Abstract
In eukaryotes, the entwined pathways of RNA transport and local translational regulation are key determinants in the spatio-temporal articulation of gene expression. One of the main advantages of this mechanism over transcriptional control in the nucleus lies in the fact that it endows local sites with independent decision-making authority, a consideration that is of particular relevance in cells with complex cellular architecture such as neurons. Localized RNAs typically contain codes, expressed within cis-acting elements, that specify subcellular targeting. Such codes are recognized by trans-acting factors, adaptors that mediate translocation along cytoskeletal elements by molecular motors. Most transported mRNAs are assumed translationally dormant while en route. In some cell types, especially in neurons, it is considered crucial that translation remains repressed after arrival at the destination site (e.g., a postsynaptic microdomain) until an appropriate activation signal is received. Several candidate mechanisms have been suggested to participate in the local implementation of translational repression and activation, and such mechanisms may target translation at the level of initiation and/or elongation. Recent data indicate that untranslated RNAs may play important roles in the local control of translation.
Keywords: cis-acting targeting element, trans-acting factor, molecular motor, activity-dependent translation
INTRODUCTION
It has been 40 years since publication of the first reports of RNA localization in eukaryotic cells (Bodian 1965, Koenig 1965a,b). Curiously, these early data were initially given little attention. Even after evidence for localized RNAs and ribosomes had emerged from several laboratories in the early 1980s (Colman et al. 1982, Jeffery et al. 1983, Palacios-Prü et al. 1981, Steward & Levy 1982), RNA localization continued to be considered something of an obscure oddity in cell biology. The notion was greeted with skepticism.
- Translational control
the regulation of gene expression at the level of translation
- Untranslated RNA
an RNA that, in contrast to an mRNA, does not encode a polypetide sequence. It typically carries codes to specify functionality, e.g., catalytic action or subcellular targeting
In the late 1980s and early 1990s, however, it became increasingly clear that protein synthetic machinery and individual RNAs can be targeted to distant sites, sometimes at considerable distances from perikaryal somatic regions. Because evidence supporting this notion was emerging from diverse cell types including Xenopus oocytes, mammalian neurons, and glial cells, and from Drosophila embryos, among others, researchers began to realize that a common principle might be at work. Today we understand that mRNAs are transported, localized, and locally translated in many eukaryotic cell types. These mechanisms are of fundamental importance in the regulation of gene expression as they allow cells to delegate control to autonomously acting local sites. In the past several years we have witnessed dynamic progress in our understanding of mechanisms that govern RNA localization and local translational control, mechanisms that underlie the spatio-temporal modulation of local protein repertoires. Hand-in-hand with this development has come the realization that small untranslated RNAs, many hitherto unappreciated (Couzin 2002), play important roles as mediators of translational control.
Why do cells localize RNAs? RNA transport and local translation have now been documented in vertebrates, invertebrates, and unicellular organisms, and it therefore appears that RNA localization is an ancient cellular mechanism. Today this mechanism is used by diverse cell types for various purposes, but one of the main benefits that cells—especially complex and highly polarized cells—derive from it is the ability to control gene expression locally. This consideration is particularly relevant in neurons where the plastic modulation of synaptic connections forms the cellular substratum for higher brain functions such as learning and memory. Long-term, experience-dependent, and input-specific synaptic remodeling requires changes of gene expression and de novo protein synthesis; given the fact that a typical neuron makes several thousand synaptic connections on its dendritic arbor, the advantages of local translational control are obvious.
The dynamic expansion of the field in recent years dictates that we focus on selected model systems. Our main emphasis here is on mammalian cells, but we refer to non-mammalian cells whenever pertinent. For the same reason, we restrict this review to literature published within the last four years. The reader is referred to a number of excellent review articles that cover earlier periods or subject areas that are outside the scope of this review. Several of these articles have appeared in the Annual Reviews series (Chartrand et al. 2001, Goldstein & Yang 2000, Kapp & Lorsch 2004, Palacios & St. Johnston 2001, Piper & Holt 2004, Steward & Schuman 2001). Piper & Holt (2004) provide a current account of local translation in axons, a topic not covered here. Other relevant reviews have appeared elsewhere (Bassell & Kelic 2004, Carson et al. 2001, Darnell 2003, Giuditta et al. 2002, Job & Eberwine 2001, Kloc & Etkin 2005, Kloc et al. 2002, Martin 2004, Smith 2004, Steward & Schuman 2003, Wang & Tiedge 2004).
RNA TRANSPORT AND LOCALIZATION
RNA localization is a widespread phenomenon that has been observed in many eukaryotic cell types, including unicellular organisms (e.g., yeast), developing germ cells and embryos (e.g., in Drosophila and Xenopus), and plant and other animal cells (e.g., mammalian neurons). Active targeting involves recognition of cis-acting RNA elements, i.e., RNA segments that contain codes to specify targeting, by trans-acting RNA-binding factors. This interaction results in the formation of ribonucleotide protein (RNP) complexes, which travel along cytoskeletal filaments with the help of motor proteins. At their destination sites, delivered transcripts are anchored and are now ready for translation.
Cis-Acting Targeting Elements
Location-coding, cis-acting targeting elements have been identified in various localized transcripts of higher eukaryotes (Table 1). In Xenopus oocytes, more than 20 RNAs are transported via three different pathways to either the vegetal pole or the animal hemisphere (King et al. 1999). Vegetal localization occurs during an early and a late phase of oogenesis. RNAs of the early METRO (messenger transport organizer) pathway (Kloc & Etkin 2005), such as Xcat-2, Xdaz1, Xpat, Xwnt-11, and Xlsirts, initially associate with the mitochondrial cloud (stages I–II) and subsequently migrate to the vegetal cortex between late stage II and early stage III. Transcripts of the late pathways, including Vg1 (Melton 1987) and VegT (Zhang & King 1996), are transported during late stage III and early stage IV and remain anchored to the vegetal cortex until the end of oogenesis. This pathway localizes germ layer determinants to the vegetal hemisphere. In Vg1 mRNAs, a 340-nucleotide (nt) motif, termed Vg1 localization element (VLE), mediates vegetal targeting (Mowry & Melton 1992). The VLE and the cis-elements of virtually all other vegetally localized transcripts contain clusters of short CAC-containing motifs (Betley et al. 2002). Slight sequence variations between these motifs appear to correlate with distinct functions during the localization process (Zhou & King 2004). However, Xvelo1 transcripts of the late vegetal pathway use a 75-nt stem-loop localization element (Claussen & Pieler 2004). Thus targeting of different mRNAs to identical cellular sites may involve diverse as well as overlapping molecular mechanisms.
Table 1.
cis-acting elements and trans-acting factors in cell types of vertebrate organisms. cis-acting elements have often been only broadly defined. Position range and length therefore represent maxima that in many cases are likely to be further narrowed down in the future
| Species, cell type, RNA |
Position | Name, length (nucleotides) |
trans-acting factor | References |
|---|---|---|---|---|
| Xenopus oocytes | ||||
| Vg1 | 3′ UTR | VLE, 340 | Vg1RBP/VERA, Prrp, VgRBP78, VgRBP69, VgRBP60, VgRBP40, VgRBP36, and VgRBP33 |
(Deshler et al. 1997, Mowry & Cote 1999, Mowry & Melton 1992, Schwartz et al. 1992, Zhao et al. 2001) |
| VegT | ?, ? | ? | (Betley et al. 2002) | |
| Xvelo1 | LE, 75 | UV-crosslinking pattern similar to VLE |
(Claussen & Pieler 2004) | |
| Chicken fibroblasts | ||||
| β-actin | 3′ UTR | zipcode, 54 | ZBP1, ZBP2 | (Kislauskis et al. 1994, Ross et al. 1997) |
| Chicken neurons (dendrites) | ||||
| β-actin | 3′ UTR | zipcode, 54 | ZBP1 | (Zhang et al. 2001) |
| Mammalian oligodendrocytes | ||||
| MBP | 3′ UTR | A2RE, 11 | hnRNP A2 | (Ainger et al. 1997, Hoek et al. 1998, Munro et al. 1999, Shan et al. 2003) |
| Mammalian neurons (dendrites) | ||||
| β-actin | 3′ UTR | zipcode, 54 | ZBP1 | (Eom et al. 2003, Tiruchinapalli et al. 2003) |
| BC1 | 5′ domain | BC1 DTE, 65 | ? | (Muslimov et al. 1997) |
| CaMKIIα, ligatin | ORF, 3′ UTR | Y element, ∼14 | translin | (Severt et al. 1999) |
| CaMKIIα, neurogranin | 3′ UTR | CNDLE, 28–30 | ? | (Mori et al. 2000) |
| CaMKIIα | 3′ UTR | CaMKIIα DTE, ∼1200 | ? | (Blichenberg et al. 2001) |
| CaMKIIα | 3′ UTR | CPE, 6 | CPEB | (Huang et al. 2003) |
| GluR2 | 3′ UTR | ? | ? | (Ju et al. 2004) |
| MAP2 | 3′ UTR | MAP2 DTE, 640 | MARTA1, MARTA2 | (Blichenberg et al. 1999; Rehbein et al. 2000, 2002) |
| MAP2 | 3′ UTR | CPE, 6 | CPEB | (Huang et al. 2003) |
| MAP2 | ORF | A2RE, 11 | hnRNP A2 | (Shan et al. 2003) |
| PKMζ | 5′ UTR, ORF | Mζ DTE1, 499 | ? | (Muslimov et al. 2004) |
| PKMζ | 3′ UTR | Mζ DTE2, 42 | ? | (Muslimov et al. 2004) |
| Vasopressin | ORF, 3′ UTR | DLS, 395 | PABP | (Mohr et al. 2002, Mohr & Richter 2001) |
| Mammalian neurons (axon hillock) | ||||
| tau | 3′ UTR | fragment H, 240 | HuD | (Aranda-Abreu et al. 1999) |
Numerous RNAs are targeted to dendrites in mammalian neurons (Eberwine et al. 2002). The dendritic delivery of some of these mRNAs is dependent on neuronal activity. Synaptic activation in vivo strongly upregulates Arc/arg3.1 gene expression (Link et al. 1995, Lyford et al. 1995) and results in the selective, N-methyl-d-aspartate (NMDA) receptor (NMDAR)–dependent recruitment of corresponding transcripts to dendritic segments in which synapses had been stimulated (Steward & Worley 2001). Similarly, epileptogenic stimuli in vivo produce increased brain-derived neurotrophic factor (BDNF) mRNA levels, coupled with an NMDAR-dependent transcript accumulation in proximal dendritic segments in hippocampus (Tongiorgi et al. 2004). A common requirement in these pathways would be retrograde synapse-to-nucleus signaling, and mechanisms underlying such signaling are now beginning to be addressed (Thompson et al. 2004).
