Abstract
Rosiglitazone (Rosi) belongs to the class of thiazolidinediones (TZDs) that are ligands for peroxisome proliferator-activated receptor γ (PPARγ). Stimulation of PPARγ suppresses bone formation and enhances marrow adipogenesis. We hypothesized that activation of PPARγ down-regulates components of the IGF regulatory system, leading to impaired osteoblast function. Rosi treatment (1 μM) of a marrow stromal cell line (UAMS-33) transfected with empty vector (U-33/c) or with PPARγ2 (U-33/γ2) were analyzed by microarray. Rosi reduced IGF-I, IGF-II, IGFBP-4, and the type I and II IGF receptor (IGF1R and IGF2R) expression at 72 h in U-33/γ2 compared with U-33/c cells (P < 0.01); these findings were confirmed by RT-PCR. Rosi reduced secreted IGF-I from U-33/γ2 cells by 75% (P < 0.05). Primary marrow stromal cells (MSCs) extracted from adult (8 months) and old (24 months) C57BL/6J (B6) mice were treated with Rosi (1 μM) for 48 h. IGF-I, IGFBP-4, and IGF1R transcripts were reduced in Rosi-treated MSCs compared with vehicle (P < 0.01) and secreted IGF-I was also suppressed (P < 0.05). B6 mice treated with Rosi (20 mg/kg·d) for short duration (i.e. 4 d), and long term (i.e. 7 wk) had reduced serum IGF-I; this was accompanied by markedly suppressed IGF-I transcripts in the liver and peripheral fat of treated animals. To determine whether Rosi affected circulating IGF-I in humans, we measured serum IGF-I, IGFBP-2, and IGFBP-3 at four time points in 50 postmenopausal women randomized to either Rosi (8 mg/d) or placebo. Rosi-treated subjects had significantly lower IGF-I at 8 wk than baseline (−25%, P < 0.05), and at 16 wk their levels were reduced 14% vs. placebo (P = 0.15). We conclude that Rosi suppresses IGF-I expression in bone and liver; these changes could affect skeletal acquisition through endocrine and paracrine pathways.
PEROXISOME PROLIFERATOR-activated receptor-γ (PPARγ) is a nuclear receptor necessary and sufficient for adipogenesis (1). There are four major transcripts for PPARγ, each generated from a separate promoter, which translate to two protein isoforms, PPARγ1 and PPARγ2. PPARγ1 is expressed in many tissues, including bone, whereas PPARγ2, is specifically expressed in adipocytes. Activation of this receptor by endogenous (e.g. saturated fatty acids, PGJ2, HEDE) or exogenous ligands promotes insulin sensitivity and influences energy homeostasis (2). Thiazolidinediones [TZDs: e.g. rosiglitazone (Rosi) and pio-glitazone] are a class of pharmacological ligands that activate PPARγ and have been widely used to treat type II diabetes mellitus.
Recent attention has focused on the effects of TZDs on cell proliferation and differentiation (3, 4). Freudlisperger et al. (5) reported that TZDs suppress the growth of human melanoma cells in vitro, and Aiello et al. (6) noted these agonists were also effective in partially reversing the epithelial mesenchymal transition of anaplastic thyroid cancer. In the latter study, the authors found that TZDs caused cell cycle arrest, due to suppression of cyclin D1, but up-regulation of p21 and p27 (6). In addition, PPARγ agonists have been reported to block the biological actions of IGF-I by increasing PTEN and suppressing the PI3K/Akt pathway. Similar findings have been noted by Kim et al. in MCF-7 breast cancer cells and He et al. in keratinocytes (7, 8).
The role of PPARγ in the acquisition and maintenance of bone mass has recently been scrutinized. In mice, Rosi reduces bone mineral density (BMD), principally by impairing osteoblast (OB) differentiation (9–11). PPARγ heterozygous-deficient mice (Pparγtm1Tka/+) mice have high trabecular bone volume (BV/TV), a notable absence of marrow fat, and protection against age-related bone loss (12). A similar relationship between PPARγ expression and high bone mass was noted in FABP4-Wnt10b transgenic mice (13). Also, in a large observational study, Schwartz et al. reported accelerated bone loss in older diabetic women receiving TZDs (14).
