Abstract
The primary objective of this study was to extend our knowledge of the geographical distribution, genetic diversity, and natural host associations of the hantaviruses indigenous to North America. Antibody to a hantavirus was found in 5 (20.8%) of 24 Coues' oryzomys (Oryzomys couesi) and none of 41 other rodents captured near the town of Catacamas in eastern Honduras, and a hantavirus was isolated from one of the antibody-positive Coues' oryzomys. Analyses of nucleotide and amino acid sequence data indicated that the viral isolate is a strain of a novel hantaviral species (proposed species name “Catacamas virus”) that is phylogenetically most closely related to Bayou virus, a hantaviral species that is principally associated with Oryzomys palustris (marsh oryzomys) in the southeastern United States. Catacamas virus is the first evidence for the occurrence of a hantaviral species in Honduras and the first evidence that a hantaviral species is naturally associated with an Oryzomys species other than O. palustis.
INTRODUCTION
Hantavirus pulmonary syndrome (HPS) is a zoonosis caused by some of the hantaviruses (family Bunyaviridae, genus Hantavirus) that are principally associated with sigmodontine rodents (family Cricetidae, subfamily Sigmodontinae). The viral species that have been causally associated with HPS include Bayou virus (BAYV), Black Creek Canal virus (BCCV), and New York virus (NYV) on the mainland United States, 1-4 Sin Nombre virus (SNV) in Canada and on the mainland United States,5,6 Choclo virus (CHOV) in Panama,7 Andes virus (ANDV) in Argentina and Chile,8-10 and Laguna Negra virus (LANV) in Paraguay and Argentina.11,12 Other hantaviral species that are naturally associated with sigmodontine rodents include El Moro Canyon virus (ELMCV) and Muleshoe virus (MULV) on the mainland United States,13,14 Rio Segundo virus (RIOSV) in Costa Rica,15 Caño Delgadito virus (CADV) and Maporal virus (MAPV) in Venezuela,16,17 and Rio Mamore virus (RIOMV) in Bolivia.18 The objectives of this study were to extend our knowledge of the geographical distribution, natural host associations, and genetic diversity of the hantaviral species indigenous to North America and to define better the taxonomical relationship of CADV to other hantaviral species that are principally associated with sigmodontine rodents.
MATERIALS AND METHODS
Study site
Small rodents were captured at a site (UTM 16-624523-1637511) located 4.0 km east of the town of Catacamas in eastern Honduras. The landscape of the study site included improved pastures, a stream, banana groves, and mango and other fruit orchards.
Capture and processing of rodents
The rodents were captured in live traps (H. B. Sherman Traps, Inc., Tallahassee, FL, model LFATDG) as described in Texas Tech University Animal Care and Use Protocol #01178X. Four hundred traps were set each night during a 2-day period in July 2001. The traps were placed within grassy areas, set at 5-m intervals, and baited with a mixture of rice, millet, cracked corn, and wheat.
Each animal was assigned a unique museum (TK) number and then identified to species level based on external morphologic features.19 Samples of blood were dried on Nobuto strips (Advantec MFS, Inc., Pleasanton, CA), samples of heart, kidney, liver, lung, skeletal muscle, and spleen were stored individually in cryovials in liquid nitrogen, and the skins and skeletons were prepared as museum voucher specimens. The tissue samples and the skins and skeletons subsequently were deposited into the Natural Science Research Laboratory, Museum, Texas Tech University.
Antibody assays
The blood samples were tested for immunoglobulin G (IgG) against the CADV prototype strain VHV-574, using an enzyme-linked immunosorbent assay (ELISA) described previously.16 The test antigen was a lysate of Vero E6 cells infected with strain VHV-574. The control (comparison) antigen was a lysate of uninfected Vero E6 cells. Serial 4-fold dilutions (from 1:80 through 1:5,120) of each blood sample were tested against the test antigen and the control antigen. Antibody bound to antigen was detected by using a mixture of a goat anti-Rat IgG peroxidase conjugate and a goat anti-Peromyscus leucopus IgG peroxidase conjugate in conjunction with the ABTS Microwell Peroxidase Substrate System (Kirkegaard and Perry Laboratories, Gaithersburg, MD). Optical densities (OD) at 405 nm (reference = 490 nm) were measured with a Dynex MRX II microplate reader (Dynatech Industries, Inc., McLean, VA). The adjusted OD (AOD) of a blood-antigen reaction was the optical density of the well coated with the test antigen less the OD of the well coated with the control antigen. A blood sample was considered to be antibody-positive if the AOD at 1:80 was ≥ 0.200, the AOD at 1:320 was ≥ 0.200, and the sum of the AODs for the series of 4-fold dilutions (from 1:80 through 1:5,120) was ≥ 0.750.20 The titer of a positive sample was the reciprocal of the highest dilution for which the AOD was ≥ 0.200.20
The blood samples were tested for IgG against hantavirus strain HV C1280001, using an indirect fluorescent antibody test (IFAT) described previously.21 (Strain HV C1280001 was isolated from a lung of rodent TK102040 in this study.) The cell spots were prepared from a suspension that contained a 1:1 mixture of Vero E6 cells infected with HV C1280001 and uninfected Vero E6 cells. Antibody (IgG) bound to cell-associated hantaviral antigen was revealed by using a goat anti-Peromyscus leucopus IgG fluorescein isothiocyanate conjugate (Kirkegaard and Perry Laboratories).
Virus assay
A lung of rodent TK102040 and the lungs of two other antibody-positive rodents, specifically TK102095 and TK102097, were tested for infectious hantavirus by cultivation in Vero E6 cells as described previously.16 Hantaviral antigen in the cultured cells was detected by using an IFAT in which the primary antibody was a hyperimmune mouse ascitic fluid raised against SNV and CADV. Mouse IgG bound to cell-associated hantaviral antigen was revealed by using a goat anti-mouse IgG fluorescein isothiocyanate conjugate (Kirkegaard and Perry Laboratories).
Genetic characterization of HV C1280001 and CADV strain VHV-574
The genomes of hantaviruses consist of 3 RNA segments, designated small (S), medium (M), and large (L), which encode the viral nucleocapsid (N) protein, glyco-protein precursor (GPC), and RNA-dependent RNA polymerase, respectively. Distinction between hantaviral species oftentimes is based on comparisons of the amino acid sequences of complete N proteins and comparisons of the amino acid sequences of complete GPCs.22
The nucleotide sequence of a 1943-nt fragment of the S segment and the nucleotide sequence of a 3616-nt fragment of the M segment of HV C1280001 were determined. The 1943-nt fragment of the S segment included the entire N protein gene. Similarly, the 3616-nt fragment of the M segment included the entire GPC gene.
