Abstract
Lewis lung carcinoma cells contain specific high-affinity binding sites for the eicosanoid 12(S)-hydroxy-5,8,10,14-eicosatetraenoic acid [12(S)-HETE]. These binding sites have a cytosolic/nuclear localization and contain the heat shock proteins hsp70 and hsp90 as components of a high molecular weight cytosolic binding complex. The ligand binding subunit of this complex is a protein with an apparent molecular mass of ≈50 kDa as judged by gel permeation chromatography. In this report, we present data showing that the 50-kDa 12(S)-HETE binding protein interacts as a homodimer with steroid receptor coactivator-1 (SRC-1) in the presence of 12(S)-HETE. Two putative interaction domains were mapped. One of these (amino acids 701–781) was within the nuclear receptor interaction domain in SRC-1 required for binding of various steroid and thyroid hormone receptors. It contains the most C-terminal of the three copies of LXXLL motif present in the nuclear receptor interaction domain. The second interaction domain was present in the N-terminal part of SRC-1 (amino acids 1–221). This region has two LXXLL motifs, one does not bind and the other binds only weakly to steroid and thyroid hormone receptors. Glutathione S-transferase (GST) pulldown experiments and far Western analyses demonstrated that the N-terminal region of SRC-1 (amino acids 1–212) alone does not bind the 50-kDa 12(S)-HETE binding protein, whereas GST/ΔSRC-11–1138 ligand-dependently pulled down a protein of ≈50 kDa in size. Our results suggest that the 50-kDa 12(S)-HETE binding protein is a receptor that may signal through interaction with a nuclear receptor coactivator protein.
The eicosanoid 12(S)-hydroxy-5,8,10,14-(Z,Z,E,Z)-eicosatetraenoic acid [12(S)-HETE] is formed from arachidonic acid by the enzyme 12-lipoxygenase (1, 2). Platelets, polymorphonuclear leukocytes, macrophages, endothelial cells, and smooth muscle cells can synthesize 12(S)-HETE (3, 4). Its biological effects are diverse and include stimulation of platelet adhesion and aggregation (5), chemotaxis of human polymorphonuclear and eosinophilic leukocytes (6), and angiogenesis (7). It also has been shown to promote the ability of certain cancer cells [e.g., Lewis lung carcinoma (LLC) and B16 melanoma], to metastasize, apparently as a consequence of increased expression of a cell adhesion glycoprotein (8).
12(S)-HETE binds specifically and with high affinity to LLC cells (9). The binding sites are predominantly cytosolic/nuclear and the cytosolic sites occur in a protein complex of high apparent molecular mass, ≈650 kDa (10). Two heat shock proteins, hsp70 and hsp90, were detected as substoichiometric components of this complex, and addition of ATP caused the 650-kDa complex to dissociate into a 50-kDa 12(S)-HETE binding subunit (11, 12). The subcellular localization, the presence of heat shock proteins in a large cytosolic complex, the size of the ligand binding subunit, and a subnanomolar dissociation constant all resemble corresponding properties of members of the steroid-thyroid hormone receptor family (13).
Nuclear receptors are ligand-dependent transcription factors that bind as dimers to specific DNA sequences in promoter regions of target genes (13, 14). The receptors either stimulate (most commonly) or repress transcription of such genes. Stimulated transcription requires binding of additional proteins to the liganded receptor, so-called coactivator proteins, which in turn interact with the basal transcriptional machinery. Steroid receptor coactivator-1 (SRC-1) is recruited by several nuclear receptors, e.g., the estrogen, glucocorticoid, progesterone, peroxisome proliferator-activated, thyroid hormone, and the 9-cis retinoic acid receptor, RXR (15). A consensus motif (LXXLL, where X is any amino acid) in SRC-1 and related proteins, which is necessary and sufficient for interaction with nuclear receptors, has been identified (16). The interaction between LXXLL-containing domains (LXDs) in SRC-1 and peroxisome proliferator-activated receptor-γ has been studied in great detail by crystallography (17).