Dendritic targeting elements (DTEs) have been identified in a number of neuronal mRNAs. Such DTEs appear to be quite diverse in length and sequence, and they may also encode different destination sites within dendritic arborizations. Protein repertoires in dendritic domains are typically mosaic and diverse. Synapses, in particular, feature highly specialized complements of macromolecular components. It has now become apparent that part of this complexity is implemented and maintained by on-site translation of locally available mRNAs. The complexity of neuronal DTEs may therefore be a reflection of the diversity of intracellular target sites, as is illustrated by the following examples.
- Cis-acting element
a segment within an RNA that contains a code to specify functionality (e.g., subcellular targeting)
- Dendritic targeting element (DTE)
a cis-acting element that directs a neuronal RNA to or along dendrites
- nt
nucleotide
- RNP
ribonucleoprotein
- Targeting code
information contained within an RNA, expressed by an RNA motif within a cis-acting element, that specifies subcellular targeting
Calcium/calmodulin-dependent protein kinase II (CaMKII) is enriched in postsynaptic microdomains. The 3′ untranslated region (UTR) of CaMKIIα mRNA has been reported to contain two distinct nonoverlapping DTEs, one of about 30 nt (Mori et al. 2000), the other one located within about 1200 nt (Blichenberg et al. 2001). In mouse brain in vivo, however, the smaller element alone is not sufficient to mediate dendritic targeting (Miller et al. 2002). In addition, the 3′ UTR of CaMKIIα mRNA contains two copies of a hexanucleotide motif called cytoplasmic polyadenylation element (CPE), which facilitate dendritic translocation (Huang et al. 2003).
In shank1 transcripts, a DTE is contained within 200 nt of the 3′ UTR (Böckers et al. 2004), whereas in vasopressin transcripts, a DTE resides within a 395-nt segment that spans part of the open reading frame (ORF) and the 3′ UTR (Prakash et al. 1997). In protein kinase Mζ (PKMζ) mRNA, one DTE is positioned within 499 nt at the interface of the 5′ UTR and the ORF (499 nt) and mediates somatic export, whereas a second one, a stem-loop structure of 42 nt in the 3′ UTR, is responsible for distal dendritic mRNA targeting (Muslimov et al. 2004). The element encoding distal targeting features a GA kink-turn (K-turn) motif. K-turns have been identified as sites for RNA-protein interactions in various RNAs (Klein et al. 2001), and the question is thus raised whether such or similar motifs play a more widespread role in RNA localization. In small untranslated BC1 RNA, a 65-nt segment in the 5′ stem-loop domain is sufficient for dendritic targeting (Muslimov et al. 1997). (Because all localized RNAs, regardless of whether they are translated, carry codes to specify their destination sites, the term noncoding should be avoided; see Brosius & Tiedge 2004.) In various other dendritic mRNAs, DTEs have not yet been mapped or identified. Taken together, however, dendritic targeting signals of different mRNAs appear to be quite diverse, differing in length, sequence, relative position, and number per RNA, and encoding differential destination sites. Therefore, one is left to wonder whether the apparent complexity of DTEs is indeed dictated solely by biological necessity or if underlying principles have been slow to emerge.
Mammalian oligodendrocytes are highly elongated glial cells that extend extensively branched processes to form insulating myelin sheaths around axons. Several mRNAs, including the one encoding myelin basic protein (MBP), are transported into these processes (Barbarese et al. 1999, Boccaccio 2000). Targeting of MBP mRNA is mediated by an 11-nt cis-acting element, referred to as the heterogeneous ribonucleoprotein (hnRNP) A2 response element (A2RE), that has also been implicated in RNP granule formation and cap-dependent translation (Carson et al. 2001). A2REs in three different transcripts of the human immunodeficiency virus 1 (HIV-1) also mediate extrasomatic targeting of microinjected mRNAs in cultured oligodendrocytes, albeit with distinct efficiencies (Mouland et al. 2001). Flanking sequences thus seem to modulate the targeting competence of A2RE. A2RE has also been shown to support dendritic targeting in neurons (Shan et al. 2003, Smith 2004). An A2RE-like element was identified in the ORF of dendritic MAP2 mRNAs; in this case, however, a DTE is located in the 3′ UTR, whereas the ORF was found targeting incompetent (Blichenberg et al. 1999).
Actin polymerization at the leading edge of moving cells regulates the extension of lamellipodia, filopodia, or pseudopodia (Rafelski & Theriot 2004). In chicken embryonic fibroblasts and myoblasts, β-actin transcripts are concentrated just proximal to the leading edge (Lawrence & Singer 1986). A 54-nt “zipcode” in the 3′ UTR mediates mRNA sorting to the leading edge (Kislauskis et al. 1994). The same zipcode also mediates targeting of β-actin messages into both growth cones of immature chicken neurons (Zhang et al. 2001) and dendrites of mature rat hippocampal neurons (Eom et al. 2003). Thus the functional role of the β-actin cis-acting element appears to be conserved in avian and mammalian species. On the other hand, restriction of avian MAP2 mRNA to neuronal somata in the chicken retina (in contrast to a somatodendritic distribution in the mammalian retina) and the absence of a DTE-like sequence in the 3′ UTR of the avian mRNA suggest that acquisition of dendritic targeting competence has been a relatively recent event in evolution (Cristofanilli et al. 2004). Taken together, the currently available data indicate that localized mRNAs use both shared and unique cis-acting elements that may have been acquired at distinct time points during evolution. Long cis-acting elements often appear to consist of multiple short motifs, sometimes cooperating, sometimes partially redundant. The physical description of such motifs will be an important future challenge.
- CPE
cytoplasmic polyadenylation element
- ORF
open reading frame
- RNA motif
a three-dimensional structural design that is contained, in identical or similar form, in a number of RNAs. Such motifs may represent codes that are recognized by trans-acting factors
- UTR
untranslated region
- KH domain
ribonucleoprotein K homology domain
- RRM
RNA recognition motif
- Trans-acting factor (TAF)
a factor, typically a protein, that mediates RNA functionality (e.g., subcellular targeting) by recognizing a code within a cis-acting element
Trans-Acting Factors
By definition, trans-acting factors (TAFs) are proteins that support RNA transport and/or localization by binding to cis-acting elements (Figure 1). In this context, TAFs are decoding devices, proteins that recognize location-specifying codes in RNAs.
Figure 1.