Pluripotent bone marrow stromal/stem cells (MSCs) become OBs under the influence of skeletal transcriptional regulators including osterix, TAZ, Msx1, Runx2, and β-catenin, whereas proteins such as PPARγ2, C/EBPα and β, and sterol regulatory element binding protein-1 stimulate MSCs to differentiate into adipocytes (1, 13, 15–19). Activation of PPARγ by TZDs can force MSCs into the adipocyte lineage, although this may not be a mutually exclusive process as previously demonstrated using ligands of different chemical structures (19–21). PPARγ2 activity is also up-regulated in the marrow of older mice and diabetic rodents, and its enhanced expression correlates with the magnitude of marrow adiposity (22, 23).
Two new observations led us to determine how PPARγ activation affects IGF-I in the context of OB differentiation. First, in an F2 population from two strains of mice, we found a quantitative trait locus (QTL) for serum IGF-I and BMD on mouse chromosome 6 in the vicinity of the Pparγ gene (24–26). Second, we noted that treatment of yellow obese agouti mice with Rosi for 8 wk lowered serum IGF-I (27). Because IGF-I stimulates both OB proliferation and differentiation, we hypothesized that pharmacological activation of PPARγ with Rosi would down-regulate IGF-I expression in pre-OBs, thereby contributing to impaired bone acquisition.
Materials and Methods
In vitro studies
Cell cultures and treatment regimens
Rosi was obtained from Tularik Inc. (San Francisco, CA). Murine marrow-derived UAMS-33 cells stably transfected with a vector expressing mRNA for PPARγ 2 (clone 28.6), referred to as U-33/γ 2 cells, and UAMS-33 cells transfected with an empty vector control (clone γ c2), referred to as U-33/c cells, have been previously described (19). Both cell lines were derived from parental UAMS-33 cells, and express endogenously PPARγ 1 isoform, whereas only U-33/γ 2 express ectopically PPARγ 2 isoform. Cells were maintained in αMEM supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 0.5 mg/ml G418 for positive selection of transfected cells, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin (Sigma) at 37 C in a humidified atmosphere containing 5% CO2. Media and additives were purchased from Life Technologies (Gaithersburg, MD). Bone marrow cultures were established from femur marrow aspirates as previously described and maintained in basal medium consisting of αMEM supplemented with 15% heat-inactivated fetal bovine serum (Hyclone), 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin at 37 C in a humidified atmosphere containing 5% CO2 (22).
The AML12 (α mouse liver 12) cell line was obtained from American Type Culture Collection (Manassas, VA). It was cultured in a 1:1 mixture of DMEM and Ham’s F12 medium with 10 μg/ml insulin, 5.5 μg/ml transferrin, 6.7 ng/ml sodium selenite, 40 ng/ml dexamethasone, and 10% fetal bovine serum. The medium also contained 4 mM glutamine,100 IU/ml penicillin, and 100 μg/ml streptomycin. AML12 cells were seeded onto 60-mm tissue culture plates and were grown to confluence, then changed to fresh medium, treated with different concentration of Rosi (Cayman Chemical, Ann Arbor, MI). After 48 h, cells were harvested, and total RNA was prepared for ribonuclease protection assay (RPA) using RNASTAT reagent.
Microarray experiment
U-33/γ2 and U-33/c cells were propagated for one passage and then seeded at the density of 3×105 cells/cm2. After 48 h of growth, when cultures achieved approximately 80% confluency, cells were treated with either 1 μM Rosi or the same volume of vehicle [dimethylsulfoxide (DMSO)] for 2, 24, and 72 h, followed by RNA isolation using RNeasy kit (QIAGEN Inc., Valencia, CA). RNA quality was assessed using Agilent 2100 Bioanalyzer (Palo Alto, CA) and conformed to the standards described by the manufacturer. The replicate experiment was performed independently on a fresh batch of cells. Two replicates were used for microarray analysis. The 2 × 2 × 3 full factorial design corresponded to: two treatments (Rosi or none), two levels of PPAR-γ2 (PPARγ2 present or not), and three levels of time (2, 24, and 72 h).