Our knowledge of the genome of VHV-574 previously was limited to a 1130-nt fragment of the S segment, a 439-nt fragment of the 5′ half of the GPC gene, and a 575-nt fragment of the 3′ half of the GPC gene (GenBank accession nos. AF000140, AY953444, and AY953442, respectively). In this study, the lengths of the complete S segment and the complete M segment of VHV-574 were 1986 nucleotides and 3675 nucleotides, respectively.
Total RNA was isolated from monolayers of infected Vero E6 cells, using TRIzol® Reagent (Invitrogen Life Technologies, Inc., Carlsbad, CA). Reverse transcription of the S segment RNA and the M segment RNA was done by using SuperScript II RNase H− Reverse Transcriptase (Invitrogen Life Technologies, Inc.) in conjunction with oligonucleotide 5′-GGTGGTTGTGGTAGTAGTAGACTCC-3′ and HTM7 (5′-TAGTAGTAGACTCCGCAAGAAGAAGCA-3′), respectively.2 Amplicons (PCR products) were generated from 3 overlapping fragments of the S segment first-strand cDNA and from 2 overlapping fragments of the M segment first-strand cDNA by using the Master Taq Kit (Eppendorf North America, Inc., Westbury, NY) in conjunction with oligonucleotides described previously or designed based on sequence data generated in this study. The sequences of the termini of the S segment and the termini of the M segment of VHV-574 were determined from amplicons generated from circularized RNA. The termini were ligated together by using T4 RNA Ligase (Promega Corp., Madison, WI). Reverse transcription through the ligation site of the S segment RNA and through the ligation site of the M segment RNA was accomplished by using SuperScript II RNase H− Reverse Transcriptase (Invitrogen Life Technologies, Inc.) in conjunction with HTS28 (5′-CAGAATCATCTCATTAGTCC-3′) and HTM29 (5′-GAACATTCAGGCTCATTTCG-3′), respectively. Amplification of a 541-nt fragment of the S segment first-strand cDNA and a 1236-nt fragment of the M segment first-strand cDNA was done by using the Master Taq Kit (Eppendorf North America, Inc.) in conjunction with oligo-nucleotides HTS22 (5′-AGATCTGCCATCTGACGCT-3′) and HTS28 and oligonucleotides HTM29 and HTM46 (5′-GTACAGCACCAGCTGCAACATG-3′), respectively. Amplicons of the expected size were purified from agarose gel slices by using the QIAquick Gel Extraction Kit (Qiagen, Inc., Valencia, CA). Both strands of each purified amplicon were sequenced directly, using the dye termination cycle sequencing technique (Applied Biosystems, Inc., Foster City, CA). The nucleotide sequences of the S and M segments of HV C1280001 and the S and M segments of VHV-574 were deposited into the GenBank nucleotide sequence database under accession nos. DQ256126 and DQ177347, and DQ285566 and DQ284451, respectively.
The nucleotide sequences of the N protein genes and GPC genes and the amino acid sequences of the N proteins and GPCs of HV C1280001 and VHV-574 were compared with the homologous sequences of ANDV strains AH-1, Chile-9717869, CHI-7913, Hu39694, Lechiguanas, and Oran, BAYV, BCCV, CHOV strain 588, ELMCV strain RM-97, LANV strain 510B, MAPV strain HV 97021050, MULV strain SH-Tx-339, NYV strain Rhode Island-1 (RI-1), RIOMV strain OM-556, SNV strain NM R11, Prospect Hill virus (PHV), Puumala virus (PUUV) strain CG1820, Hantaan virus (HTNV) strain 76-118, and Seoul virus (SEOV) strain SR11 (GenBank accession nos. AF324902 and AF324901, AF291702 and AF291703, AY228237 and AY228238, AF482711 and AF028023, AF482714 and AF028022, AF482715 and AF028024, L36929 and L36930, L39949 and L39950, DQ285046 and DQ285047, U11427 and U26828, AF005727 and AF005728, AY267347 and AY363179, U54575, U09488 and U36801, U5213600, L37904 and L37903, M34011 and X55129, M32750 and M29979, M14626 and M14627, and M34881 and M34882, respectively). Prospect Hill virus and PUUV are principally associated with arvicoline rodents (family Cricetidae, subfamily Arvicolinae) and HTNV and SEOV are principally associated with murine rodents (family Muridae, subfamily Murinae). The hantaviral species principally associated with sigmodontine rodents (New World rats and mice) are phylogenetically distinct from the hantaviral species principally associated with arvicoline rodents (voles and lemmings) and from the hantaviral species principally associated with murine rodents (Old World rats and mice).23 The amino acid sequences were aligned by using the computer program CLUSTALW (1.7).24 The nucleotide sequences were aligned manually based on the computer-generated multiple amino acid sequence alignments. The analyses of the multiple sequence alignments were done by using programs in the computer software packages MEGA2 and PAUP*, version 4.0b10.25,26 The neighbor-joining (NJ) analyses were carried out on uncorrected p model distances, with all 3 nucleotide positions included in the distance calculations. In the maximum parsimony (MP) analyses, third position nucleotides and uninformative first and second position nucleotides were excluded, and the characters were weighted in accordance with the transition/transversion (ti/tv) ratio calculated from the data. Bootstrap support for the NJ analyses was based on 1000 repetitions of the heuristic search and bootstrap support for the MP analyses was based on 500 repetitions of the heuristic search.27
Genetic characterization of rodents