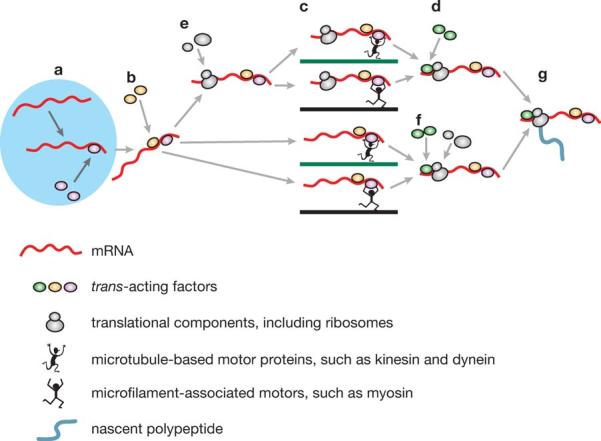
Active transport of mRNAs from the nucleus (left, light blue)to peripheral cytoplasmic destination sites in eukaryotic cells. Transport proceeds through a number of sequential phases as follows: (a) Recognition of cis-acting elements by nuclear trans-acting factors (TAFs) (purple ellipses) and formation of ribonucleotide protein (RNP) complexes; (b) nuclear export of RNP particles and recruitment of cytoplasmic TAFs ( yellow ellipses); (c) motor-based active transport of cytoplasmic RNP particles along cytoskeletal filaments (green and blue lines); (d, f ) association with additional cytoplasmic TAFs, such as anchoring proteins, at destination sites (light green ellipses); (e, f ) recruitment of ribosomes and other translational components (not shown) either before (e)or after ( f ) cytoplasmic translocation; ( g) locally controlled translation.
Vg1RBP/VERA and ZBP1, TAFs that interact with the Vg1-VLE and the β-actin cis-acting element, respectively, represent two members of a family of RNA-binding proteins that contain two RNA recognition motifs (RRMs) and four hnRNP K homology (KH) domains (Bassell & Kelic 2004, Yaniv & Yisraeli 2002). Zipcode mutations reduce ZBP1 association with β-actin mRNA and transcript localization to the leading edge of fibroblasts (Ross et al. 1997). In developing neurons, disruption of the interaction between the zipcode and ZBP1 inhibits neurotrophin-induced β-actin mRNA localization into growth cones and reduces growth cone motility (Zhang et al. 1999). In mature neurons, ZBP1 and β-actin mRNA granules reside in dendritic shafts and spines (Tiruchinapalli et al. 2003). Reduced ZBP1 levels diminish dendritic β-actin mRNA concentrations and impair the growth of dendritic filopodia after neurotrophin stimulation, whereas overexpression of zipcode-containing mRNAs increases the density of dendritic filopodia (Eom et al. 2003). Members of the Vg1RBP/ZBP1 family of RNA-binding proteins are thus mediators of cytoplasmic mRNA targeting in different species and cell systems.
Two TAFs that interact with dendritic MAP2 mRNA, MARTA1 and MARTA2, belong to the FUSE-binding protein (FBP) sub-family of KH-domain proteins (Duncan et al. 1994, Rehbein et al. 2002; K. Zivraj, M. Rehbein, F. Buck, M. Schweizer, D. Richter, S. Kindler, manuscript in preparation). MARTA1 is the rodent ortholog of human KH-type splicing regulatory protein (KSRP/FBP2), a component of a multiprotein complex that has been implicated in neuron-specific splicing (Min et al. 1997). ZBP2, the chicken ortholog of MARTA1, interacts with the zipcode of β-actin mRNA (Gu et al. 2002). Located predominantly in the nucleus, ZBP2 undergoes nucleo-cytoplasmic shuttling, and overexpression of truncated ZBP2 partially disrupts β-actin mRNA targeting to both lamellipodia of polarized chicken fibroblasts and growth cones of neurons in culture.
In contrast to MARTA1/ZBP2, MARTA2/FBP3 predominantly resides in the somatodendritic compartment of neurons where it was found associated with polysomes. When expressed in cultured neurons, exogenous MARTA2 partially colocalizes with recombinant DTE-containing mRNAs in granules along dendrites. Overexpression of a truncated MARTA2 version completely disrupts dendritic targeting of endogenous MAP2 mRNA granules. Thus two members of the FBP family appear to participate in RNA localization in different cell types.
Testis/brain RNA-binding protein (TB-RBP; known as translin in primates) binds to conserved Y and H elements of several testis and brain mRNAs and represses their translation (Han et al. 1995, Kwon & Hecht 1993). TB-RBP attaches translationally repressed mRNAs to the microtubule cytoskeleton (Han et al. 1995). Its presence in intercellular cytoplasmic bridges between spermatids suggests that TB-RBP may mediate mRNA transport between male germ cells (Morales et al. 1998). These data indicate an interplay between RNA transport and translational repression.
Other proteins may also play dual roles in RNA targeting and translational control. One example is provided by the Staufen family of double-stranded (ds) RNA-binding proteins (Roegiers & Jan 2000). In Drosophila oocytes and early embryos, Staufen is essential for localization and translational control of different maternal transcripts and for the establishment of the antero-posterior body pattern. Two Staufen orthologs, Stau1 and Stau2, have been described in mammals (Duchaine et al. 2002, Monshausen et al. 2001, Tang et al. 2001). Stau1 is found in most tissues, whereas Stau2 is preferentially present in brain (Duchaine et al. 2002, Monshausen et al. 2001). In neurons, both proteins are located in dendrites, associate with dendritic microtubules, and reside in granules (Duchaine et al. 2002, Monshausen et al. 2001, Tang et al. 2001). At the same time, enrichment in polysome fractions may suggest an additional role in translation (see below). Similarly, the cytoplasmic polyadenylation element (CPE), contained in a subset of dendritic mRNAs, performs the dual functions as a translational control element (TCE) and as a DTE. Its binding protein, CPEB, mediates both cytoplasmic polyadenylation-induced translation (see below) and transport of CPE-containing mRNAs to dendrites (Huang et al. 2003). In summary, it appears that localized RNAs may recruit binding proteins for the dual purpose of RNA transport and local control of translation, underscoring once again that the two mechanisms are functionally interdependent.
- Translational control element (TCE)
a segment within an RNA that contains a code to instruct regulation of its translation
Cytoplasmic mRNA targeting may already be initiated in the nucleus, as newly accumulating evidence from various cell types has been suggesting. Several TAFs are nucleocytoplasmic shuttling proteins that first appear to associate with transcript in the nucleus and subsequently direct mRNA targeting in the cytoplasm (Farina & Singer 2002). In Drosophila oocytes, the nuclear processing history of oskar transcripts is a determinant of their cytoplasmic fate (Hachet & Ephrussi 2004). Similarly, transcript recognition and formation of a specific RNP complex in the nucleus is an early event in Vg1 mRNA localization in Xenopus oocytes (Kress et al. 2004). The RNP is remodeled after its export to the cytoplasm, and additional transport factors are recruited into the complex. Several other TAFs also seem to shuttle between the nucleus and the cytoplasm (Bassell & Kelic 2004). The notion thus emerges that RNA targeting in eukaryotic cells is often a multi-step process in which an RNA, from the time of its transcription in the nucleus to the anchoring at its final destination site, enlists a set of TAFs in a sequentially orchestrated manner. Relay-type targeting, with multiple TAFs cooperating in the delivery of the cargo, may be advantageous especially when long and complex transport routes have to be negotiated, e.g., in dendrites with thousands of potential synaptic destination sites.
Cytoskeletal Elements and Molecular Motors
Different cytoskeletal elements have been shown to support RNA transport and local anchoring (Figure 1). In fibroblasts, microfilaments are used to localize particles containing β-actin transcripts and ZBP1. In neurons, on the other hand, ZBP1 and its β-actin mRNA target seem to move predominantly along microtubules (Bassell & Kelic 2004). Thus ZBP1 may serve as an adaptor between mRNA and either microfilament- or microtubule-based molecular motors. It is plausible that in neurons, long-range RNA transport is mediated by microtubules, whereas localization in the destination microdomain is supported by actin filaments (Muslimov et al. 2002). Neuronal CPEB granules contain both kinesin and dynein motors, and their bidirectional movement in dendrites is microtubule dependent (Huang et al. 2003). In addition, kinesin 1 complexes from mammalian brain were shown to contain several of the aforementioned TAFs (Kanai et al. 2004). In the axon-like processes of P19 embryonic carcinoma cells, knock-down of another kinesin family member, KIF3A, impairs the sorting of tau mRNA (Aronov et al. 2002). In oligodendrocytes, inhibition of kinesin disrupts targeting of MBP mRNAs (Carson et al. 1997); in testis, kinesin KIF17b associates with TB-RBP, suggesting that a microtubule-dependent RNA transport system operates in mammalian male germ cells (Chennathukuzhi et al. 2003). Evidence is thus accumulating that long-range RNA transport in various mammalian cell types is mediated by microtubule-based kinesin- and dynein-type molecular motors.
LOCAL CONTROL OF TRANSLATION
In this section, we discuss mechanisms of local regulation of translation, using selected examples to highlight common principles. We probe the significance of translational repression en route and at destination sites. A brief synopsis of the translational pathway in eukaryotes serves as an introduction.
