Summary
Epigenetic gene silencing suppresses transposon activity and is critical for normal development [1, 2]. Two common epigenetic gene silencing marks are DNA methylation and histone H3 lysine 9 dimethylation (H3K9me2) [1]. In Arabidopsis thaliana, H3K9me2, catalyzed by the methyltransferase KRYPTONITE (KYP/SUVH4), is required for maintenance of DNA methylation outside of the standard CG sequence context [3, 4]. Additionally, loss of DNA methylation in the met1 mutant correlates with a loss of H3K9me2[5, 6]. Here we show that KYP dependent H3K9me2 is found at non-CG methylation sites in addition to those rich in CG methylation. Furthermore, we show that the SRA domain of KYP binds directly to methylated DNA, and SRA domains with missense mutations found in loss-of-function kyp mutants have reduced binding to methylated DNA in vitro. These data suggest that DNA methylation is required for the recruitment or activity of KYP, and suggest a self-reinforcing loop between histone and DNA methylation. Lastly, we found that SRA domains from two Arabidopsis SRA-RING proteins also bind methylated DNA, and that the SRA domains from KYP and SRA-RING proteins prefer methylcytosines in different sequence contexts. Hence, unlike the methyl-binding-domain (MBD), which only binds methylated-CpG sequences, the SRA domain is a versatile new methyl-DNA-binding motif.
Results and Discussion
Histone H3K9 dimethylation is correlated with levels of DNA methylation
We utilized chromatin immunoprecipitation (ChIP) and genomic bisulfite sequencing to further investigate the relationship between histone methylation and DNA methylation. In Arabidopsis, DNA methylation (5-methyl cytosine) occurs not only at CG residues, but also at CNG and asymmetric cytosines (CNN) [2]. METHYLTRANSFERASE 1 (MET1), a homolog of mammalian Dnmt1, maintains methylation at CG residues [7, 8], while CHROMOMETHYLASE3 (CMT3) maintains the majority of CNG methylation, and a small amount of the CNN methylation [9, 10]. A third enzyme, DOMAINS REARRANGED METHYLASE 2 (DRM2) is mainly responsible for maintaining CNN methylation, plays a minor role in the maintenance of CNG methylation, and is also required for de novo methylation of cytosines in all sequence contexts [2, 11]. Genomic regions that are enriched in methylated DNA are also found associated with H3K9me2 [12-14].
It has previously been shown that mutations in KYP lead to reduction of non-CG DNA methylation, but not CG methylation, suggesting that non-CG methylation is dependent on histone H3K9me2 [3, 4]. CMT3 is responsible for this non-CG methylation and has been shown in vitro to bind directly to H3 tails trimethylated at lysine 9 and lysine 27 [9, 10, 15]. Conversely, the loss of DNA methylation in met1 mutants is also associated with decreased H3K9me2 at many loci [5, 6, 12, 16]. One possible explanation for this observation is that KYP is in a complex with MET1, such that loss of MET1 causes a loss of KYP activity. A second possibility is that DNA methylation itself directly affects KYP activity or targeting.
To test these ideas, we examined H3K9me2 levels in different DNA methyltransferase mutants and at two loci with very different patterns of CG or non-CG methylation (Figure S1). AtSN1 is a 170-bp SINE retrotransposon heavily methylated at non-CG and to a lesser extent at CG sites [~13 vs 3 per element; Figure 1A, see also [17]]. AtSN1 is also marked by H3K9me2, which is dependent on KYP, illustrated by its loss in a kyp null line, kyp-6 (Figure 1B). Elimination of most of the DNA methylation in the drm1 drm2 cmt3 triple mutant also results in significant loss of H3K9me2 methylation, showing a strong correlation between levels of DNA and histone methylation (Figure 1B). In contrast, the met1-3 mutation showed complete elimination of CG methylation from AtSN1, but only a minor effect on non-CG methylation (Figure 1A), and H3K9me2 was observed at wild-type levels (Figure 1B). This suggests that KYP activity is not affected by loss of MET1 per se, and that histone H3 methylation can be maintained at genomic regions containing only non-CG methylation.
Figure 1.