Rodents TK102040 and TK102014 were identified at the study site as Coues' oryzomys (Oryzomys couesi). The nucleotide sequences of the cytochrome b genes of TK102040 and TK102014, three other Coues' oryzomys (TK93218, TK93244, and TK72660), and 4 marsh oryzomys (Oryzomys palustris–TK91240, TK51628, TK111000, and TK27995) were determined to improve our knowledge of the taxonomical relationship between O. couesi and O. palustris, the principal host of BAYV.28 Rodents TK93218 and TK93244 were captured at Las Minas in Oaxaca, Mexico, TK72660 was captured in Cameron County, Texas, TK91240 in Galveston County, Texas, TK51628 in Calhoun County, Texas, TK111000 in Brazoria County, Texas, and TK27995 in Okmulgee County, Oklahoma. Mitochondrial DNA was isolated from liver as described previously.29 The complete cytochrome b gene (1143-bp) was amplified by PCR, using Taq DNA Polymerase (Promega Corp.) in conjunction with oligonucleotides MVZ05 and MVZ14 or MVZ05 and 1115.30,31 Amplicons of the expected size were purified from agarose gel slices by using the QIAquick PCR Purification Kit (Qiagen, Inc.). Both strands of each purified amplicon were sequenced directly, using the dye termination cycle sequencing technique (Applied Biosystems, Inc.) in conjunction with oligonucleotides MVZ05 and MVZ14,30 1115,31 L14841,32 F1,33 400R,34 700H and 700L,35 or 870R.36 The nucleotide sequences of the cytochrome b genes of TK102040, TK102014, TK91240, TK93218, TK93244, TK51628, TK27995, TK111000, and TK72660 were deposited into the GenBank nucleotide sequence database under accession nos. DQ185383, DQ185384, DQ185382, DQ185385, DQ185386, DQ370031, DQ370032, DQ370033, and DQ370034, respectively, and then compared with the nucleotide sequences of the cytochrome b genes of a marsh oryzomys from Collier County, Florida, 4 russet oryzomys (Oryzomys russatus) captured at different localities in Brazil, 2 Emmon's oryzomys (Oryzomys emmonsae) captured at a single locality in Brazil, 2 Azara's broad-headed oryzomys (Oryzomys megacephalus) captured at different localities in Brazil, 1 Azara's broad-headed oryzomys captured in Guyana, 1 Azara's broad-headed oryzomys captured in Suri-name, 2 Atlantic Forest oryzomys (Oryzomys laticeps) captured at different localities in Brazil, and 1 Ucayali spiny rat (Scolomys ucayalensis) captured in Brazil (GenBank accession nos. L37388, AF181271, AF181272, AF251523, and AF251524, AF251525 and AF251526, AF251516 and AF251519, AF251518, AF251517, AF251521 and AF251522, and AF527420, respectively).37,38 A previous study established that O. russatus and O. emmonsae are sister species and that O. megacephalus and O. laticeps are sister species.38 The O. russatus, O. emmonsae, O. megacephalus, and O. laticeps sequences were included in the analyses to provide a metric for interpretation of the genetic distances between the Coues' oryzomys and the marsh oryzomys. The analyses of the multiple sequence alignment were done by using programs in the computer software package PAUP*, version 4.0b10, and the computer program MrBayes 3.1.2.26,39 Sequence nonidentities were equivalent to uncorrected p model distances. The S. ucayalensis sequence was the designated outgroup taxon in the Bayesian analysis.39 A GTR+I+G model with a site-specific gamma distribution was used with the following options in MrBayes 3.1.2: two simultaneous runs of 4 Markov-chains, 10 million generations, and sample frequency = every 100th generation.40 The first 500 trees were discarded after review of the likelihood scores and the consensus tree (50% majority rule) was constructed from the remaining trees.
RESULTS
Sixty-five rodents were captured during the 2-night trapping period in July 2001. The captured rodents included 24 Coues' oryzomys, 2 Salvin's spiny pocket mice (Liomys salvini), 1 Alfaro's oryzomys (Oryzomys alfaroi), 1 Mexican harvest mouse (Reithrodontomys mexicanus), and 37 southern cotton rats (Sigmodon hirsutus). Antibody reactive against CADV strain VHV-574 was detected in 3 Coues' oryzomys (TK102040, TK102095, and TK102097) and none of the 62 other rodents. The antibody titers in the positive animals ranged from 320 (TK102040 and TK102097) to 1280 (TK102095). Antibody reactive against HV C1280001 (the hantavirus isolated from TK102040) was detected in TK102040, TK102095, TK102097, two other Coues' oryzomys, and none of the 60 other rodents.
Infectious hantavirus (strain HV C1280001) was isolated from the sample of lung tissue from rodent TK102040. Hantaviral antigen was not detected in the Vero E6 cells inoculated with the original suspension of lung tissue from TK102040 but was observed in 40% of the Vero E6 cells inoculated with first-passage (Vero E6 + 1) material. The attempts to isolate infectious hantavirus from rodents TK102095 and TK102097 were unsuccessful.
The alignment of complete N protein amino acid sequences was 433 characters in length. In pairwise comparisons, nonidentities (uncorrected p model distances) among the sigmodontine rodent-associated hantaviral species ranged from 4.7–17.8%, HV C1280001 exhibited the lowest nonidentity (4.7%) with BAYV, SNV exhibited the lowest nonidentity (6.3%) with NYV, and CADV strain VHV-574 exhibited the lowest nonidentity (13.8%) with MAPV (Table 1).
Table 1.