Mechanisms of Local Translational Control
The translation of an mRNA into a cognate protein proceeds in the three sequential steps of initiation, elongation, and termination (reviewed in Sonenberg et al. 2000). Regulation can occur at any of these steps, but initiation is typically rate-limiting and thus often a target for regulation (Gingras et al. 1999, Kapp & Lorsch 2004).
During initiation, a 43S preinitiation complex is formed by the binding of an eIF2•GTP•Met-tRNAi ternary complex to the 40S ribosomal subunit (Figure 2). The 43S complex is then recruited to the initiator codon of the mRNA to form a stable 48S complex. This step requires participation of a set of factors from the eIF4 family. These factors subsequently dissociate, the 60S ribosomal subunit joins the 40S subunit to form an 80S complex, and elongation ensues (Figure 2; Hershey & Merrick 2000). During elongation, eEF1A guides aminoacyl-tRNAs to the A site on the ribosome; following peptide bond formation, the ribosome is translocated by one codon along the mRNA, a step that is catalyzed by eEF2. Finally, termination at a stop codon is mediated by a set of release factors.
Figure 2.
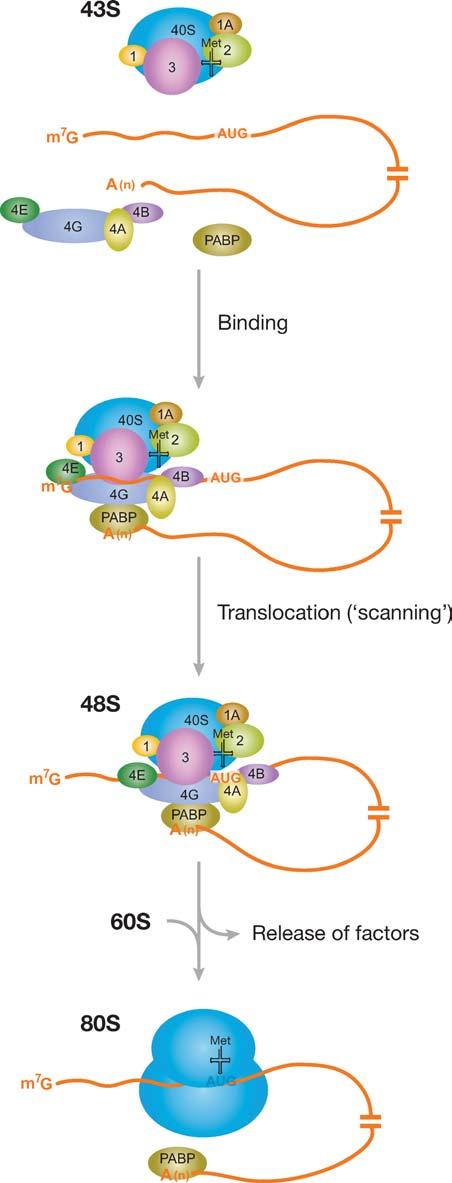
Cap-dependent translation initiation in eukaryotes. Recruitment of the 43S preinitiation complex to the mRNA, mediated by factors of the eIF4 family and poly(A) binding protein (PABP), results in the formation of a 48S complex at the AUG initiator codon. Alternatively to the scenario shown here, eIF4E may remain bound to the cap during scanning (resulting in a looping-out of the 5′ UTR; Jackson 2000); in general, it remains to be established at which point after initial cap-binding the eIF4 factors dissociate from the mRNA and from PABP. Translation may also be initiated in cap-independent fashion, for instance by binding of the 43S complex to an internal ribosome entry site (IRES), which is typically located directly upstream of the AUG start codon (not shown; see Hellen & Sarnow 2001). Some factors (e.g., eIF5) have been omitted for clarity.
Because of its relevance for the long-term, experience-dependent modulation of synaptic strength (and thus for neuronal plasticity), translational control of gene expression in synapto-dendritic domains has increasingly become a subject of general interest. Therefore, we use this system as a case in point to exemplify some of the emerging general principles of local translational control (Figure 3).
Figure 3.
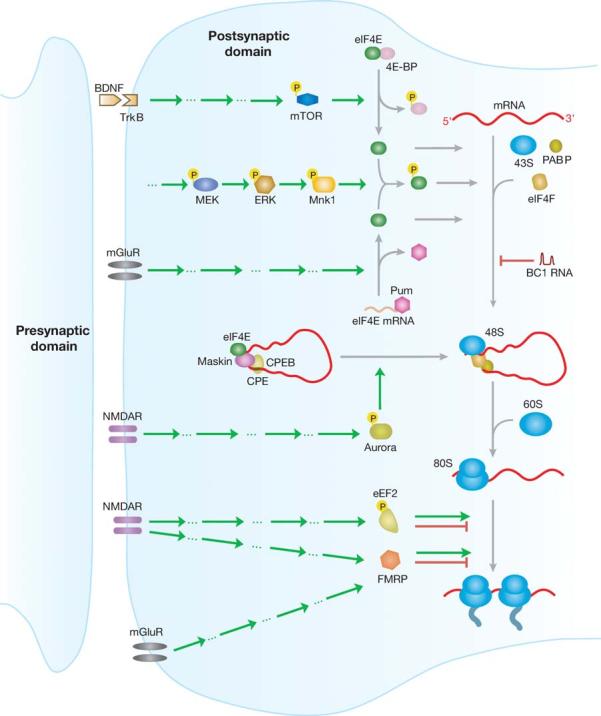
Many roads lead to Rome: possible pathways of translational control at the synapse. Translational control can be implemented by activation of various receptor systems (left) and can be mediated at the level of initiation and elongation (right). Mechanisms in addition to those summarized here have been discussed; for example, FMRP has also been reported to repress translation at the level of initiation (Laggerbauer et al. 2001). Green arrows, stimulation; red arrows, inhibition. Aspects of this diagram remain speculative; note that not all factors shown are currently known to be active at the synapse. Factors not drawn to scale.
Translation initiation factor eIF4E, a subunit of the eIF4F complex, binds to the 5′ cap of mRNAs and promotes recruitment of the 43S preinitiation complex (Figure 2; Gingras et al. 1999, Kapp & Lorsch 2004). Several signaling pathways have been reported in neurons that modulate translation by targeting eIF4E. One involves BDNF and the mammalian target of rapamycin (mTOR), a serine/threonine kinase. Activated mTOR phosphorylates eIF4E-binding proteins (eIF4EBPs), resulting in their dissociation from eIF4E and consequently in the activation of translation initiation (Raught et al. 2000). This pathway has been implicated in translational regulation in mammalian neurons (Takei et al. 2001, 2004; Tang et al. 2002). Key components of the mTOR pathway, such as mTOR, eIF4E, and eIF4E-BP, have been identified in postsynaptic domains (Tang et al. 2002). BDNF, a member of the neurotrophin family, has been shown to be involved in long-term synaptic potentiation (Korte et al. 1995). Application of rapamycin, an inhibitor of the mTOR pathway, to hippocampal slices prevents BDNF-induced synaptic potentiation (Tang et al. 2002). BDNF induces phosphorylation of eIF4E-BP, eIF4E, and mTOR in cortical neurons, as well as in isolated dendrites (Takei et al. 2001, 2004), indicating that the mTOR pathway is essential for BDNF to activate translation in dendrites. A subset of mRNAs has been identified as targets of the BDNF-activated mTOR pathway (Schratt et al. 2004).
- Eukaryotic initiation factor (eIF)
a factor that promotes translation initiation, i.e., the sequence of events that results in the formation of an 80S ribosomal complex at the AUG initiator codon
The mitogen-activated protein kinase (MAPK) signaling pathway has also been reported to activate local translation through its downstream target eIF4E (Kelleher et al. 2004). Upon phosphorylation by MAPK/ERK kinase (MEK), extracellular signal-regulated kinase (ERK), a member of the MAPK family, activates its downstream substrate MAP kinase-interacting kinase 1 (Mnk1) (Fukunaga & Hunter 1997, Waskiewicz et al. 1997). Activated Mnk subsequently phosphorylates eIF4E, thus promoting translation initiation (Pyronnet et al. 1999, Waskiewicz et al. 1999). These combined findings suggest a coregulation of local translation by targeting eIF4E through the MEK and mTOR signaling pathways (Gingras et al. 1999; Figure 3).