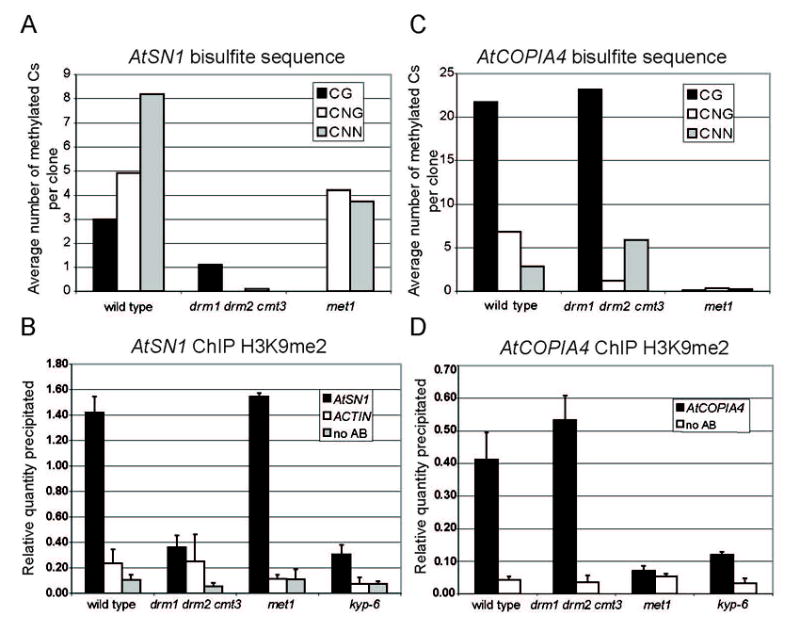
Correlation of DNA and histone methylation in Arabidopsis. (A) Bisulfite sequencing data for wild type, drm1 drm2 cmt3 and met1 at AtSN1, expressed as the average number of methylated CG, CNG or CNN residues per clone. (B) ChIP analysis using antibodies specific for H3K9me2 from the wild type, drm1 drm2 cmt3, met1 and kyp-6. AtSN1 was amplified from the IP and no antibody control (no AB). ACTIN was amplified from the IP. The relative quantity precipitated for each sample was derived from a standard curve of input DNA. The average of two biological replicas done in duplicate is shown with standard deviation. (C) Bisulfite sequencing data from AtCOPIA4. The remaining non-CG methylation in the drm1 drm2 cmt3 triple mutant is discussed in reference [19]. (D) ChIP analysis of H3K9me2 at AtCOPIA4. The average of two biological replicas done in duplicate is shown with standard deviation.
Unlike AtSN1, AtCOPIA4 is a CG-rich retrotransposon [18]. Bisulfite sequencing of a 500 nucleotide segment revealed an average of 22 methylated CG residues, 6 methylated CNGs, and 3 methylated asymmetric cytosines in wild-type plants (Figure 1C, see also [19]). This element is also associated with KYP-dependent H3K9me2 (Figure 1D; see also [18]). In drm1 drm2 cmt3 the CNG methylation was greatly reduced, while the CG and histone methylation levels were unchanged (Figure 1C and 1D). However, in met1-3 all DNA methylation was eliminated, as was most of the H3K9me2. Taking both the results at AtSN1 and AtCOPIA4 into account, we conclude that mutations in particular DNA methyltransferases may not directly affect histone methylation, but rather the level of either CG or non-CG DNA methylation is correlated with H3K9me2. One explanation for these results is that DNA methylation is important for recruitment or activity of KYP.
The SRA domain from KYP binds methylated DNA.
The only recognizable domain in KYP besides the catalytic SET domain is the YDG (named for three conserved amino acids)/SRA (SET and RING associated) domain [20]. We isolated a new allele of KYP (kyp-5) with a missense mutation in the SRA domain (E208K) as part of a previously described screen for suppressors of silencing of the SUPERMAN locus (Figure 2A) [9]. Like null alleles [3, 5, 13], this allele decreased H3K9me2 in chromocenters (Figure 2B). In addition, two previously reported mutant kyp alleles contained missense mutations (S200F and R260H) in the SRA domain, and transcriptional silencing due to overexpression of the KYP-related gene SUVH2 was alleviated by mutation of its SRA domain [4, 21]. These results show that the SRA domain is critical for KYP function.
Figure 2.