Nonidentities among the predicted amino acid sequences of the nucleocapsid proteins and among the nucleotide sequences of the nucleocapsid protein genes of 18 hantaviral strains naturally associated with sigmodontine rodents*
| Amino acid sequence nonidentity (%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species† | HV C1280001 | BAYV | BCCV | CHOV | ELMCV | MULV | NYV | SNV | ANDV | CADV | MAPV | LANV | RIOMV |
| HV C1280001 | – | 4.7 | 8.2 | 12.9 | 14.5 | 7.5 | 12.4 | 13.6 | 11.4–12.4 | 16.1 | 12.4 | 12.1 | 11.9 |
| Bayou virus | 16.8 | – | 7.7 | 11.4 | 15.4 | 7.2 | 12.4 | 13.1 | 11.0–12.1 | 16.6 | 11.9 | 12.9 | 11.9 |
| Black Creek Canal virus | 18.1 | 19.2 | – | 14.5 | 16.6 | 9.8 | 14.5 | 16.1 | 13.6–14.0 | 17.3 | 13.6 | 14.5 | 13.3 |
| Choclo virus | 24.5 | 23.8 | 23.4 | – | 16.1 | 15.4 | 11.4 | 12.1 | 9.3–10.0 | 14.7 | 10.5 | 12.1 | 11.2 |
| El Moro Canyon virus | 23.9 | 23.6 | 24.8 | 24.5 | – | 17.3 | 15.7 | 15.4 | 17.1–17.5 | 17.3 | 16.1 | 17.8 | 17.3 |
| Muleshoe virus | 19.2 | 19.3 | 19.5 | 24.2 | 24.3 | – | 16.4 | 17.3 | 14.3–15.2 | 17.3 | 14.3 | 14.7 | 14.0 |
| New York virus | 23.1 | 22.6 | 24.5 | 23.7 | 23.1 | 23.9 | – | 6.3 | 12.1–12.6 | 15.4 | 13.1 | 12.9 | 13.3 |
| Sin Nombre virus | 23.7 | 23.3 | 25.0 | 24.0 | 23.4 | 24.8 | 16.7 | – | 12.9–14.5 | 16.4 | 14.0 | 14.7 | 15.4 |
| Andes virus ‡ | 21.3–23.0 | 21.7–23.8 | 21.8–24.5 | 20.6–21.8 | 22.4–23.6 | 23.0–24.2 | 22.3–23.6 | 22.7–24.4 | – | 14.3–14.7 | 8.4–9.1 | 9.6–10.3 | 8.9–9.3 |
| Caño Delgadito virus | 24.3 | 24.1 | 25.5 | 23.4 | 23.7 | 24.4 | 23.8 | 23.9 | 22.0–22.9 | – | 13.8 | 15.9 | 15.4 |
| Maporal virus | 22.1 | 22.1 | 22.7 | 22.2 | 23.8 | 23.4 | 22.9 | 24.6 | 19.9–21.3 | 23.3 | – | 10.5 | 8.4 |
| Laguna Negra virus | 21.1 | 22.5 | 23.1 | 21.3 | 25.3 | 24.1 | 23.6 | 23.7 | 20.2–21.5 | 25.2 | 22.1 | – | 6.8 |
| Rio Mamore virus | 22.7 | 22.6 | 22.9 | 22.0 | 23.4 | 22.9 | 23.1 | 23.8 | 19.0–20.4 | 23.4 | 18.3 | 17.6 | – |
|
Nucleotide sequence nonidentity (%) |
|||||||||||||
Abbreviations: BAYV = Bayou virus; BCCV = Black Creek Canal virus; CHOV = Choclo virus; ELMCV = El Moro Canyon virus; MULV = Muleshoe virus; NYV = New York virus; SNV = Sin Nombre virus; ANDV = Andes virus; CADV = Caño Delgadito virus; MAPV = Maporal virus; LANV = Laguna Negra virus; RIOMV = Rio Mamore virus.
Numbers above and below the diagonal line are the amino acid sequence nonidentities and nucleotide sequence nonidentities, respectively.
Catacamas virus strain HV C1280001 was isolated from a Coues' oryzomys in this study.
Six strains of ANDV were included in the pairwise comparisons of predicted amino acid sequences and the pairwise comparisons of nucleotide sequences: AH-1, Chile-9717869, CHI-7913, Hu39694, Lechiguanas, and Oran. Nonidentities among the amino acid sequences and among the nucleotide sequences of these six strains ranged from 0.0 to 4.0% and from 2.5 to 16.7%, respectively.
The alignment of complete GPC amino acid sequences was 1153 characters in length. In pairwise comparisons, nonidentities among the sigmodontine rodent-associated hantaviral species ranged from 5.2–27.4%, HV C1280001 exhibited the lowest nonidentity (6.4%) with BAYV, SNV exhibited the lowest nonidentity (5.2%) with NYV, and CADV strain VHV-574 exhibited the lowest nonidentity (22.8%) with SNV (Table 2).
Table 2.
Nonidentities among the predicted amino acid sequences of the glycoprotein precursors and among the nucleotide sequences of the glycoprotein precursor genes of 16 hantaviral strains naturally associated with sigmodontine rodents*
| Amino acid sequence nonidentity (%) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species† | HV C1280001 | BAYV | BCCV | CHOV | ELMCV | NYV | SNV | ANDV | CADV | LANV | MAPV |
| HV C1280001 | – | 6.4 | 11.1 | 23.2 | 23.8 | 20.8 | 19.7 | 24.3 – 25.1 | 24.7 | 24.5 | 25.0 |
| Bayou virus | 20.9 | – | 11.5 | 22.6 | 23.4 | 21.1 | 19.1 | 23.9 – 24.8 | 24.2 | 24.4 | 25.2 |
| Black Creek Canal virus | 22.5 | 22.3 | – | 23.8 | 23.8 | 21.4 | 19.9 | 24.1 – 25.1 | 25.1 | 24.3 | 25.5 |
| Choclo virus | 28.6 | 29.0 | 29.2 | – | 25.2 | 23.5 | 21.5 | 17.0 – 18.1 | 23.6 | 18.4 | 17.2 |
| El Moro Canyon virus | 28.9 | 29.5 | 28.8 | 30.3 | – | 21.7 | 20.4 | 26.3 – 27.1 | 26.4 | 25.8 | 27.4 |
| New York virus | 27.7 | 28.3 | 27.4 | 29.5 | 28.7 | – | 5.2 | 22.8 – 23.8 | 24.4 | 24.2 | 24.1 |
| Sin Nombre virus | 27.6 | 27.2 | 26.7 | 28.7 | 28.0 | 19.1 | – | 21.7 – 22.4 | 22.8 | 23.2 | 23.1 |
| Andes virus ‡ | 29.4 – 30.4 | 29.1 – 30.1 | 28.7 – 30.5 | 26.1 – 26.7 | 31.0 – 31.8 | 29.0 – 30.4 | 28.4 – 29.0 | – | 25.7 – 26.5 | 13.2 – 13.7 | 14.6 – 15.7 |
| Caño Delgadito virus | 30.1 | 30.2 | 30.4 | 29.7 | 30.0 | 29.5 | 28.7 | 30.1 – 30.6 | – | 26.1 | 25.3 |
| Laguna Negra virus | 29.0 | 30.1 | 28.8 | 27.5 | 30.3 | 28.5 | 28.9 | 24.1 – 24.9 | 29.9 | – | 18.6 |
| Maporal virus | 29.1 | 29.0 | 29.6 | 26.4 | 31.7 | 28.7 | 28.8 | 24.6 – 25.2 | 30.3 | 25.9 | – |
|
Nucleotide sequence nonidentity (%) |
|||||||||||
Abbreviations: BAYV = Bayou virus; BCCV = Black Creek Canal virus; CHOV = Choclo virus; ELMCV = El Moro Canyon virus; NYV = New York virus; SNV = Sin Nombre virus; ANDV = Andes virus; CADV = Caño Delgadito virus; LANV = Laguna Negra virus; MAPV = Maporal virus.
Numbers above and below the diagonal line are the amino acid sequence nonidentities and nucleotide sequence nonidentities, respectively.