In this report, we present data showing that the 50-kDa 12(S)-HETE binding protein interacts with SRC-1 in a strictly ligand-dependent manner. Our data suggest that the 50-kDa 12(S)-HETE binding subunit is a receptor that signals through interaction with a nuclear receptor coactivator protein.
Materials and Methods
[5,6,8,9,11,12,14,15(n)-3H]12(S)-HETE (specific activity 219 Ci/mmol; 1 Ci = 37 GBq) was from New England Nuclear, and unlabeled 12(S)-HETE was obtained from Cayman Chemicals (Ann Arbor, MI). Cell medium, antibiotics, FCS, restriction enzymes, and T4 ligase were from Life Technologies (Paisley, Scotland). A reagent kit (TnT kit) for in vitro transcription/translation was purchased from Promega.
Construction of SRC-1 Deletion Mutants and Glutathione S-Transferase (GST) Fusion Proteins.
pcDNA3 constructs.
pcDNA3 constructs were made from full-length SRC-1 cDNA subcloned into the NotI and ApaI sites in pcCNA3 (pcDNA3/SRC-1). SRC-1/BamHI was constructed by fusing the BamHI fragment of SRC-1 with pcDNA3 digested in the same way. SRC-1/EcoRV was constructed by fusing the EcoRV fragment of SRC-1 with pcDNA3 digested in the same way. SRC-1/HindIII was constructed by fusing the HindIII fragment (nucleotides 638 to 3,414) of SRC-1 with the HindIII cut vector. This construct does not include the HindIII fragment (nucleotides −127 to 637) upstream of the SRC-1 encoding region. These constructs were used as templates for in vitro transcription/translation reactions. pcDNA3/SRC-1 was linearized by digestion with AatII to produce pcDNA3/ΔSRC-1 (amino acids 1 to 1,393) or by digestion with NheI to form pcDNA3/ΔSRC-1 (amino acids 1 to 947) and used as template for in vitro transcription/translation experiments.
pGEX constructs.
Two fragments of SRC-1 cDNA were subcloned in-frame with the GST gene in the pGEX-JDK vector. pcDNA3/SRC-1, digested by XbaI and HindIII and a fragment (nucleotides −44 to 637), was ligated to pGEX-JDK vector cut by XbaI and HindIII to give pGEX-JDK/ΔSRC-11–212. The other fragment (nucleotides 638 to 3,414) was ligated to HindIII-cut pGEX-JDK/ΔSRC-11–212 to give pGEX-JDK/ΔSRC-11–1,138. These constructs were transformed into Y1090 cells for production of GST fusion proteins. Luria broth-ampicillin was inoculated with an overnight culture of transformed Y1090 bacteria and grown for 4 h at 37°C. Protein production was induced by adding 1 mM isopropyl-β-d-thiogalactoside and the cells were allowed to grow 3 h more. Bacteria were collected by centrifugation, resuspended in TEDG buffer (50 mM Tris⋅HCl, pH 7.4/1.5 mM EDTA/10% glycerol/0.4 M NaCl), lysed by sonication, and centrifuged at 25,000 rpm at 4°C in a Beckman Coulter SW41 rotor.
Cell Culture.
LLC cells were obtained from the American Type Culture Collection (Rockville, MD). The cells were grown in Dulbecco's modified Eagle's medium supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 4 mM l-glutamine, and 10% (vol/vol) heat-inactivated (30 min at 56°C) FCS at 37°C in a humidified atmosphere containing 8% (vol/vol) CO2. Cultures were passaged by scraping with a rubber policeman after treatment with 0.54 mM EDTA in buffered isotonic saline. Cells were not used beyond 20 consecutive passages.
Preparation of Cytosol.