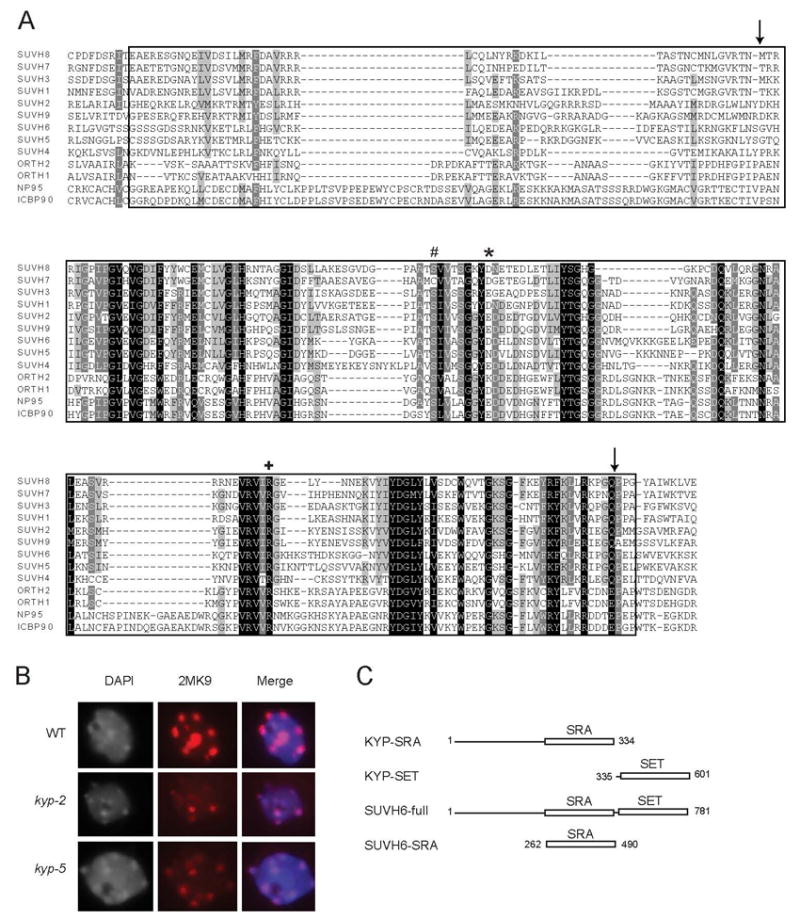
The SRA domain. (A) ClustalW alignment of the SRA domains from the nine SUVH genes, two Arabidopsis SRA- and RING-domain-containing proteins, mouse np95, and human ICBP90. The site of the KYP S200F[4] mutation is indicated with “#”, the E208K mutation is indicated with a “*”, and the R260H[4] mutation is indicated with a “+”. Arrows show the SRA domain identified by SMART and the boxed in amino acids are those included in the GST-SUVH6-SRA fusion protein. The association of SRA domains with SET domain proteins is unique to the plant kingdom. (B) Immunofluorescence of H3K9me2 compared to DAPI in nuclei isolated from wild type, kyp-2 (null allele), and kyp-5 (SRA mutation E208K). (C) Regions of the SUVH genes contained in the various constructs analyzed in Figure 3.
The role of the SRA domain is unclear, but biochemical studies of a class of mammalian SRA- and RING-domain-containing proteins have been reported. In the murine protein, Np95, the SRA domain was shown to bind histone H3 in vitro and be required for chromatin association in vivo [22]. However, a second study demonstrated that a human SRA-RING protein, ICBP90, bound methylated DNA in crude extracts, suggesting that the SRA either binds methylated DNA directly or binds indirectly through a methyl-DNA-binding domain protein present in the extracts [23].
We therefore tested whether the SRA domain from KYP was sufficient for binding to either histone H3 or methylated DNA. An in vitro translated SRA domain did not bind histone H3 (data not shown). However, analysis of DNA binding using mobility shift assays with a purified GST (glutathione S-transferase)-tagged KYP-SRA fusion protein (amino acids 1-334, Figure 2C) revealed preferential binding to double-stranded oligonucleotides containing 5-methyl cytosine residues in all sequence contexts (all-mC)] compared to unmethylated oligonucleotides with the same sequence (all-C) (Figure 3A). In contrast, a GST-tagged KYP-SET fusion protein (amino acids 335-601) did not bind to methylated or unmethylated DNA. The addition of an anti-GST antibody to the all-mC binding reaction blocked DNA binding (last two lanes in Figure 3A). These results indicate that the amino-terminus of KYP containing the SRA domain binds methylated but not unmethylated DNA.
Figure 3.