Gene expression was analyzed using GeneChip Mouse Genome 430 2.0 Array covering over 39,000 transcripts on a single array (Affymetrix, Inc., Santa Clara, CA). The procedures of converting RNA to cDNA, labeling, microarray hybridization, and GeneChips scanning were performed by the Microarray Facility at the University of Iowa according to the standardized protocol. The probe set used for IGF-I were three recently confirmed sets covering exon 6 of the IGF-I gene (probe nos. 1419519, 1437401, and 1434413).
IGF-1 gene expression analysis using quantitative real-time RT-PCR
Total RNA from U-33/γ2 and U-33/c cells as well as liver and peripheral fat was isolated using RNAeasy kit according to the manufacturer’s protocol (QIAGEN, Inc). The gene-specific primer sequences were selected using the Taqman Probe and Primer Design function of the Primer Express version 1.5 software (Applied Biosystems, Foster City, CA). RT reactions were carried using 2 μg RNA, subjected previously to deoxyribonuclease digestion, and a TaqMan Reverse Transcription Reagents (Applied Biosystems), followed by PCR in real-time using a SYBR Green PCR Master Mix (Applied Biosystems) and an ABI Prism 7700 Sequence Detection System (Applied Biosystems). The reactions were performed using the following cycling conditions: 95 C for 10 min, then 40 cycles of 95 C for 15 sec followed by 60 C for 1 min. The optimal concentrations of primers and templates that were used in each reaction were established based on the standard curve created before the reaction and corresponded to approximately 100% reaction efficiency. PCR results were then normalized to the expression of 18S rRNA in the same samples. Gene expression was analyzed using the following pairs of primers: IGF-I (specific for exon 6) (forward, GCTCTGCTTGCTCAC-CTTCAC; reverse, CACACGAACTGAAGAGCATCCA), IGF-1 (specific for exon 4) (forward, GCCCCACTGAAGCCTACAAA; reverse, TGA-GTCTTGGGCATGTCAGTGT), IGF1R (forward, CTTCGGCCTCCTC-AGCCT; reverse, CAGCCTTGTGTCCTGAGTGTCT), IGF binding protein 4 (IGFBP-4) (forward, CCCCTGCGTACATTGATGC; reverse, CGATTTCCGACAGCTCCGT) and 18S rRNA (forward, TTCGAACG-TCTGCCCTATCAA; reverse, ATGGTAGGCACGGCGACTA).
Samples of liver were extracted for RNA and processed as previously described (26). For each PCR, 1 μl of diluted cDNA was added to 5 μl of 2× iTaq SYBR Green Supermix with ROX (catalog no. 170-8851; Bio-Rad, Hercules, CA), and 100 nM of each forward and reverse primer in a total reaction volume of 10 μl. Cycling conditions were 2 min hold at 50 C, 3 min hold at 95 C, 40 cycles of 95 C for 15 sec followed by 60 C for 1 min, and all reactions were run on the ABI 7900HT Sequence Detection System (Applied Biosystems, Warrington, UK). Each of the 48 sets of gene-specific primers was analyzed by real-time PCR to assess transcript levels, with each primer pair run once per biological replicate. Four biological replicates were run per strain. Differential expression was assessed using a global pattern recognition algorithm as previously described (28–30). A 127-bp fragment of mouse IGF-I exon 4 was amplified by PCR and subcloned into pGEM 4Z. Antisense 32P-labeled probe was prepared and used in an RPA using reagents from Ambion (Austin, TX) as described previously (31). The 28S rRNA probe was obtained from Ambion.
Collection of conditioned media
For measurements of IGF-I protein, conditioned media were collected as follows: U-33/γ2 and U-33/c were plated in replicates at the density 9 × 103 cells/cm2 and were grown to 70% confluency (2–3 d). Primary bone marrow cultures from B6 mice were seeded at the density 2.5 × 105/cm2 and were grown for 10 d, the period of time necessary for mesenchymal cells to exhaust their proliferative potential in these conditions. Next, cells were treated for 3 d with 1 μM Rosi (Tularik Inc., San Francisco, CA) in the presence of growth media followed by change to the serum-free media, and conditioned media were collected at 24, 48, and 72 h and assayed for IGF-I protein. Remaining cells were lysed, and total protein content was measured using BCA Protein Assay Kit (Pierce, Rockford, IL).