Cells were washed and sedimented by centrifugation at 200 × g for 5 min. The pellet was suspended in a hypotonic solution (1 mM NaHCO3/2 mM CaCl2/5 mM MgCl2). After swelling for 2 min, the cells were homogenized with the loose-fitting pestle of a Dounce homogenizer. These substances were added to their indicated final concentrations: 0.25 M sucrose, 50 mM Tris⋅HCl (pH 7.5), 25 mM KCl, and 5 mM MgCl2. The homogenate was centrifuged at 100,000 × g for 1 h in a Beckman Coulter TLA 100.2 rotor.
Gel Permeation Chromatography.
Cytosol from LLC cells was chromatographed on Superdex 200 (Amersham Pharmacia), eluted at 1 ml/min with 15 mM Tris⋅HCl, pH 8.0, by using an FPLC instrument (Amersham Pharmacia). Fractions of 0.2 or 1 ml were collected, and the radioactivity was measured in a liquid scintillation counter (LKB Rackbeta 1214; Turku, Finland). The void volume was determined with blue dextran, and the column was calibrated with aldolase (Mr = 158,000), catalase (Mr = 232,000), ferritin (Mr = 440,000), and thyroglobulin (Mr = 669,000) (all obtained from Amersham Pharmacia).
Isoelectric Focusing.
LLC cells were incubated with 0.1 nM 12(S)-[3H]HETE, and cytosol was prepared as described above. A preparative scale isoelectric focusing cell (Rotofor System; Bio-Rad) was used. A pH gradient of 1.5% ampholyte (Biolyte 5/8) and 10% glycerol was preformed for 1 h, and the sample was then loaded at a pH of ≈6.5 and run for 2 h at a constant power of 11 W. The chamber was evacuated by using a vacuum pump, and the gradient was automatically divided into 20 samples. The pH in each sample was recorded, and the radioactivity was measured in a liquid scintillation counter.
Isolation of 50-kDa 12(S)-HETE Binding Subunit.
LLC cytosol was treated with ATP (10 mM addition at 0, 5, and 10 min) to obtain the 50-kDa 12(S)-HETE binding protein. The sample was chromatographed on Superdex 200, and the fraction containing the 50-kDa 12(S)-HETE binding protein was collected.
Assay of Interaction Between the 50-kDa 12(S)-HETE Binding Protein and SRC-1 and SRC-1 Deletion Mutants.
The 50-kDa 12(S)-HETE binding protein was incubated with 1 nM unlabeled 12(S)-HETE (added as an ethanol solution) or with the same amount of vehicle ethanol [0.8% (vol/vol)] for 1 h at 4°C. [35S]methionine-labeled, in vitro-translated SRC-1 (6.5 nCi) was added to both samples and incubated for another hour at 4°C. As a control, [3H]-labeled 12(S)-HETE (2 nM) was incubated with the 50-kDa 12(S)-HETE binding protein for 1 h at 4°C before addition of unlabeled SRC-1. The samples were then analyzed on Superdex 200, and bound radioactivity was measured in a liquid scintillation counter. The deletion mutants were analyzed following the same protocol, but the incubation time was reduced to 30 min.
GST Pulldown Assay and Far Western Blotting (18, 19).
GSH-agarose (25 μl) was added to a 1-ml aliquot of bacterial lysate in an Eppendorf tube and incubated for 30 min at 4°C to immobilize the GST/SRC-1 fusion protein on the agarose beads. Noninteracting proteins were removed by washing. ATP-treated LLC cytosol was incubated with these beads for 1 h at 4°C in the presence or absence of 1 nM 12(S)-HETE. Alternatively, the 50-kDa 12(S)-HETE binding protein, purified by gel permeation chromatography, was incubated with these beads for 1 h at 4°C in the presence or absence of 1 μM 12(S)-HETE. The beads were then washed with NETN buffer (0.5% Nonidet P-40/20 mM Tris, pH 8/100 mM NaCl/1 mM EDTA) and boiled in SDS/PAGE sample buffer. Interacting proteins were separated on SDS/PAGE gels, electrotransferred to nitrocellulose membranes, and renatured by guanidinium-hydrochloride treatment. Interacting proteins were detected by incubating the nitrocellulose membrane with 32P-labeled GST/ΔSRC-11–1138 fusion protein in the presence of 5 nM (cytosol) or 1 μM (50-kDa subunit) 12(S)-HETE followed by autoradiography.