Work with Drosophila has recently indicated that expression of eIF4E itself is subject to regulation. Pumilio (Pum), a translational repressor, plays an important role in determining the Drosophila anterior-posterior body pattern during early embryogenesis (Johnstone & Lasko 2001). Pum has been found to control dendritic morphogenesis (Ye et al. 2004) and to be involved in long-term memory formation (Dubnau et al. 2003). At the Drosophila neuromuscular junction, post-synaptic Pum downregulates eIF4E expression (Menon et al. 2004). Pum selectively interacts with the 3′ UTR of eIF4E mRNA, indicating that Pum may modulate synaptic function through direct control of eIF4E expression (Menon et al. 2004).
eIF4F, the heterotrimeric protein that mediates recruitment of the 43S complex to the mRNA, is composed of eIF4E, the cap-binding protein; eIF4G, a large scaffolding protein; and eIF4A, an ATP-dependent RNA helicase that unwinds secondary structure elements in the 5′ UTR prior to recruitment of the 43S complex (Figure 2; Gingras et al. 1999, Kapp & Lorsch 2004). The activity of eIF4F is significantly enhanced by its interaction with PABP, a protein that binds to the poly(A) tails of mRNAs. This interaction has been shown to be the target of dendritic BC1 RNA (Wang et al. 2002). BC1 RNA is a small, untranslated RNA that is selectively transported to dendrites (Muslimov et al. 1997). It represses translation initiation by inhibiting formation of the 48S preinitiation complex (Wang et al. 2002). BC1-mediated repression is effective in cap-dependent initiation and in one subtype of IRES-mediated initiation. Because IRES-mediated initiation may be the preferred mode at the synapse, particularly in response to stimulation (Dyer et al. 2003, Pinkstaff et al. 2001), BC1 RNA is well positioned for a modulatory role in synaptic translation.
Also targeting translation initiation, the CPE pathway has first been analyzed in Xenopus oocytes. Some mRNAs remain translationally dormant until their short poly(A) tails are extended in the cytoplasm (Mendez & Richter 2001). Cytoplasmic polyadenylation is dependent on the presence of a CPE in the 3′ UTR of the mRNA. Upon phosphorylation of CPEB by the kinase aurora, CPEB binds to CPE and recruits poly(A) polymerase to polyadenylate the mRNA. CPEB is in turn recognized by two CPEB-binding proteins, Symplekin, a scaffold protein, and xGLD-2, a poly(A) polymerase (Barnard et al. 2004). Polyadenylation promotes the dissociation of maskin from eIF4E, thus “demasking” eIF4E and allowing it to participate in translation initiation (Richter 2000).
Dendritic CaMKIIα mRNA contains two CPE-like sequences in its 3′ UTR (Wu et al. 1998). Translation of CaMKIIα mRNA depends on the same polyadenylation mechanism that has previously been described in Xenopus oocytes, and factors such as aurora, CPEB, poly(A) polymerase, and maskin have been identified at synaptic sites of hippocampal neurons (Huang et al. 2002, Wells et al. 2001, Wu et al. 1998). NMDAR activation was found essential for CPE-dependent synthesis of CaMKIIα (Wells et al. 2001). NMDAR stimulation induced phosphorylation of aurora, thus activating local translation of CaMKIIα mRNA (Huang et al. 2002). Dendritic plasminogen activator (tPA) mRNA has recently also been shown to interact with CPEB; it was rapidly polyadenylated and translated into cognate protein after metabotropic glutamate receptor (mGluR) activation (Shin et al. 2004). Cytoplasmic polyadenylation thus provides a mechanism of translational control that is selective for CPE-bearing mRNAs.
Elongation may also be a target for local translational control, if not as frequently as initiation. One such target is eEF2 (Scheetz et al. 1997, 2000). In the amphibian tectum, phosphorylation of eEF2 is dependent on the activation of NMDARs and can be induced by visual stimulation (Scheetz et al. 1997). Phosphorylation of eEF2 results in reduced overall protein synthesis in eukaryotes (Nairn et al. 2001) but may enhance translation of some mRNAs in developing neurons (Scheetz et al. 2000). This mechanism may thus provide an alternative to the typically initiation-targeted local translational control (Figure 3).
FMRP has been implicated in the regulation of dendritic translation, but the exact mode of action has not yet been resolved (Antar & Bassell 2003, O'Donnell & Warren 2002). The protein inhibits translation whereas a disease-causing mutant FMRP does not (Laggerbauer et al. 2001). FMRP has been suggested to inhibit translation through interaction with mRNAs (Li et al. 2001). At the synapse, NMDAR, as well as mGluR activation, has been implicated in the FMRP pathway (Antar et al. 2004, Gabel et al. 2004, Weiler et al. 1997). FMRP has also been suggested to interact with untranslated BC1 RNA to repress translation of such mRNAs that base-pair with the 5′ BC1 domain (Zalfa et al. 2003). However, because FMRP associates with polyribosomes (Corbin et al. 1997; Eberhart et al. 1996; Feng et al. 1997; Khandjian et al. 1996, 2004; Stefani et al. 2004; Weiler et al. 2004), whereas BC1 RNA is enriched in the lighter RNP fractions (Krichevsky & Kosik 2001), it appears more likely that FMRP and BC1 RNA operate in translational control without directly interacting with each other. Furthermore, the functional role of FMRP in translation may be more complex in vivo—inhibitory or stimulatory, depending on the target mRNA—as polysome loading was found differentially altered in the absence of FMRP (Brown et al. 2001). The phosphorylation status of FMRP may be another determinant of translational regulation: Whereas nonphosphorylated FMRP was found associated with actively translating polyribosomes, phosphorylated FMRP tended to be associated with apparently stalled polyribosomes (Ceman et al. 2003). In this scenario, FMRP would modulate translation at the level of elongation (Figure 3). Another twist is added by the fact that FMRP binds eEF1A mRNA and represses its translation in vivo (Sung et al. 2003). Another proposed mechanism links FMRP with microRNAs (miRNAs) (Jin et al. 2004). miRNAs are small RNAs (21–24 nt) that function as translation repressors through partial base-paring with their target mRNAs (Ambros 2003, Ruvkun 2001). miRNAs have been identified from mammalian brain preparations where they were found enriched in polyribosome fractions (Kim et al. 2004). dFXR, the Drosophila ortholog of FMRP, has also been suggested to interact in the RNAi pathway (Caudy et al. 2002, Ishizuka et al. 2002), supporting the hypothesis that FMRP regulates translation of target mRNAs through miRNAs and/or small interfering RNAs (siRNAs).
- IRES
internal ribosome entry site
Translational Repression During Transport
It is generally assumed that mRNAs will have to be translationally repressed while en route to prevent ectopic expression (Kwon et al. 1999). This principle has been confirmed in several cell types.
During oogenesis and early embryogenesis, Drosophila oskar mRNA is guided by its 3′ UTR from nurse cells into the anterior margin of the oocytes to be subsequently anchored at the posterior pole (Johnstone & Lasko 2001). During transport, the translation of oskar mRNA is repressed by several trans-acting factors, including Bruno and Apontic proteins (Lie & MacDonald 1999, Webster et al. 1997). oskar mRNA is translated into cognate protein only after it reaches the posterior pole; in fact, ectopic expression of oskar mRNA during transport results in severe developmental defects (Kim-Ha et al. 1995). Support for this model comes from recent work with Cup, a newly identified component of the oskar RNP complex (Wilhelm et al. 2003). Cup, an eIF4E-binding protein, is required for both transport and translational repression of oskar mRNA, suggesting again a close coordination between the two processes (Wilhelm et al. 2003).
A similar coordination is provided by CPEB in neurons. As discussed above, CPEB has been shown to facilitate dendritic transport of CPE-containing mRNAs (Huang et al. 2003). During transport, CPEB is colocalized with the translational repressor maskin in transport particles. An interplay between both proteins may therefore play a role in the transport of translationally dormant mRNAs in dendrites (Huang et al. 2003).
In many cell types, RNP complexes have been identified as the actual motile transport units. These macromolecular complexes appear to contain multiple RNAs, associated TAFs, and/or components of the translational machinery. Work with MBP mRNA showed that transport granules were formed within minutes after microinjection of the RNA into oligodendrocytes (Ainger et al. 1997). Translation of MBP mRNA can be activated only when it arrives within the myelinating processes, thereby avoiding inappropriate localization of MBP (Boccaccio 2000). RNA transport granules have been reported to lack essential translational components and to be unable to incorporate radioactive amino acids, indicating that they are translationally incompetent (Krichevsky & Kosik 2001). The translational repressor FMRP has recently been found to interact with the human ZBP1 ortholog IMP1 in granules (Rackham & Brown 2004). Similarly, the RNA-binding protein RNG105 is a translational repressor that has been found associated with neuronal RNA granules (Shiina et al 2005). These data suggest further functional links between RNA transport and translational repression.
In summary, translational repression seems to be important during RNA transport to prevent ectopic expression, and—at least in neurons—also after arrival at the destination site. Inappropriately controlled protein synthesis at the synapse may upset a delicate neuronal excitation-inhibition balance, and therefore, it may be essential to have multiple systems in place to keep local translation in the “off” state until a valid “go” signal is received. The nature of such a signal, and the mechanism by which it is transduced to the local translational machinery, will be a major challenge for future work.