Catacamas virus strain HV C1280001 was isolated from a Coues' oryzomys in this study. Muleshoe virus and Rio Mamore virus were not included in the pairwise comparisons because the GenBank nucleotide sequence database did not contain the sequence of the complete glycoprotein precursor gene of a strain of Muleshoe virus or Rio Mamore virus.
Six strains of ANDV were included in the pairwise comparisons of predicted amino acid sequences and the pairwise comparisons of nucleotide sequences: AH-1, Chile-9717869, CHI-7913, Hu39694, Lechiguanas, and Oran. Nonidentities among the amino acid sequences and among the nucleotide sequences of these six strains ranged from 0.9 to 8.1% and from 2.8 to 21.3%, respectively.
The NJ analysis of uncorrected p model distances generated from the alignment of N protein gene sequences separated the hantaviral species associated with sigmodontine rodents from the hantaviral species associated with arvicoline or murine rodents, indicated that HV C1280001 is phylogenetically most closely related to BAYV, and placed the HV C1280001-BAYV lineage in a sister relationship to a lineage represented by BCCV (Fig. 1A). Note that HV C1280001 and BAYV are phylogenetically more closely related to BCCV and MULV than to ANDV, CHOV, or MAPV and that BCCV and MULV are phylogenetically more closely related to HV C1280001 and BAYV than to CADV. Also note that ANDV, CHOV, and MAPV are principally associated with Oligoryzomys species, BCCV, MULV, and CADV are principally associated with Sigmodon species, and—as indicated previously—BAYV is principally associated with Oryzomys palustris.7,8,10,14,16,17,28,41-43 The results of the NJ analysis of uncorrected p model distances generated from the alignment of N protein amino acid sequences, the MP analysis of the alignment of N protein gene sequences, and the MP analysis of the alignment of N protein amino acid sequences were essentially the same as the results of the NJ analysis of uncorrected p model distances generated from the alignment of N protein gene sequences.
Figure 1.
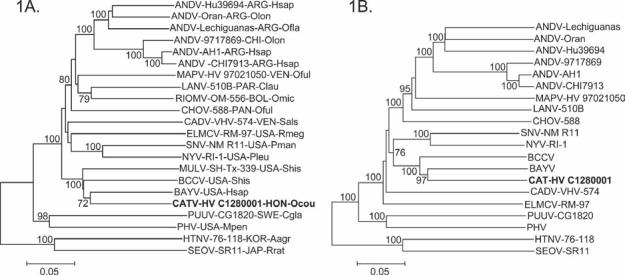
Phylogenetic relationships among hantaviruses based on neighbor-joining analyses of genetic distances generated from comparisons of (1A) complete nucleocapsid protein gene sequences and (1B) complete glycoprotein precursor gene sequences. The scale bar indicates a sequence divergence of 0.05. The numerical value at the node indicates the percentage of 1000 bootstrap replicates that supported the interior branch. Bootstrap support values less than 70% are not listed. The branch labels include (in the following order) viral species, strain (if known), country, and host species. ANDV, Andes virus; BAYV, Bayou virus; BCCV, Black Creek Canal virus; CADV, Caño Delgadito virus; CATV, Catacamas virus; CHOV, Choclo virus; ELMCV, El Moro Canyon virus; HTNV, Hantaan virus; LANV, Laguna Negra virus; MAPV, Maporal virus; MULV, Muleshoe virus; NYV, New York virus; PHV, Prospect Hill virus; PUUV, Puumala virus; RIOMV, Rio Mamore virus; SEOV, Seoul virus; SNV, Sin Nombre virus. ARG, Argentina; BOL, Bolivia; CHI, Chile; HON, Honduras; JAP, Japan; KOR, Korea; PAN, Panama; PAR, Paraguay; SWE, Sweden; USA, United States of Amerca; VEN, Venezuela. Aagr, Apodemus agrarius (striped field mouse); Cgla, Clethrionomys glareolus (bank vole); Clau, Calomys laucha (small vesper mouse); Hsap, Homo sapiens (human); Mpen, Microtus pennsylvanicus (meadow vole); Ocou, Oryzomys couesi (Coues' oryzomys); Ofla, Oligoryzomys flavescens (yellow pygmy colilargo); Oful, Oligoryzomys fulvescens (fulvous colilargo); Olon, Oligoryzomys longicaudatus (long-tailed colilargo); Omic, Oligoryzomys microtis (small-eared colilargo); Pleu, Peromyscus leucopus (white-footed mouse); Pman, Peromyscus maniculatus (deer mouse); Rmeg, Reithrodontomys megalotis (western harvest mouse); Rrat, Rattus rattus (house rat); Shis, Sigmodon hispidus (hispid cotton rat).
The phylogenetic relationships reconstructed by NJ analysis of uncorrected p model distances generated from the alignment of GPC gene sequences were very similar to the results of the NJ analysis of uncorrected p model distances generated from the alignment of N protein gene sequences (Figure 1B and Figure 1A, respectively). The results of the NJ analysis of uncorrected p model distances generated from the alignment of GPC amino acid sequences, the MP analysis of the alignment of GPC gene sequences, and the MP analysis of the alignment of GPC amino acid sequences were essentially the same as the results of the NJ analysis of uncorrected p model distances generated from the alignment of GPC gene sequences.
In pairwise comparisons of cytochrome b gene sequences, nonidentities (uncorrected p model distances) among the 5 Coues' oryzomys ranged from 0.5–5.3%, nonidentities among the 5 marsh oryzomys ranged from 0.1–6.3%, nonidentities between the Coues' oryzomys and the marsh oryzomys ranged from 9.9–11.3%, and nonidentities between the russet oryzomys and Emmon's oryzomys and between the Azara's broad-headed oryzomys and Atlantic Forest oryzomys ranged from 10.7–11.7% and from 9.7–11.5%, respectively. The results of the Bayesian analysis of cytochrome b gene sequences indicated that the Coues' oryzomys and marsh oryzomys represent different phylogenetic lineages and placed the Coues' oryzomys lineage in a sister relationship to the marsh oryzomys lineage relative to the O. russatus–O. emmonsae lineage and to the O. megacephalus–O. laticeps lineage. Bootstrap support for monophyly of the Coues' oryzomys and for monophyly of the marsh oryzomys in the Bayesian analysis was 100%.