Results
Purification of the 12(S)-HETE Binding Complex by Isoelectric Focusing.
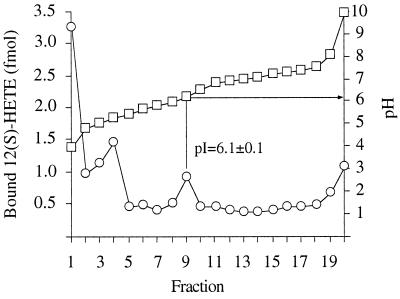
Cytosol, prelabeled with 12(S)-[3H]HETE, was loaded onto a linear pH gradient ranging from pH 5 to 8 (Fig. 1). Free 12(S)-[3H]HETE emerged between pH 4 and 5.2, whereas specifically bound 12(S)-HETE appeared at pH 6.1 ± 0.1 (mean value ± SE; n = 4).
Figure 1.
Purification by isoelectric focusing of cytosolic 12(S)-HETE binding complex. LLC cells were incubated with 0.1 nM 12(S)-[3H]HETE. Cytosol was prepared and subjected to isoelectric focusing in a Rotofor system. Fractions were withdrawn and the radioactivity (○) and pH (□) were measured. In the presence of a thousandfold excess of unlabeled 12(S)-HETE, the peak in fraction 9 was undetectable.
Ligand-Induced Interaction Between the 50-kDa 12(S)-HETE Binding Subunit and SRC-1.
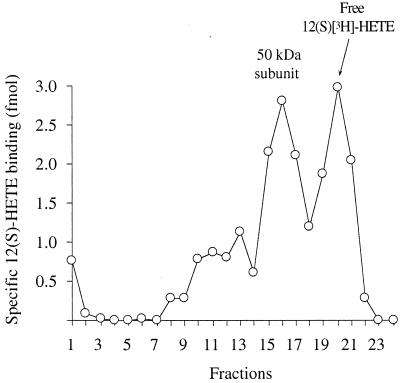
Cytosol from LLC cells was treated with ATP as described in Materials and Methods to dissociate the 650-kDa 12(S)-HETE binding complex into subunits. After isolating the 50-kDa 12(S)-HETE binding subunit by gel permeation chromatography, it was incubated with 1 nM 12(S)-[ 3H]HETE in the presence or absence of 1 μM unlabeled 12(S)-HETE. Fig. 2 shows the binding of 12(S)-HETE to the 50-kDa component (fractions 15–17). The radioactivity in fractions 19–21 represents free 12(S)-[ 3H]HETE.
Figure 2.
Preparation of liganded 50-kDa 12(S)-HETE binding protein. Cytosol from 108 LLC cells was treated with 10 mM ATP. The fraction containing the 50-kDa 12(S)-HETE binding component was incubated with 1 nM 12(S)-[3H]HETE in the presence or absence of 1 μM unlabeled 12(S)-HETE for 1 h at 4°C. The samples were then rechromatographed on Superdex 200. The results are shown as specific binding (total minus nonspecific binding in corresponding chromatography fractions).