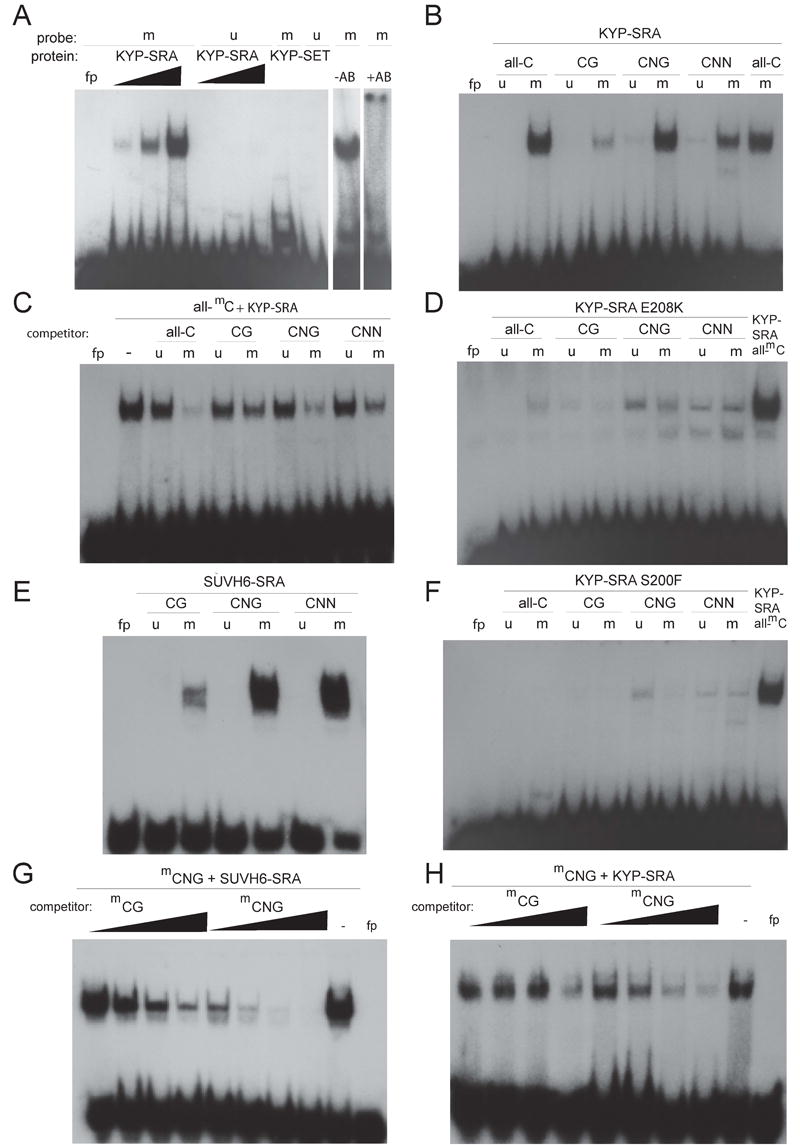
KYP SRA domain preferentially binds methylated DNA. (A) Mobility shift assays using increasing amounts of GST-KYP-SRA (KYP-SRA) (25, 50 and 100 nM) added to a binding reaction with either a methylated (m) or unmethylated (u) all-C double stranded oligonucleotide probe. GST-KYP-SET (KYP-SET) was added at 100 nM in the indicated lanes. Anti-GST antibodies (AB) were added to the binding reaction shown in the last lane to yield a super-shifted complex. KYP-SRA (B), KYP-SRA(E208K) (D) or KYP-SRA(S200F) (F) was bound to oligonucleotides with cytosines methylated in all sequences contexts (all-mC), or in a CG, CNG, or CNN context. In D and F the last lane is GST-KYP-SRA bound to the all-mC probe used as a positive control. (C) Competition assays were performed by addition of the indicated unlabeled DNA, added in 1000 fold excess to the all-mC probe. (E) GST-SUVH6-SRA (10 nM) was bound to the indicated probes. (G-H) GST-SUVH6-SRA (10 nM) or GST-KYP-SRA (100 nM) was bound to the methylated CNG probe with either no competitor (-), or 100x, 300x, 1000x, or 3000x unlabeled mCG or mCNG oligonucleotides. The lane indicated by fp (free probe) has no added protein.
Further analysis using various double-stranded oligonucleotides containing only CG, CNG, or CNN methylation revealed that the SRA domain in KYP binds to DNA with methylated cytosines in all contexts with a preference for CNG and CNN sequences (Figure 3B). This preference was supported by competition experiments (Figure 3C and 3H).
To provide additional evidence that the SRA domain was indeed responsible for methyl-DNA binding, we engineered two different SRA mutations into the GST-KYP SRA construct (E208K and S200F). We observed weaker overall binding and a reduced ability of the mutant proteins to discriminate between methylated and unmethylated oligonucleotides (Figure 3D and 3F). These results strongly suggest that the SRA domain is responsible for methyl-DNA binding in the KYP amino-terminus.