DISCUSSION
The isolation of HV C1280001 from rodent TK102040 is the first evidence for association of the genus Hantavirus with an Oryzomys species other than O. palustris. The nonidentity between the amino acid sequences of the N proteins of HV C1280001 and BAYV is similar in magnitude to the nonidentity between the amino acid sequences of the N proteins of SNV and NYV. Likewise, the nonidentity between the amino acid sequences of the GPCs of HV C1280001 and BAYV is similar in magnitude to the nonidentity between the amino acid sequences of the GPCs of SNV and NYV. Thus, the viral species represented by HV C1280001 should be considered distinct from BAYV. The name “Catacamas virus” is proposed to distinguish this novel species from BAYV and from all other hantaviral species.
The Oryzomys species known to occur in Honduras are O. alfari (cane oryzomys), O. alfaroi, O. bolivaris (Bolivar oryzomys), O. couesi, and O. rostratus (long-nosed oryzomys).19,44 Oryzomys couesi can be distinguished from the 4 other species on the basis of external morphologic features.19
Rodent TK102040 and the 4 other antibody-positive rodents in this study were Coues' oryzomys. Antibody to HV C1280001 was found in 5 (20.8%) of the 24 Coues' oryzomys captured at the study site. Collectively, the high prevalence of antibody to HV C1280001 in the Coues' oryzomys, the absence of antibody to HV C1280001 in the other rodents captured at the study site, and the isolation of HV C1280001 from TK102040 indicate that O. couesi is the principal host of Catacamas virus (CATV).
The geographical range of O. palustris, the principal host of BAYV, extends from Kansas, New Jersey, and Florida to northeastern Mexico, and the geographical range of O. couesi extends from southern Texas to Colombia.28,45-47 A previous study of oryzomys captured in Texas established that there are measurable differences between O. couesi and O. palustris in adult body size, pelage coloration, other anatomic features, and chromosomal morphology.48 The results of the analyses of cytochrome b gene sequence data in the present study support the separation of O. couesi from O. palustris and indicate that substantial evolutionary divergence has occurred between these rodent species and, as such, that CATV is ecologically as well as phylogenetically distinct from BAYV.22
As noted previously, 3 hantaviral species are naturally associated with Sigmodon (cotton rat) species. Black Creek Canal virus is principally associated with S. hispidus (hispid cotton rat) in southern Florida,41 MULV is naturally and perhaps principally associated with S. hispidus in northern Texas,14 and CADV is principally associated with Sigmodon alstoni (Alston's cotton rat) in western Venezuela.16 The results of the present study confirm the specific status of CADV and indicate that BCCV and MULV are phylogenetically more closely related to CATV and BAYV than to CADV (Fig. 1A).
The present-day principal host relationships of many hantaviral species appear to be a consequence of a long-term shared evolutionary relationship between the genus Hantavirus and the rodent families Cricetidae and Muridae. Evidence for this ancient virus-host relationship includes the association of phylogenetically closely related hantaviral species with phylogenetically closely related rodent species, for example—SNV with P. maniculatus and NYV with P. leucopus,49-51 and ANDV, CHOV and MAPV, and RIOMV with Oligoryzomys longicaudatus (long-tailed colilargo),10,42,43 Oligoryzomys fulvescens (fulvous colilargo),7,17 and Oligoryzomys microtis (small-eared colilargo),18 respectively. The results of analyses of cytochrome b gene sequences in an earlier study indicated that Oryzomys species are phylogenetically more closely related to Oligoryzomys species than to Sigmodon species.29 The results of the phylogenetic analyses in the present study, specifically the monophyly of CATV, BAYV, BCCV, and MULV and the separation of the CATV-BAYV-BCCVMULV lineage from the ANDV-CHOV-MAPV-RIOMV lineage, suggest that the association of CADV with S. alstoni or the association of CATV with O. couesi and BAYV with O. palustris is not the result of an ancient virus-host relationship. A plausible explanation for the discordance between the phylogeny of the viruses and the phylogeny of their rodent hosts is that the present-day association of CATV with O. couesi and BAYV with O. palustris is a consequence of inter-specific (cotton rat-to-oryzomys) virus transmission at some time after the last common ancestor of S. alstoni and S. hispidus. Alternatively, the association of CADV with S. alstoni was established after the last common ancestor of S. alstoni and S. hispidus.
The human health significance of CATV has not been investigated. The results of a recent study in Panama indicated that persons infected with CHOV usually do not develop severe HPS.52 In contrast, persons infected with SNV, ANDV, or some of the other sigmodontine rodent-associated hantaviruses frequently develop severe (if not fatal) HPS. Studies are needed to assess the risk of CATV infection in persons who live or work in rural areas of eastern Honduras and the severity of disease caused by CATV in these persons.
Acknowledgments
Members of the 2001 Field Methods Class, Texas Tech University, and members of the 2001 Sowell Expedition captured and processed the rodents. Mary Louise Milazzo, Maria N. B. Cajimat, and J. Delton Hanson contributed equally to this study.
Footnotes
Financial support: A grant from James Sowell to Robert J. Baker (Texas Tech University) and Robert D. Bradley provided financial support for the field component of this study. National Institutes of Health Grant AI-41435 (“Ecology of emerging arenaviruses in the southwestern United States”) provided financial support for the laboratory work at UTMB and at Texas Tech University.