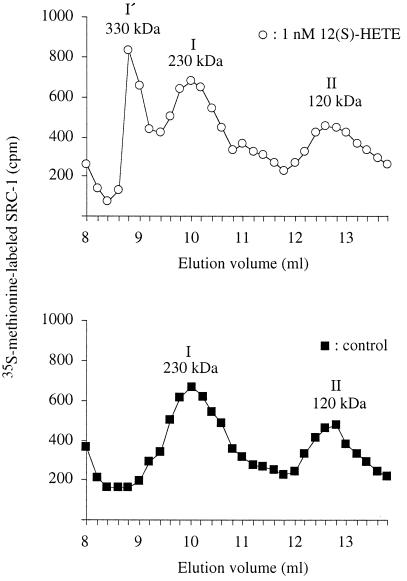
[35S]Methionine-labeled SRC-1 was prepared by in vitro transcription and translation. Gel permeation chromatography on Superdex 200 (Fig. 3, filled squares) showed that [35S]SRC-1 (molecular mass, 157 kDa) in the reticulocyte lysate mixture eluted corresponding to a complex of 230 kDa (peak I). An incomplete (120 kDa) [35S]SRC-1 fragment (peak II) also was observed. When liganded 50-kDa 12(S)-HETE binding protein was added, the 230-kDa peak shifted to 330 kDa (peak I′ in Fig. 3). No shift was observed when the 50-kDa 12(S)-HETE binding protein had been preincubated with ethanol instead of 12(S)-HETE. SRC-1 also was incubated with or without 12(S)-HETE in the absence of the 50-kDa binding protein. Both of these experiments showed only peaks I and II, and peak I′ was not detectable. To verify that the peak I′ contained both SRC-1 and the 50-kDa binding protein, 12(S)-[3H]HETE was preincubated with the 50-kDa protein and then with reticulocyte lysate containing unlabeled SRC-1 (Fig. 4B). The combined data are consistent with the binding of two molecules of liganded 50-kDa 12(S)-HETE binding protein per SRC-1 molecule.
Figure 3.
Ligand-dependent interaction between 50-kDa 12(S)-HETE binding protein and SRC-1. Cytosol from LLC cells was treated with 10 mM ATP and chromatographed on Superdex 200. The fraction containing the 50-kDa 12(S)-HETE binding protein was collected and incubated with (○) or without (■) 1 nM 12(S)-HETE for 1 h at 4°C. This incubation was followed by an incubation with 6.5 nCi [35S]methionine-labeled SRC-1 for 1 h at 4°C. The samples were rechromatographed on Superdex 200, and fractions were collected for radioactivity measurements.
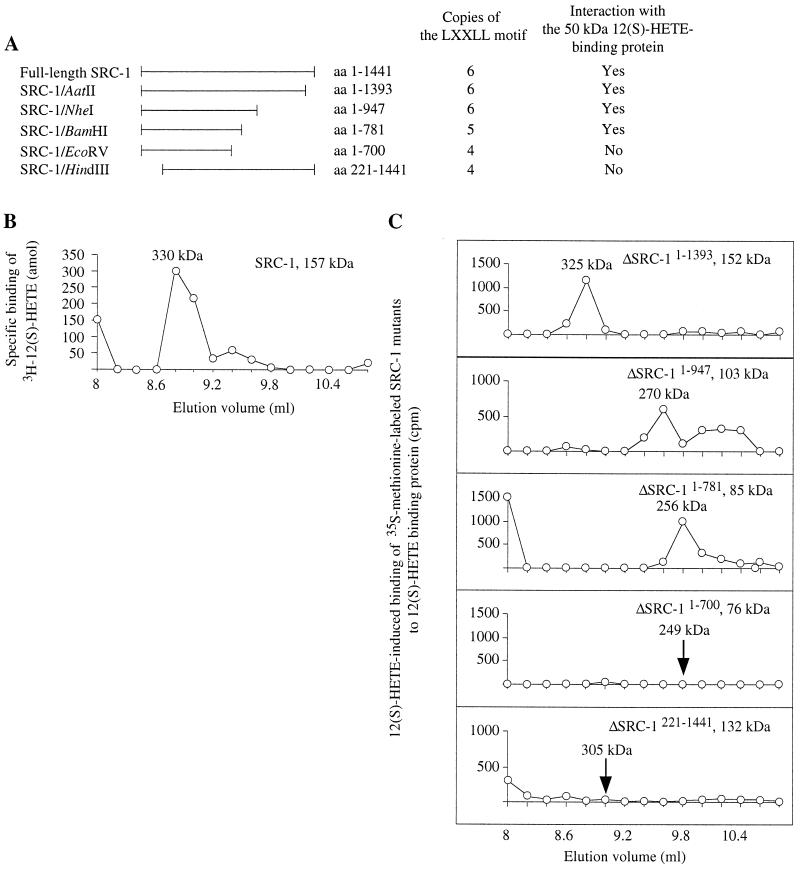
Figure 4.
