SM, Pi JH, Lee HK, Kim HS, Jang JJ, Kim CS, Kim SH, Lee JY, Yoo NJ: Alterations of Fas (APO-1/CD95) gene in transitional cell carcinomas of urinary bladder. Cancer Res 1999, 59:3068-3072 [PubMed] [Google Scholar]
- 33.Lee SH, Shin MS, Park WS, Kim SY, Kim HS, Han JY, Park GS, Dong SM, Pi JH, Kim CS, Kim SH, Lee JY, Yoo NJ: Alterations of Fas (Apo-1/CD95) gene in non-small cell lung cancer. Oncogene 1999, 18:3754-3760 [DOI] [PubMed] [Google Scholar]
- 34.Muschen M, Re D, Betz B, Moers C, Wolf J, Niederacher D, Diehl V, Beckmann MW: Resistance to CD95-mediated apoptosis in breast cancer is not due to somatic mutation of the CD95 gene. Int J Cancer 2001, 92:309-310 [DOI] [PubMed] [Google Scholar]
- 35.Park WS, Oh RR, Kim YS, Park JY, Lee SH, Shin MS, Kim SY, Kim PJ, Lee HK, Yoo NY, Lee JY: Somatic mutations in the death domain of the Fas (Apo-1/CD95) gene in gastric cancer. J Pathol 2001, 193:162-168 [DOI] [PubMed] [Google Scholar]
- 36.Shin MS, Park WS, Kim SY, Kim HS, Kang SJ, Song KY, Park JY, Dong SM, Pi JH, Oh RR, Lee JY, Yoo NJ, Lee SH: Alterations of Fas (Apo-1/CD95) gene in cutaneous malignant melanoma. Am J Pathol 1999, 154:1785-1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Apostolou S, De Rienzo A, Murthy SS, Jhanwar SC, Testa JR: Absence of BCL10 mutations in human malignant mesothelioma. Cell 1999, 97:684-686 [DOI] [PubMed] [Google Scholar]
- 38.Dyer MJ: Bcl10 mutationsin malignancy. Br J Cancer 1999, 80:1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fakruddin JM, Chaganti RS, Murty VV: Lack of BCL10 mutations in germ cell tumors and B cell lymphomas. Cell 1999, 97:683-684 [DOI] [PubMed] [Google Scholar]
- 40.Takahashi H, Maeda Y, Seto M, Hosokawa Y: Nucleotide insertions and deletions within the homopolymeric runs of adenines and thymidines of BCL10 cDNAs in normal peripheral blood leukocytes. Blood 2000, 95:2728-2729 [PubMed] [Google Scholar]
- 41.Cascino I, Fiucci G, Papoff G, Ruberti G: Three functional soluble forms of the human apoptosis-inducing Fas molecule are produced by alternative splicing. J Immunol 1995, 154:2706-2713 [PubMed] [Google Scholar]
- 42.Cheng J, Zhou T, Liu C, Shapiro JP, Brauer MJ, Kiefer MC, Barr PJ, Mountz JD: Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science 1994, 263:1759-1762 [DOI] [PubMed] [Google Scholar]
- 43.Liu C, Cheng J, Mountz JD: Differential expression of human Fas mRNA species upon peripheral blood mononuclear cell activation Biochem J 1995, 310:957-963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato K, Ohshima K, Ishihara S, Anzai K, Suzumiya J, Kikuchi M.: Elevated serum soluble Fas ligand in natural killer cell proliferative disorders. Br J Haematol 1998, 103:1164-1166 [DOI] [PubMed] [Google Scholar]
- 45.Tanaka M, Suda T, Haze K, Nakamura N, Sato K, Kimura F, Motoyoshi K, Mizuki M, Tagawa S, Ohga S, Hatake K, Drummond AH, Nagata S: Fas ligand in human serum. Nat Med 1996, 2:317-322 [DOI] [PubMed] [Google Scholar]
- 46.Siegel RM, Frederiksen JK, Zacharias DA, Chan FK, Johnson M, Lynch D, Tsien RY, Lenardo MJ: Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science 2000, 288:2354-2357 [DOI] [PubMed] [Google Scholar]
- 47.O’Connell J, Houston A, Bennett MW, O’Sullivan GC, Shanahan F: Immune privilege or inflammation? Insights into the Fas ligand enigma Nat Med 2001, 7:271-274 [DOI] [PubMed] [Google Scholar]