PERSPECTIVE
Impressive as it may be, recent progress in the field of RNA transport and local translational control can not distract from the fact that we remain ignorant of some very fundamental aspects. Various cis-acting RNA elements have been described that encode subcellular destination sites, but we do not understand how such codes are expressed in the structural design of RNA motifs. We are therefore not yet able to read such codes, and we do not know how they are decoded by TAFs. What is urgently needed is a physical description—in conjunction with a functional dissection—of code-carrying RNA motifs that specify targeting. The number of such motifs may in fact be limited (Moore 1999). In evolutionary terms, RNA has predated proteins, and at least some functional RNA motifs may therefore have evolved early. It will be interesting to see how such motifs have been recruited and adapted by RNAs to encode subcellular destination sites, and how they have evolved to differ from one another in the various aspects of RNA targeting. A physical description of RNA target-encoding motifs will also be essential for us to understand how transport RNPs are assembled around cis-acting elements, how such RNP formation shapes the molecular architecture of transport-competent particles that have been described in various systems, and how these particles are delivered to their final destination sites.
Similarly, although much has been learned about translational control mechanisms, our understanding of how translation is repressed during transport and at the destination site remains rudimentary. It is poorly understood how repression is implemented in transport granules, and how mRNAs are released from granules. When and how are released RNAs activated (i.e., derepressed) for translation, and what are the roles of untranslated RNAs in these mechanisms? These questions are particularly relevant at synaptic sites in neurons where translational repression/activation is assumed to be controlled by the local activity status (Smith et al. 2005).
Activity-dependent local protein synthesis is considered one of the key mechanisms in the long-term modulation of synaptic connections, and the pathway is thus seen underlying higher brain functions, including learning and memory. This area will certainly attract more attention, in part because it appears that neurological diseases such as the Fragile X Syndrome are causally related to RNA localization and/or translation at synaptic sites. However, we are still far from arriving at a clear picture of the underlying molecular mechanism and the way it is disrupted in the disease. To understand the disease, it appears, it will be necessary that we first understand the codes.
SUMMARY POINTS.
Both mRNAs and untranslated RNAs are localized in various eukaryotic cell types.
Cis-acting elements contain codes, likely to be expressed as RNA motifs, that specify RNA targeting.
Targeting codes are recognized by trans-acting factors, proteins that bind cis-acting elements and act as adaptors in the assembly of larger ribonucleoprotein (RNP) particles.
RNPs are translocated along cytoskeletal elements by molecular motors.
Microtubules and microfilaments have been implicated in subcellular RNA localization; long-range transport in neurons is typically mediated by microtubules.
Most transported mRNAs appear to be translationally dormant while en route.
In neurons, mRNAs at their destination sites will often have to remain translationally repressed until an activation signal is received.
Untranslated RNAs may participate in the local control of translation.
ACKNOWLEDGMENTS
Work in the authors' laboratories has been supported by the Deutsche Forschungsgemeinschaft (Ki488/2-6 to S.K., Ri192/24-1 to D.R.); the Human Frontier Science Program Organization (RG0120/1999-B to S.K.); the Volkswagenstiftung (i/74,041 to D.R.); the European Commission (QLG3-CT-1999-00,908 to D.R.); the U.S. Department of Defense (DAMD17-02-1-0520 to H.T.); and the National Institutes of Health (NS34158 and NS46769 to H.T.). H.T. thanks Jun Zhong for comments on the manuscript, and Jürgen Brosius and Christopher Hellen for stimulating discussions.
Footnotes
The authors declare that they have no conflicting financial interests.
LITERATURE CITED
- Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, Carson JH. Transport and localization elements in myelin basic protein mRNA. J. Cell Biol. 1997;138:1077–87. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–76. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 2004;24:2648–55. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Bassell GJ. Sunrise at the synapse: the FMRP mRNP shaping the synaptic interface. Neuron. 2003;37:555–58. doi: 10.1016/s0896-6273(03)00090-4. [DOI] [PubMed] [Google Scholar]
- Aranda-Abreu GE, Behar L, Chung S, Furneaux H, Ginzburg I. Embryonic lethal abnormal vision-like RNA-binding proteins regulate neurite outgrowth and tau expression in PC12 cells. J. Neurosci. 1999;19:6907–17. doi: 10.1523/JNEUROSCI.19-16-06907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J. Cell Sci. 2002;115:3817–27. doi: 10.1242/jcs.00058. [DOI] [PubMed] [Google Scholar]
- Barbarese E, Brumwell C, Kwon S, Cui H, Carson JH. RNA on the road to myelin. J. Neurocytol. 1999;28:263–70. doi: 10.1023/a:1007097226688. [DOI] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–51. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Kelic S. Binding proteins for mRNA localization and local translation, and their dysfunction in genetic neurological disease. Curr. Opin. Neurobiol. 2004;14:574–81. doi: 10.1016/j.conb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Betley JN, Frith MC, Graber JH, Choo S, Deshler JO. A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 2002;12:1756–61. doi: 10.1016/s0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- Blichenberg A, Rehbein M, Muller R, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur. J. Neurosci. 2001;13:1881–88. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- Blichenberg A, Schwanke B, Rehbein M, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J. Neurosci. 1999;19:8818–29. doi: 10.1523/JNEUROSCI.19-20-08818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaccio GL. Targeting of mRNAs within the glial cell cytoplasm: how to hide the message along the journey. J. Neurosci. Res. 2000;62:473–79. doi: 10.1002/1097-4547(20001115)62:4<473::AID-JNR1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Böckers TM, Segger-Junius M, Iglauer P, Bockmann J, Gundelfinger ED, et al. Differential expression and dendritic transcript localization of Shank family members: identification of a dendritic targeting element in the 3′ untranslated region of Shank1 mRNA. Mol. Cell. Neurosci. 2004;26:182–90. doi: 10.1016/j.mcn.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Bodian D. A suggestive relationship of nerve cell RNA with specific synaptic sites. Proc. Natl. Acad. Sci. USA. 1965;53:418–25. doi: 10.1073/pnas.53.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J, Tiedge H. RNomenclature. RNA Biol. 2004;1:81–83. doi: 10.4161/rna.1.2.1228. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–87. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Carson JH, Cui H, Krueger W, Schlepchenko B, Brumwell C, Barbarese E. RNA trafficking in oligodendrocytes. Results Probl. Cell Differ. 2001;34:69–81. doi: 10.1007/978-3-540-40025-7_5. [DOI] [PubMed] [Google Scholar]
- Carson JH, Worboys K, Ainger K, Barbarese E. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil. Cytoskelet. 1997;38:318–28. doi: 10.1002/(SICI)1097-0169(1997)38:4<318::AID-CM2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–96. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 2003;12:3295–305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Chartrand P, Singer RH, Long RM. RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol. 2001;17:297–310. doi: 10.1146/annurev.cellbio.17.1.297. [DOI] [PubMed] [Google Scholar]
- Chennathukuzhi V, Morales CR, El-Alfy M, Hecht NB. The kinesin KIF17b and RNA-binding protein TB-RBP transport specific cAMP-responsive element modulator-regulated mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA. 2003;100:15566–71. doi: 10.1073/pnas.2536695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen M, Pieler T. Xvelo1 uses a novel 75-nucleotide signal sequence that drives vegetal localization along the late pathway in Xenopus oocytes. Dev. Biol. 2004;266:270–84. doi: 10.1016/j.ydbio.2003.09.043. [DOI] [PubMed] [Google Scholar]
- Colman DR, Kreibich G, Frey AB, Sabatini DD. Synthesis and incorporation of myelin polypeptides into CNS myelin. J. Cell Biol. 1982;95:598–608. doi: 10.1083/jcb.95.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin F, Bouillon M, Fortin A, Morin S, Rousseau F, Khandjian EW. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Mol. Genet. 1997;6:1465–72. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- Couzin J. Small RNAs make big splash. Science. 2002;298:2296–97. doi: 10.1126/science.298.5602.2296. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M, Thanas S, Brosius J, Kindler S, Tiedge H. Neuronal MAP2 mRNA: species-dependent differential dendritic targeting competence. J. Mol. Biol. 2004;341:927–34. doi: 10.1016/j.jmb.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Darnell RB. Memory, synaptic translation, and…prions? Cell. 2003;115:767–68. doi: 10.1016/s0092-8674(03)01022-5. [DOI] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–31. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003;13:286–96. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Duchaine TF, Hemraj I, Furic L, Deitinghoff A, Kiebler MA, DesGroseillers L. Staufen2 isoforms localize to the somatodendritic domain of neurons and interact with different organelles. J. Cell Sci. 2002;115:3285–95. doi: 10.1242/jcs.115.16.3285. [DOI] [PubMed] [Google Scholar]
- Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, et al. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–80. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- Dyer JR, Michel S, Lee W, Castellucci VF, Wayne NL, Sossin WS. An activity-dependent switch to cap-independent translation triggered by eIF4E dephosphorylation. Nat. Neurosci. 2003;6:219–20. doi: 10.1038/nn1018. [DOI] [PubMed] [Google Scholar]
- Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 1996;5:1083–91. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Belt B, Kacharmina JE, Miyashiro K. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem. Res. 2002;27:1065–77. doi: 10.1023/a:1020956805307. [DOI] [PubMed] [Google Scholar]
- Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 2003;23:10433–44. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina KL, Singer RH. The nuclear connection in RNA transport and localization. Trends Cell Biol. 2002;12:466–72. doi: 10.1016/s0962-8924(02)02357-7. [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997;1:109–18. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–33. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel LA, Won S, Kawai H, McKinney M, Tartakoff AM, Fallon JR. Visual experience regulates transient expression and dendritic localization of fragile X mental retardation protein. J. Neurosci. 2004;24:10579–83. doi: 10.1523/JNEUROSCI.2185-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Giuditta A, Kaplan BB, van Minnen J, Alvarez J, Koenig E. Axonal and presynaptic protein synthesis: new insights into the biology of the neuron. Trends Neurosci. 2002;25:400–4. doi: 10.1016/s0166-2236(02)02188-4. [DOI] [PubMed] [Google Scholar]
- Goldstein LS, Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu. Rev. Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J. Cell Biol. 2002;156:41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This study demonstrates that nuclear RNA processing and RNP assembly regulate cytoplasmic mRNA localization events. Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–63. doi: 10.1038/nature02521.