Mapping of SRC-1 interaction domains for 50-kDa 12(S)-HETE binding protein. (A) Preparation and properties of SRC-1 deletion mutants (for details see Material and Methods). (B) Cytosol from LLC cells was treated with 10 mM ATP and chromatographed on Superdex 200. The fraction containing the 50-kDa 12(S)-HETE binding protein was incubated with 2 nM 12(S)-[3H]HETE for 1 h before the addition of unlabeled SRC-1. This incubation was allowed to proceed additionally for1 h at 4°C. The sample was rechromatographed on Superdex 200, and fractions were collected for radioactivity measurements. (C) Interaction assays. Cytosol from LLC cells was treated with 10 mM ATP and chromatographed on Superdex 200 to isolate the 50-kDa 12(S)-HETE binding protein. This protein was incubated with or without 1 nM 12(S)-HETE for 30 min at 4°C before addition of [35S]methionine-labeled deletion mutants of SRC-1 (30 min at 4°C). Samples were analyzed by gel permeation chromatography on Superdex 200, and radioactivity was measured in the fractions collected. The chromatograms show radioactivity in 12(S)-HETE-incubated fractions minus radioactivity in fractions from control experiments without ligand.
Mapping of SRC-1 Regions That Interact with the 50-kDa 12(S)-HETE Binding Protein.
One N-terminal and four C-terminal deletion mutants of SRC-1 (Fig. 4A) were prepared as described in Materials and Methods. Cytosol from LLC cells was treated with ATP, and the 50-kDa 12(S)-HETE binding protein, isolated by gel permeation chromatography, was incubated with 1 nM 12(S)-HETE (or ethanol in control samples) for 30 min at 4°C. Full-length SRC-1 or SRC-1 deletion mutants were added, and incubations were allowed to proceed for 30 min longer. The samples were analyzed by Superdex 200 chromatography (Fig. 4C). Three C-terminal deletion mutants [SRC-1/AatII (amino acids 1–1,393), SRC-1/NheI (amino acids 1–947), and SRC-1/BamHI (amino acids 1–781)] all interacted with the 50-kDa 12(S)-HETE binding protein. Another C-terminal deletion mutant, SRC-1/EcoRV (amino acids 1–700), and the N-terminal deletion mutant, SRC-1/HindIII (amino acids 221–1,441), did not interact with the 50-kDa 12(S)-HETE binding protein.