REFERENCES
- 1.Khan AS, Spiropoulou CF, Morzunov S, Zaki SR, Kohn MA, Nawas SR, McFarland L, Nichol ST. Fatal illness associated with a new hantavirus in Louisiana. J Med Virol. 1995;46:281–286. doi: 10.1002/jmv.1890460320. [DOI] [PubMed] [Google Scholar]
- 2.Morzunov SP, Feldmann H, Spiropoulou CF, Semenova VA, Rollin PE, Ksiazek TG, Peters CJ, Nichol ST. A newly recognized virus associated with a fatal case of hantavirus pulmonary syndrome in Louisiana. J Virol. 1995;69:1980–1983. doi: 10.1128/jvi.69.3.1980-1983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan AS, Gaviria M, Rollin PE, Hlady WG, Ksiazek TG, Armstrong LR, Greenman R, Ravkov E, Kolber M, Anapol H, Sfakianaki ED, Nichol ST, Peters CJ, Khabbaz RF. Hantavirus pulmonary syndrome in Florida: association with the newly identified Black Creek Canal virus. Am J Med. 1996;100:46–48. doi: 10.1016/s0002-9343(96)90010-8. [DOI] [PubMed] [Google Scholar]
- 4.Hjelle B, Lee SW, Song W, Torrez-Martinez N, Song JW, Yanagihara R, Gavrilovskaya I, Mackow ER. Molecular linkage of hantavirus pulmonary syndrome to the white-footed mouse, Peromyscus leucopus: genetic characterization of the M genome of New York virus. J Virol. 1995;69:8137–8141. doi: 10.1128/jvi.69.12.8137-8141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drebot MA, Gavrilovskaya I, Mackow ER, Chen Z, Lindsay R, Sanchez AJ, Nichol ST, Artsob H. Genetic and serotypic characterization of Sin Nombre-like viruses in Canadian Peromyscus maniculatus mice. Virus Res. 2001;75:75–86. doi: 10.1016/s0168-1702(01)00227-1. [DOI] [PubMed] [Google Scholar]
- 6.Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 7.Vincent MJ, Quiroz E, Gracia F, Sanchez AJ, Ksiazek TG, Kitsutani PT, Ruedas LA, Tinnin DS, Caceres L, Garcia A, Rollin PE, Mills JN, Peters CJ, Nichol ST. Hantavirus pulmonary syndrome in Panama: identification of novel hantaviruses and their likely reservoirs. Virology. 2000;277:14–19. doi: 10.1006/viro.2000.0563. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez Della Valle M, Edelstein A, Miguel S, Martinez V, Cortez J, Cacace ML, Jurgelenas G, Estani SS, Padula P. Andes virus associated with hantavirus pulmonary syndrome in northern Argentina and determination of the precise site of infection. Am J Trop Med Hyg. 2002;66:713–720. doi: 10.4269/ajtmh.2002.66.713. [DOI] [PubMed] [Google Scholar]
- 9.López N, Padula P, Rossi C, Lázaro ME, Franze-Fernández MT. Genetic identification of a new hantavirus causing severe pulmonary syndrome in Argentina. Virology. 1996;220:223–226. doi: 10.1006/viro.1996.0305. [DOI] [PubMed] [Google Scholar]
- 10.Meissner JD, Rowe JE, Borucki MK, St Jeor SC. Complete nucleotide sequence of a Chilean hantavirus. Virus Res. 2002;89:131–143. doi: 10.1016/s0168-1702(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 11.Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, Bryan RT, Mills JN, Peters CJ, Nichol ST. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology. 1997;238:115–127. doi: 10.1006/viro.1997.8840. [DOI] [PubMed] [Google Scholar]
- 12.Levis S, Garcia J, Pini N, Calderon G, Ramirez J, Bravo D, St Jeor S, Ripoll C, Bego M, Lozano E, Barquez R, Ksiazek TG, Enria D. Hantavirus pulmonary syndrome in northwestern Argentina: circulation of Laguna Negra virus associated with Calomys callosus. Am J Trop Med Hyg. 2004;71:658–663. [PubMed] [Google Scholar]
- 13.Torrez-Martinez N, Song W, Hjelle B. Nucleotide sequence analysis of the M genomic segment of El Moro Canyon hantavirus: antigenic distinction from Four Corners hantavirus. Virology. 1995;211:336–338. doi: 10.1006/viro.1995.1413. [DOI] [PubMed] [Google Scholar]
- 14.Rawlings JA, Torrez-Martinez N, Neill SU, Moore GM, Hicks BN, Pichuantes S, Nguyen A, Bharadwaj M, Hjelle B. Cocirculation of multiple hantaviruses in Texas, with characterization of the small (S) genome of a previously undescribed virus of cotton rats (Sigmodon hispidus) Am J Trop Med Hyg. 1996;55:672–679. doi: 10.4269/ajtmh.1996.55.672. [DOI] [PubMed] [Google Scholar]
- 15.Hjelle B, Anderson B, Torrez-Martinez N, Song W, Gannon WL, Yates TL. Prevalence and geographic genetic variation of hantaviruses of New World harvest mice (Reithrodontomys): identification of a divergent genotype from a Costa Rican Reithrodontomys mexicanus. Virology. 1995;207:452–459. doi: 10.1006/viro.1995.1104. [DOI] [PubMed] [Google Scholar]
- 16.Fulhorst CF, Monroe MC, Salas RA, Duno G, Utrera A, Ksiazek TG, Nichol ST, de Manzione NMC, Tovar D, Tesh RB. Isolation, characterization, and geographic distribution of Caño Delgadito virus, a newly discovered South American hantavirus (family Bunyaviridae) Virus Res. 1997;51:159–171. doi: 10.1016/s0168-1702(97)00091-9. [DOI] [PubMed] [Google Scholar]
- 17.Fulhorst CF, Cajimat MNB, Utrera A, Milazzo ML, Duno GM. Maporal virus, a hantavirus associated with the fulvous pygmy rice rat (Oligoryzomys fulvescens) in western Venezuela. Virus Res. 2004;104:139–144. doi: 10.1016/j.virusres.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Bharadwaj M, Botten J, Torrez-Martinez N, Hjelle B. Rio Mamore virus: genetic characterization of a newly recognized hantavirus of the pygmy rice rat, Oligoryzomys microtis, from Bolivia. Am J Trop Med Hyg. 1997;57:368–374. doi: 10.4269/ajtmh.1997.57.368. [DOI] [PubMed] [Google Scholar]
- 19.Hall ER. The Mammals of North America. Volume II. John Wiley & Sons, Inc.; New York: 1981. pp. 601–1181. [Google Scholar]
- 20.Fulhorst CF, Milazzo ML, Duno G, Salas RA. Experimental infection of the Sigmodon alstoni cotton rat with Caño Delgadito virus, a South American hantavirus. Am J Trop Med Hyg. 2002;67:107–111. doi: 10.4269/ajtmh.2002.67.107. [DOI] [PubMed] [Google Scholar]
- 21.Tesh RB, Wilson ML, Salas R, de Manzione NM, Tovar D, Ksiazek TG, Peters CJ. Field studies on the epidemiology of Venezuelan hemorrhagic fever: implication of the cotton rat Sigmodon alstoni as the probable rodent reservoir. Am J Trop Med Hyg. 1993;49:227–235. doi: 10.4269/ajtmh.1993.49.227. [DOI] [PubMed] [Google Scholar]