- Han JR, Yiu GK, Hecht NB. Testis/brain RNA-binding protein attaches translationally repressed and transported mRNAs to microtubules. Proc. Natl. Acad. Sci. USA. 1995;92:9550–54. doi: 10.1073/pnas.92.21.9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC. The pathway and mechanism of initiation of protein synthesis. 2000:33–88. See Sonenberg et al. 2000. [Google Scholar]
- Hoek KS, Kidd GJ, Carson JH, Smith R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–29. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–53. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligand binding to NMDARs triggers activation of Aurora kinase, resulting in the phosphorylation of CPEB, which in turn is required for the polyadenylation of CPE-containing dormant mRNAs and thus for their translational activation. Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-d-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–48. doi: 10.1093/emboj/21.9.2139.
- This paper and the related paper by Caudy et al. (2002) show that the Drosophila fragile X protein is associated with components of the RNAi pathway. FMRP may also interact with components of the miRNA pathway (Jin et al. 2004). Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–508. doi: 10.1101/gad.1022002.
- Jackson RJ. A comparative view of initiation site selection mechanisms. 2000:127–83. See Sonenberg et al. 2000. [Google Scholar]
- Jeffery WR, Tomlinson CR, Brodeur RD. Localization of actin messenger RNA during early ascidian development. Dev. Biol. 1983;99:408–17. doi: 10.1016/0012-1606(83)90290-7. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004;7:113–17. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Job C, Eberwine J. Localization and translation of mRNA in dendrites and axons. Nat. Rev. Neurosci. 2001;2:889–98. doi: 10.1038/35104069. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, et al. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat. Neurosci. 2004;7:244–53. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–25. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116:467–79. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat. Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Huot ME, Tremblay S, Davidovic L, Mazroui R, Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc. Natl. Acad. Sci. USA. 2004;101:13357–62. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. USA. 2004;101:360–65. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, MacDonald PM. Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–12. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- King ML, Zhou Y, Bubunenko M. Polarizing genetic information in the egg: RNA localization in the frog oocyte. Bioessays. 1999;21:546–57. doi: 10.1002/(SICI)1521-1878(199907)21:7<546::AID-BIES3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–51. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–21. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. RNA localization mechanisms in oocytes. J. Cell Sci. 2005;118:269–82. doi: 10.1242/jcs.01637. [DOI] [PubMed] [Google Scholar]
- Kloc M, Zearfoss NR, Etkin LD. Mechanisms of subcellular mRNA localization. Cell. 2002;108:533–44. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- Koenig E. Synthetic mechanisms in the axon. I. Local axonal synthesis of acetylcholinesterase. J. Neurochem. 1965a;12:343–55. doi: 10.1111/j.1471-4159.1965.tb04235.x. [DOI] [PubMed] [Google Scholar]
- Koenig E. Synthetic mechanisms in the axon. II. RNA in myelin-free axons of the cat. J. Neurochem. 1965b;12:357–61. doi: 10.1111/j.1471-4159.1965.tb04236.x. [DOI] [PubMed] [Google Scholar]
- Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhöffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc. Natl. Acad. Sci. USA. 1995;92:8856–60. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TL, Yoon YJ, Mowry KL. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J. Cell Biol. 2004;165:203–11. doi: 10.1083/jcb.200309145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–96. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Kwon S, Barbarese E, Carson JH. The cis-acting RNA trafficking signal from myelin basic protein mRNA and its cognate trans-acting ligand hnRNP A2 enhance cap-dependent translation. J. Cell Biol. 1999;147:247–56. doi: 10.1083/jcb.147.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YK, Hecht NB. Binding of a phosphoprotein to the 3′ untranslated region of the mouse protamine 2 mRNA temporally represses its translation. Mol. Cell. Biol. 1993;13:6547–57. doi: 10.1128/mcb.13.10.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 2001;10:329–38. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45:407–15. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–83. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie YS, MacDonald PM. Apontic binds the translational repressor Bruno and is implicated in regulation of oskar mRNA translation. Development. 1999;126:1129–38. doi: 10.1242/dev.126.6.1129. [DOI] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, et al. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc. Natl. Acad. Sci. USA. 1995;92:5734–38. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, et al. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–45. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Martin KC. Local protein synthesis during axon guidance and synaptic plasticity. Curr. Opin. Neurobiol. 2004;14:305–10. doi: 10.1016/j.conb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Melton DA. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 1987;328:80–82. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–29. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, et al. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–76. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Via mutation of the 3′ UTR of the endogenous CaMKIIα gene, this study showed that local synthesis of CaMKIIα contributes to synaptic and behavioral plasticity. Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–19. doi: 10.1016/s0896-6273(02)00978-9.
- Min H, Turck CW, Nikolic JM, Black DL. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–36. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- Mohr E, Kachele I, Mullin C, Richter D. Rat vasopressin mRNA: a model system to characterize cis-acting elements and trans-acting factors involved in dendritic mRNA sorting. Prog. Brain Res. 2002;139:211–24. doi: 10.1016/s0079-6123(02)39018-6. [DOI] [PubMed] [Google Scholar]
- Mohr E, Richter D. Messenger RNA on the move: implications for cell polarity. Int. J. Biochem. Cell Biol. 2001;33:669–79. doi: 10.1016/s1357-2725(01)00047-4. [DOI] [PubMed] [Google Scholar]
- Monshausen M, Putz U, Rehbein M, Schweizer M, DesGroseillers L, et al. Two rat brain Staufen isoforms differentially bind RNA. J. Neurochem. 2001;76:155–65. doi: 10.1046/j.1471-4159.2001.00061.x. [DOI] [PubMed] [Google Scholar]
- Moore PB. Structural motifs in RNA. Annu. Rev. Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- Morales CR, Wu XQ, Hecht NB. The DNA/RNA-binding protein, TB-RBP, moves from the nucleus to the cytoplasm and through intercellular bridges in male germ cells. Dev. Biol. 1998;201:113–23. doi: 10.1006/dbio.1998.8967. [DOI] [PubMed] [Google Scholar]
- Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat. Neurosci. 2000;3:1079–84. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- Mouland AJ, Xu H, Cui H, Krueger W, Munro TP, et al. RNA trafficking signals in human immunodeficiency virus type 1. Mol. Cell. Biol. 2001;21:2133–43. doi: 10.1128/MCB.21.6.2133-2143.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry KL, Cote CA. RNA sorting in Xenopus oocytes and embryos. FASEB J. 1999;13:435–45. doi: 10.1096/fasebj.13.3.435. [DOI] [PubMed] [Google Scholar]
- Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–94. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, et al. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 1999;274:34389–95. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- Muslimov IA, Nimmrich V, Hernandez AI, Tcherepanov A, Sacktor TC, Tiedge H. Dendritic transport and localization of protein kinase Mζ mRNA: implications for molecular memory consolidation. J. Biol. Chem. 2004;279:52613–22. doi: 10.1074/jbc.M409240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov IA, Santi E, Homel P, Perini S, Higgins D, Tiedge H. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J. Neurosci. 1997;17:4722–33. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov IA, Titmus M, Koenig E, Tiedge H. Transport of neuronal BC1 RNA in Mauthner axons. J. Neurosci. 2002;22:4293–301. doi: 10.1523/JNEUROSCI.22-11-04293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairn AC, Matsushita M, Nastiuk K, Horiuchi A, Mitsui K, et al. Elongation factor-2 phosphorylation and the regulation of protein synthesis by calcium. Prog. Mol. Subcell. Biol. 2001;27:91–129. doi: 10.1007/978-3-662-09889-9_4. [DOI] [PubMed] [Google Scholar]
- O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu. Rev. Neurosci. 2002;25:315–38. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Palacios IM, Johnston D. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol. 2001;17:569–614. doi: 10.1146/annurev.cellbio.17.1.569. [DOI] [PubMed] [Google Scholar]
- Palacios-Prü EL, Palacios L, Mendoza RV. Synaptogenetic mechanisms during chick cerebellar cortex development. Submicrosc. Cytol. 1981;13:145–67. [PubMed] [Google Scholar]
- Pinkstaff J, Chappell SA, Mauro VP, Edelman G, Krushel LA. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc. Natl. Acad. Sci. USA. 2001;98:2770–75. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Holt C. RNA translation in axons. Annu. Rev. Cell Dev. Biol. 2004;20:505–23. doi: 10.1146/annurev.cellbio.20.010403.111746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash N, Fehr S, Mohr E, Richter D. Dendritic localization of rat vasopressin mRNA: ultrastructural analysis and mapping of targeting elements. Eur. J. Neurosci. 1997;9:523–32. doi: 10.1111/j.1460-9568.1997.tb01629.x. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–79. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Using a novel approach, this study found that the TAF ZBP1/IMP1 and the translational inhibitor FMRP interacted in an RNA-independent way, which suggests a functional link between mRNA localization and translational repression. Rackham O, Brown CM. Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs. EMBO J. 2004;23:3346–55. doi: 10.1038/sj.emboj.7600341.