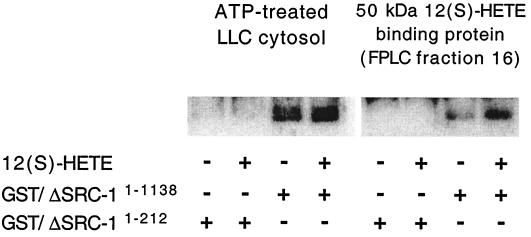
To determine whether the N-terminal part of SRC-1, eliminated in the mutant just described, was capable of interacting with the 50-kDa 12(S)-HETE binding protein, GST pulldown assay and far Western blot detection were performed (Fig. 5). The GST fusion protein constructs used encoded either SRC-1 amino acids 1–1,138 or 1–212. When immobilized SRC-11–1,138 was incubated with ATP-treated LLC cytosol, a large background of SRC-1-interacting proteins with molecular masses 50–60 kDa (probably liganded cytosolic steroid receptors) was detected in the absence of 12(S)-HETE (lane 3). Therefore, the amount of SRC-1 interacting proteins was only marginally increased by addition of 12(S)-HETE (lane 4). The experiment therefore was repeated with FPLC-purified 50-kDa 12(S)-HETE binding protein instead of cytosol. The background was now considerably lower (lane 7), and there was a marked increase in a 50-kDa protein when 12(S)-HETE was added (lane 8). No interacting proteins were detected when immobilized SRC-11–212 was used under the same conditions, either in the presence or absence of 12(S)-HETE, indicating that this part of SRC-1 is not capable of interacting with the 50-kDa 12(S)-HETE binding protein by itself.
Figure 5.
GST pulldown and far Western detection of the 50-kDa 12(S)-HETE binding protein. (Left) ATP-treated cytosol was incubated with GST/ΔSRC-11–212 (lanes 1 and 2) or GST/ΔSRC-11–1,138 (lanes 3 and 4) immobilized on GSH-agarose in the presence or absence of 1 nM 12(S)-HETE. (Right) The 50-kDa 12(S)-HETE binding protein (FPLC fraction 16) was incubated with GST/ΔSRC-11–212 (lanes 5 and 6) or GST/ΔSRC-11–1,138 (lanes 7 and 8) immobilized on GSH-agarose in the presence or absence of 1 μM 12(S)-HETE. Interacting proteins were electrotransferred to nitrocellulose membranes and detected by incubating with 32P-labeled GST/ΔSRC-11–1,138 in the presence of 5 nM 12(S)-HETE for ATP-treated cytosol and 1 μM 12(S)-HETE for 50-kDa 12(S)-HETE binding protein.
Discussion
We previously have demonstrated specific high-affinity (dissociation constant Kd < 1 nM) binding sites for 12(S)-HETE in cytosol and nuclei of LLC cells. The heat shock proteins hsp70 and hsp90 occur as substoichiometric components of a 650-kDa cytosolic complex that contains an unknown number of 50-kDa 12(S)-HETE binding subunits (9–12).
Steroid hormone receptors are ligand-dependent transcription factors that bind to response elements in promoter regions of target genes (13, 14). Transcriptional activation is mediated by two domains called activation functions, AF-1 and AF-2 (19), present in the N-terminal and C-terminal parts of the receptor, respectively. AF-2 partly overlaps the hormone (ligand)-binding domain. AF-1 and AF-2 also occur in the thyroid hormone and in the retinoid receptors. The nuclear coactivator protein SRC-1 (15) increases ligand-dependent transcriptional activation by several nuclear receptors in cotransfection assays (20). It has been shown that a conformational change induced by an agonist is necessary for the receptor to interact with SRC-1 (17). Furthermore, the inability of an antagonist-bound receptor to bind efficiently to SRC-1 has been suggested as a mechanism for hormonal antagonism (15).
Our finding that a dimer of the 50-kDa 12(S)-HETE binding protein interacts with SRC-1 in the presence of ligand is similar to observations made for several nuclear receptors (16, 17).
Human SRC-1 (21) has six copies of the LXXLL motif at amino acids 46–50 (LXD1), 112–116 (LXD2), 633–637 (LXD3), 690–694 (LXD4), 749–753 (LXD5), and 913–917 (LXD6). LXDs 3–5 form the nuclear receptor interaction domain. LXD1 binds weakly to liganded estrogen receptor, whereas LXDs 2 and 6 do not bind liganded estrogen receptor (16). Our C-terminal deletion mutants showed that removal of LXD6 had no effect on the interaction of SRC-1 with the 12(S)-HETE binding protein, whereas further removal of LXD5 prevented it. Similarly, our N-terminal deletion mutant lacking LXDs 1 and 2 had lost its ability to interact. Phrased differently, truncated SRC-1 molecules with five or six LXDs interacted, whereas two molecules with only four LXDs did not interact with the 12(S)-HETE binding protein (Fig. 4A).