SUVH6 SRA domain has a binding specificity similar to KYP
In addition to KYP, SRA domains are found in the eight other histone methyltransferase genes (SUVH class) in Arabidopsis [20]. We tested one of these, SUVH6, to determine if it too could discriminate between methylated and unmethylated DNA. GST-SUVH6-full length protein (amino acids 1-781) bound methylated, but not unmethylated oligonucleotides (Figure S2A). Furthermore, competition experiments showed that GST- SUVH6 had a higher affinity for methylated CNG and CNN substrates than methylated CG, a preference very similar to that of KYP (Figure S2B).
In order to engineer a fusion protein with just the SRA domain, we first tried cloning the SRA domain predicted by SMART [24] (delineated by arrows in Figure 2A) into vectors containing a His-tag and found the tagged proteins were insoluble. GST or MBP (maltose binding protein) fusions were soluble, but inactive in binding DNA. We re-analyzed the SRA domain by aligning the nine different SUVH SRAs and two SRA-RINGs (see below) to determine whether any additional conserved regions outside the annotated SRA domain existed that could be important for its function. As shown in Figure 2A, a short stretch of hydrophobic amino acids upstream of the annotated SRA domain was found to be conserved. When this extended SRA domain from SUVH6 was fused to GST (SUVH6-SRA), it was completely soluble and highly active in binding methylated DNA. Importantly, like full-length SUVH6, SUVH6-SRA preferred methylated CNG and CNN over CG substrates (Figure 3E). Such a preference was confirmed by competition experiments using methylated CNG substrates and methylated CG or CNG oligonucleotides as competitors (Figure 3G).
SRA Domains Associated with Arabidopsis Ring Proteins Preferentially Bind Methylated CG
In addition to the histone methyltransferases, eight other Arabidopsis proteins contain the SRA domain. Two proteins consist of a single SRA domain, whereas a class of six proteins homologous to Np95 contains the SRA as well as PHD and RING domains. To determine if the SRA domain found in the SRA-RING proteins also binds methylated DNA, we examined two Arabidopsis SRA-RING genes, which we named (ORTHRUS1) ORTH1 (At5g39550) and ORTH2 (At1g57820) (Figure 2A). Three GST fusion constructs were made from ORTH1: PNS contains the PHD domain, the N-terminal RING, and the SRA domain; PN contains the PHD and N-terminal RING domains; and SC contains the SRA and C-terminal RING domains (Figure 4A). In addition, a GST-tagged, full-length ORTH2 was made. Each of the proteins containing an SRA domain (PNS, SC, ORTH2) was able to bind methylated CG, CNG or CNN substrates, but significantly less binding was observed to the unmethylated substrates (Figure 4B and Figure S3A-B). Interestingly, unlike the KYP and SUVH6 SRA domains that prefer CNG and CNN methylation, ORTH1 and ORTH2 show preferential binding to methylated CG substrates (Figure 4B-D, Figure S3A-D). Two different mutations introduced into the SRA domain in the PNS construct, S292F and R362H [corresponding to amino acids 200 and 260 based on the KYP sequence (Figure 2A)], resulted in loss of DNA binding (Figure S4A-B). These results implicate SRA-RING proteins in functions related to DNA methylation and further support the generality of the function of the SRA as a methyl-cytosine-binding domain. The SRA domain differs from the methyl-CpG-binding domain (MBD) found in several mammalian proteins such as MeCP2, since the MBD only binds methylated CG [25], but the SRA domain can recognize methylated DNA in any sequence context. Further, our data suggest that different SRA domains display preference for methylated cytosines in different sequence contexts.
Figure 4.