- Rafelski SM, Theriot JA. Crawling toward a unified model of cell mobility: spatial and temporal regulation of actin dynamics. Annu. Rev. Biochem. 2004;73:209–39. doi: 10.1146/annurev.biochem.73.011303.073844. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, Sonenberg N. Regulation of ribosomal recruitment in eukaryotes. 2000:245–93. See Sonenberg et al. 2000. [Google Scholar]
- Rehbein M, Kindler S, Horke S, Richter D. Two trans-acting rat-brain proteins, MARTA1 and MARTA2, interact specifically with the dendritic targeting element in MAP2 mRNAs. Brain Res. Mol. Brain Res. 2000;79:192–201. doi: 10.1016/s0169-328x(00)00114-5. [DOI] [PubMed] [Google Scholar]
- Rehbein M, Wege K, Buck F, Schweizer M, Richter D, Kindler S. Molecular characterization of MARTA1, a protein interacting with the dendritic targeting element of MAP2 mRNAs. J. Neurochem. 2002;82:1039–46. doi: 10.1046/j.1471-4159.2002.01058.x. [DOI] [PubMed] [Google Scholar]
- Richter JD. Influence of polyadenylation-induced translation on metazoan development and neuronal synaptic function. 2000:785–805. See Sonenberg et al. 2000. [Google Scholar]
- Roegiers F, Jan YN. Staufen: a common component of mRNA transport in oocytes and neurons? Trends Cell Biol. 2000;10:220–24. doi: 10.1016/s0962-8924(00)01767-0. [DOI] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 1997;17:2158–65. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. Glimpses of a tiny RNA world. Science. 2001;294:797–99. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- Scheetz AJ, Nairn AC, Constantine-Paton M. N-methyl-d-aspartate receptor activation and visual activity induce elongation factor-2 phosphorylation in amphibian tecta: a role for N-methyl-d-aspartate receptors in controlling protein synthesis. Proc. Natl. Acad. Sci.USA. 1997;94:14770–75. doi: 10.1073/pnas.94.26.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat. Neurosci. 2000;3:211–16. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J. Neurosci. 2004;24:9366–77. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SP, Aisenthal L, Elisha Z, Oberman F, Yisraeli JK. A 69-kDa RNA-binding protein from Xenopus oocytes recognizes a common motif in two vegetally localized maternal mRNAs. Proc. Natl. Acad. Sci. USA. 1992;89:11895–99. doi: 10.1073/pnas.89.24.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severt WL, Biber TU, Wu X, Hecht NB, DeLorenzo RJ, Jakoi ER. The suppression of testis-brain RNA binding protein and kinesin heavy chain disrupts mRNA sorting in dendrites. J. Cell Sci. 1999;112:3691–702. doi: 10.1242/jcs.112.21.3691. [DOI] [PubMed] [Google Scholar]
- Shan J, Munro TP, Barbarese E, Carson JH, Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J. Neurosci. 2003;23:8859–66. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, Shinkura K, Tokunaga M. A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J. Neurosci. 2005;25:4420–34. doi: 10.1523/JNEUROSCI.0382-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This study shows that the RNA-binding protein CPEB appears to regulate tPA mRNA translation via polyadenylation. Local synthesis and release of tPA at synapses may therefore be effectors of synaptic plasticity. Shin CY, Kundel M, Wells DG. Rapid, activity-induced increase in tissue plasminogen activator is mediated by metabotropic glutamate receptor-dependent mRNA translation. J. Neurosci. 2004;24:9425–33. doi: 10.1523/JNEUROSCI.2457-04.2004.
- Smith R. Moving molecules: mRNA trafficking in mammalian oligodendrocytes and neurons. Neuroscientist. 2004;10:495–500. doi: 10.1177/1073858404266759. [DOI] [PubMed] [Google Scholar]
- Smith WB, Starck SR, Roberts RW, Schuman EM. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron. 2005;45:765–79. doi: 10.1016/j.neuron.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Lab. Press; Cold Spring Harbor, NY: 2000. p. 1020. [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 2004;24:9272–76. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J. Neurosci. 1982;2:284–91. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu. Rev. Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003;40:347–59. doi: 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- In dentate gyrus neurons, newly synthesized Arc mRNA is selectively delivered to synapses that have recently been activated. NMDAR activation is required for Arc mRNA targeting. Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30:227–40. doi: 10.1016/s0896-6273(01)00275-6.
- Sung YJ, Dolzhanskaya N, Nolin SL, Brown T, Currie JR, Denman RB. The fragile X mental retardation protein FMRP binds elongation factor 1A mRNA and negatively regulates its translation in vivo. J. Biol. Chem. 2003;278:15669–78. doi: 10.1074/jbc.M211117200. [DOI] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, et al. Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J. Neurosci. 2004;24:9760–69. doi: 10.1523/JNEUROSCI.1427-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N, Kawamura M, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor enhances neuronal translation by activating multiple initiation processes: comparison with the effects of insulin. J. Biol. Chem. 2001;276:42818–25. doi: 10.1074/jbc.M103237200. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–75. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc. Natl. Acad. Sci. USA. 2002;99:467–72. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KR, Otis KO, Chen DY, Zhao Y, O'Dell TJ, Martin KC. Synapse to nucleus signaling during long-term synaptic plasticity; a role for the classical active nuclear import pathway. Neuron. 2004;44:997–1009. doi: 10.1016/j.neuron.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, et al. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 2003;23:3251–61. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E, Armellin M, Giulianini PG, Bregola G, Zucchini S, et al. Brain-derived neurotrophic factor mRNA and protein are targeted to discrete dendritic laminas by events that trigger epileptogenesis. J. Neurosci. 2004;24:6842–52. doi: 10.1523/JNEUROSCI.5471-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, et al. Dendritic BC1 RNA: functional role in regulation of translation initiation. J. Neurosci. 2002;22:10232–41. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tiedge H. Translational control at the synapse. Neuroscientist. 2004;10:456–66. doi: 10.1177/1073858404265866. [DOI] [PubMed] [Google Scholar]
- Waskiewicz A, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–20. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 1999;19:1871–80. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster PJ, Liang L, Berg CA, Lasko P, MacDonald PM. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes. Dev. 1997;11:2510–21. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl. Acad. Sci. USA. 1997;94:5395–400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler IJ, Spangler CC, Klintsova AY, Grossman AW, Kim SH, et al. Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc. Natl. Acad. Sci. USA. 2004;101:17504–9. doi: 10.1073/pnas.0407533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, et al. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J. Neurosci. 2001;21:9541–48. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J. Cell Biol. 2003;163:1197–204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, et al. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron. 1998;21:1129–39. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Yaniv K, Yisraeli JK. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene. 2002;287:49–54. doi: 10.1016/s0378-1119(01)00866-6. [DOI] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 2004;14:314–21. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–27. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, et al. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–75. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J. Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, King ML. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 1996;122:4119–29. doi: 10.1242/dev.122.12.4119. [DOI] [PubMed] [Google Scholar]
- Zhao WM, Jiang C, Kroll TT, Huber PW. A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J. 2001;20:2315–25. doi: 10.1093/emboj/20.9.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, King ML. Sending RNAs into the future: RNA localization and germ cell fate. IUBMB Life. 2004;56:19–27. doi: 10.1080/15216540310001658886. [DOI] [PubMed] [Google Scholar]