- 22.Nichol ST, Beaty BJ, Elliott RM, Goldbach R, Plyusnin A, Schmaljohn CS, Tesh RB. Family Bunyaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; San Diego, CA: 2005. pp. 695–716. [Google Scholar]
- 23.Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: genome structure, expression, and evolution. J Gen Virol. 1996;77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W (1.7): improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choices. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 26.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4. Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- 27.Felsentein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution Int J Org Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 28.Ksiazek TG, Nichol ST, Mills JN, Groves MG, Wozniak A, McAdams S, Monroe MC, Johnson AM, Martin ML, Peters CJ, Rollin PE. Isolation, genetic diversity, and geographic distribution of Bayou virus (Bunyaviridae: hantavirus) Am J Trop Med Hyg. 1997;57:445–448. doi: 10.4269/ajtmh.1997.57.445. [DOI] [PubMed] [Google Scholar]
- 29.Smith MF, Patton JL. Phylogenetic relationships and the radiation of sigmodontine rodents in South America: evidence from cytochrome b. J Mammal Evol. 1999;6:89–128. [Google Scholar]
- 30.Smith MF, Patton JL. The diversification of South American rodents: evidence from mitochondrial sequence data for the akodontine tribe. Biol J Linn Soc. 1993;50:149–177. [Google Scholar]
- 31.Mendez-Harclerode FM, Hanson JD, Fulhorst CF, Milazzo ML, Ruthven DC, III, Bradley RD. Genetic diversity within the southern plains woodrat (Neotoma micropus) in southern Texas. J Mammal. 2005;86:180–190. doi: 10.1644/1545-1542(2005)086<0180:gdwtsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irwin DM, Kocher TD, Wilson AC. Evolution of the cytochrome b gene in mammals. J Mol Evol. 1991;32:138–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- 33.Whiting AS, Bauer AM, Sites JW., Jr Phylogenetic relationships and limb loss in sub-Saharan African scincine lizards (Squamata: Scinicidae) Mol Phylogenet Evol. 2003;29:582–598. doi: 10.1016/s1055-7903(03)00142-8. [DOI] [PubMed] [Google Scholar]
- 34.Tiemann-Boege I, Kilpatrick CW, Schmidly DJ, Bradley RD. Molecular phylogenetics of the Peromyscus boylii species group (Rodentia: Muridae) based on mitochondrial cytochrome b sequences. Mol Phylogenet Evol. 2000;16:366–378. doi: 10.1006/mpev.2000.0806. [DOI] [PubMed] [Google Scholar]
- 35.Peppers LL, Bradley RD. Cryptic speciation in Sigmodon hispidus: evidence from DNA sequences. J Mammal. 2000;81:332–343. [Google Scholar]
- 36.Peppers LL, Carroll DS, Bradley RD. Molecular systematics of the genus Sigmodon (Rodentia: Muridae): evidence from the mitochondrial cytochrome b gene. J Mammal. 2002;83:396–407. [Google Scholar]
- 37.Myers P, Lundrigan B, Tucker PK. Molecular phylogenetics of oryzomyine rodents: the genus Oligoryzomys. Mol Phylogenet Evol. 1995;4:372–382. doi: 10.1006/mpev.1995.1035. [DOI] [PubMed] [Google Scholar]
- 38.Bonivicino CR, Moreira MAM. Molecular phylogeny of the genus Oryzomys (Rodentia: Sigmodontinae) based on cytochrome b DNA sequences. Mol Phylogenet Evol. 2001;18:282–292. doi: 10.1006/mpev.2000.0878. [DOI] [PubMed] [Google Scholar]
- 39.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 40.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 41.Ravkov EV, Rollin PE, Ksiazek TG, Peters CJ, Nichol ST. Genetic and serologic analysis of Black Creek Canal virus and its association with human disease and Sigmodon hispidus infection. Virology. 1995;210:482–489. doi: 10.1006/viro.1995.1366. [DOI] [PubMed] [Google Scholar]
- 42.Bohlman MC, Morzunov SP, Meissner J, Taylor MB, Ishibashi K, Rowe J, Levis S, Enria D, St Jeor SC. Analysis of hantavirus genetic diversity in Argentina: S segment-derived phylogeny. J Virol. 2002;76:3765–3773. doi: 10.1128/JVI.76.8.3765-3773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Padula P, Figueroa R, Navarrete M, Pizarro E, Cadiz R, Bellomo C, Jofre C, Zaror L, Rodríguez E, Murúa R. Transmission study of Andes hantavirus infection in wild sigmodontine rodents. J Virol. 2004;78:11972–11979. doi: 10.1128/JVI.78.21.11972-11979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reid FA. A Field Guide to the Mammals of Central America and Southeast Mexico. Oxford University Press; New York: 1997. p. 334. [Google Scholar]
- 45.Schmidly DJ. Mammals of Texas. Sixth Edition University of Texas Press; Austin, TX: 2004. p. 501. [Google Scholar]
- 46.Schmidt CA, Engstrom MD. Genic variation and systematics of rice rats (Oryzomys palustris species group) in southern Texas and northeastern Tamaulipas, Mexico. J Mammal. 1994;75:914–928. [Google Scholar]
- 47.Wilson DE, Reeder DM. Mammal Species of the World. A Taxonomic and Geographic Reference. Third Edition Johns Hopkins University Press; Baltimore, MD: 2005. p. 2142. [Google Scholar]
- 48.Benson DL, Gehlbach FR. Ecological and taxonomic notes on the rice rat (Oryzomys couesi) in Texas. J Mammal. 1979;60:225–228. [Google Scholar]
- 49.Childs JE, Ksiazek TG, Spiropoulou CF, Krebs JW, Morzunov S, Maupin GO, Gage KL, Rollin PE, Sarisky J, Enscore RE. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J Infect Dis. 1994;169:1271–1280. doi: 10.1093/infdis/169.6.1271. [DOI] [PubMed] [Google Scholar]
- 50.Morzunov SP, Rowe JE, Ksiazek TG, Peters CJ, St Jeor SC, Nichol ST. Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America. J Virol. 1998;72:57–64. doi: 10.1128/jvi.72.1.57-64.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song JW, Baek LJ, Gajdusek DC, Yanagihara R, Gavrilovskaya L, Luft BJ, Mackow ER, Hjelle B. Isolation of pathogenic hantavirus from white-footed mouse (Peromyscus leucopus) Lancet. 1994;344:1637. doi: 10.1016/s0140-6736(94)90430-8. [DOI] [PubMed] [Google Scholar]
- 52.Armien B, Pascale JM, Bayard V, Munoz C, Mosca I, Guerrero G, Armien A, Quiroz E, Castillo Z, Zaldivar Y, Gracia F, Hjelle B, Koster F. High seroprevalence of hantavirus infection on the Azuero peninsula of Panama. Am J Trop Med Hyg. 2004;70:682–687. [PubMed] [Google Scholar]