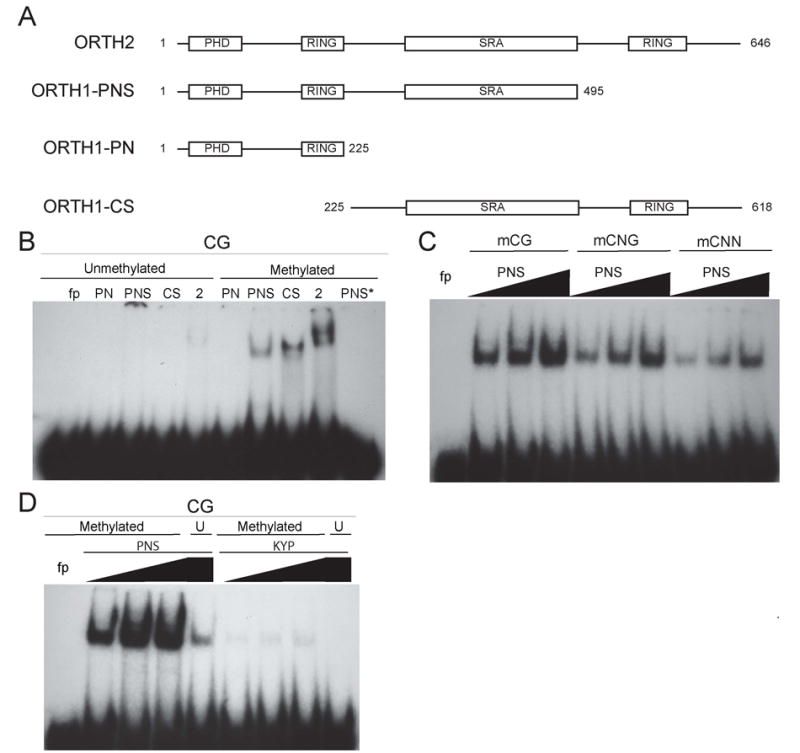
Binding of the ORTH proteins to methylated DNA is dependent on the SRA domain. (A) Diagram of the different regions of ORTH1 or ORTH2 fused to GST and used in mobility shift assays. (B) Mobility shift assays were done with the indicated protein using either unmethylated or methylated CG probes. *The last line in each panel contains binding of GST-PNS to methylated CG super-shifted by addition of anti-GST antibody. (C) Mobility shift assays were performed with increasing amounts (35, 70, and 140 nM) of GST-PNS binding to mCG, mCNG, or mCNN. (D). Mobility shift assays with increasing amounts of GST-PNS (70, 140, and 280 nM) or GST-KYP (77, 154, and 308 nM) with methylated (Methylated) or unmethylated (U) CG. The first lane in all panels is the free probe (fp).
Conclusions
Our findings extend our understanding of DNA and histone methylation pathways in Arabidopsis thaliana. The observation that the SRA domain can bind directly to methylated DNA suggests that regions rich in either CG or non-CG DNA methylation may attract KYP, leading to H3K9 methylation. Alternatively, KYP could be targeted to heterochromatin by other mechanisms and the binding of methylated DNA to the SRA domain may be somehow required for KYP activity. KYP dependent histone methylation would then provide a binding site for CMT3 leading to CNG methylation, which would promote further action by KYP. This model suggests a self-reinforcing feedback loop for the maintenance of DNA and histone methylation and may help to explain the remarkable stability of epigenetic silent states.
The potential targeting of KYP to methylated DNA is similar to the interaction of the histone methyltransferase SETDB1 with MBD1, which links DNA methylation to histone methylation in mammalian chromatin [26]. Thus, while the mechanisms are clearly different, the general phenomenon of DNA methylation targeting histone methylation appears to be conserved between plants and animals.
Supplementary Material
Acknowledgments
We thank Sriharsa Pradhan for oligonucleotides, Daniel Zilberman for sequencing kyp-5, Ana Marie Palanca and Wiam Turki-Judeh for technical assistance. M.B. was funded by American Heart Association Postdoctoral Fellowship 0625014Y. E. K. was supported by NIH training grant GM0007377-27. Funded by NIH Grant GM60398 to S.E.J and NSF-2010 MCB-0519970 to J. C. S.E.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental Data
Supplemental data include Experimental Procedures, four figures and one table and can be found with this article online at:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stancheva I. Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin. Biochem Cell Biol. 2005;83:385–395. doi: 10.1139/o05-043. [DOI] [PubMed] [Google Scholar]
- 2.Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 3.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 4.Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. Embo J. 2002;21:6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. Embo J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci U S A. 2003;100:8823–8827. doi: 10.1073/pnas.1432939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci U S A. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- 9.Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 10.Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci U S A . 2002;99(Suppl 4):16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 13.Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, Cheng X, Schubert I, Jenuwein T, Jacobsen SE. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- 14.Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- 15.Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, Jenuwein T, Khorasanizadeh S, Jacobsen SE. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. Embo J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathieu O, Probst AV, Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. Embo J. 2005;24:2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 20.Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naumann K, Fischer A, Hofmann I, Krauss V, Phalke S, Irmler K, Hause G, Aurich AC, Dorn R, Jenuwein T, Reuter G. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. Embo J. 2005;24:1418–1429. doi: 10.1038/sj.emboj.7600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 24.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 26.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