Based on the results presented in this paper, we propose that the 50-kDa 12(S)-HETE binding protein is a receptor that may influence gene transcription through its interaction with SRC-1.
Acknowledgments
This work was supported by the Swedish Medical Research Council (project 03X-5914), the Swedish Cancer Society, and the Swedish Society for Medical Research.
Abbreviations
- 12(S)-HETE
12(S)-hydroxyeicosatetraenoic acid
- LLC
Lewis lung carcinoma
- LXD
domain having one LXXLL motif (X meaning any amino acid)
- SRC-1
steroid receptor coactivator-1
- GST
glutathione S-transferase
References
- 1.Yamamoto S. Biochim Biophys Acta. 1992;1128:117–131. doi: 10.1016/0005-2760(92)90297-9. [DOI] [PubMed] [Google Scholar]
- 2.Brash A R. J Biol Chem. 1999;274:23679–23682. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- 3.Hopkins N K, Oglesby T D, Bundy G L, Gorman R R. J Biol Chem. 1984;259:14048–14053. [PubMed] [Google Scholar]
- 4.Larrue J, Rigaud M, Razaka G, Daret D, Demond-Henri J, Bricaud H. Biochem Biophys Res Commun. 1983;112:242–249. doi: 10.1016/0006-291x(83)91822-3. [DOI] [PubMed] [Google Scholar]
- 5.Sekiya F, Takagi J, Usui T, Kawajiri K, Kobayashi Y, Sato F, Saito Y. Biochem Biophys Res Commun. 1991;179:345–351. doi: 10.1016/0006-291x(91)91376-n. [DOI] [PubMed] [Google Scholar]
- 6.Goetzl E J, Woods J M, Gorman R R. J Clin Invest. 1977;59:179–183. doi: 10.1172/JCI108617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang D G, Renaud C, Stojakovic S, Diglio C A, Porter A, Honn K V. Biochem Biophys Res Commun. 1995;211:462–468. doi: 10.1006/bbrc.1995.1836. [DOI] [PubMed] [Google Scholar]
- 8.Tang D G, Honn K V. Ann NY Acad Sci. 1994;744:199–215. doi: 10.1111/j.1749-6632.1994.tb52738.x. [DOI] [PubMed] [Google Scholar]
- 9.Herbertsson H, Hammarström S. FEBS Lett. 1992;298:249–252. doi: 10.1016/0014-5793(92)80069-s. [DOI] [PubMed] [Google Scholar]
- 10.Herbertsson H, Hammarström S. Biochim Biophys Acta. 1995;1244:191–197. doi: 10.1016/0304-4165(94)00223-k. [DOI] [PubMed] [Google Scholar]
- 11.Herbertsson H, Kühme T, Evertsson U, Wigren J, Hammarström S. J Lipid Res. 1998;39:237–244. [PubMed] [Google Scholar]
- 12.Herbertsson H, Kühme T, Hammarström S. Arch Biochem Biophys. 1999;367:33–38. doi: 10.1006/abbi.1999.1233. [DOI] [PubMed] [Google Scholar]
- 13.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu V C, Näär A M, Rosenfeld M G. Curr Opin Biotechnol. 1992;3:597–602. doi: 10.1016/0958-1669(92)90002-z. [DOI] [PubMed] [Google Scholar]
- 15.Oñate S A, Tsai S Y, Tsai M-J, O'Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 16.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 18.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 19.Cavailles V, Dauvois S, Danielian P S, Parker M G. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 21.Oñate S A, Boonyaratanakornkit V, Spencer T E, Tsai S Y, Tsai M-J, Edwards D P, O'Malley B W. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]