IGF-I in the culture media and in serum from B6 mice was measured by a RIA (ALPCO, Windham, NH). IGFBPs were first separated from the IGF-I by an acid dissociation step. This was followed by the addition of a neutralization buffer containing excess recombinant human IGF-II, allowing the IGF-II to bind to the IGFBPs before immunoassay with a human anti-IGF-I polyclonal antibody. The sensitivity of the assay is 0.01 ng/ml IGF-I. The interassay coefficient of variation based on normal standards was approximately 6% for pooled serum from B6 mice and 8% for CM. There was no cross-reactivity with IGF-II. Standards were included in each assay from a single pool of 16 wk B6 female mice. IGF-I concentrations in serum were noted in nanograms per milliliter and in CM were normalized to total protein content.
In vivo studies
Mice
All mice used in this study were raised at The Jackson Laboratory (Bar Harbor, ME), and all studies and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of The Jackson Laboratory. Mice were maintained in groups of two to three in poly-carbonate boxes (130 cm2) on bedding of sterilized white pine shavings under conditions of a 14-h light, 10-h dark cycle, with free access to acidified water (pH 2.5 with HCl to retard bacterial growth) that contains 0.4 mg/ml of vitamin K (menadione Na bisulfite). Mice were fed an NIH31 diet supplemented or not supplemented with Rosi. In each of the three studies, mice were not fasted. There were two different sets of in vivo experiments; in each, Rosi was administered to all mice at the dose 20 mg/kg·d. In the first series of experiments, Rosi was administered to 9-wk-old female B6 mice (n = 14) for 4 d and compared with B6 controls; the animals were then killed and levels of serum IGF-I in blood collected from heart were measured. IGF-I pathway gene expression was measured in mRNA isolates from liver and fat using real-time PCR. Alternatively, 6-month-old female B6 were either sham or ovariectomized (OVX) (n = 6 each) and treated with either saline or Rosi 20 mg/kg·d for 4 d. Serum IGF-I was measured on d 4 in sham and OVX mice that were treated or not treated with Rosi. The second set of experiments was longitudinal in nature; first, 8-wk-old female B6 mice (n = 10) were treated for 4 d with Rosi, and serum IGF-I measured at beginning and end of that period. In the second experiment 3-wk-old female B6 mice were treated for 9 wk with Rosi, and levels of circulating IGF-I protein were measured in the blood collected from retro-orbital veins at the beginning and the end of each treatment.
Humans
To determine the effect of PPARγ agonism on circulating levels of IGF-I, we used archival samples from a recently completed randomized, placebo-controlled trial, designed to assess the effects of Rosi on biochemical markers of bone turnover. In brief, 50 healthy women, more than 5 yr postmenopausal, were randomized to receive either Rosi (GlaxoSmithKline, Philadelphia, PA) 8 mg daily (2 × 4 mg tablets) or placebo for 16 wk. Subjects took one study tablet daily for the first 2 wk, then two tablets daily for the remainder of the study. Women with illnesses or receiving therapies likely to affect bone were ineligible, as were those with osteoporosis (BMD T score at lumbar spine or total hip ≤ −2.5), and those with any other major systemic disease or with contraindications to the use of TZDs. Fasting blood samples were collected between 0800 and 1000 at baseline, 2, 4, 8, and 16 wk. Serum was stored at −70 C until analyzed. The study was approved by the Auckland Ethics Committee, and written informed consent was provided by each participant. The trial was registered at the Australian Clinical Trials Register, ACTRN 012605000218695 (www.actr.org.au).
Statistical analyses
Statistical assessment of differential gene expression
Probe intensity data from 24 Mouse 430 version 2 Affymetrix GeneChip arrays was read into the R software environment (http://www.R-project.org) directly from .CEL files using the R/affy package (32). Probe level data quality was assessed by image reconstruction, histograms of raw signal intensities and MvA plots. Normalization was carried out using the robust multiarray average method using all probe intensity data sets together to form one expression measure per gene per array (33). Briefly, the robust multiarray average method was used to adjust the background of perfect match probes, apply a quantile normalization of the corrected perfect match values, and calculate final expression measures using the Tukey median polish algorithm. An ANOVA-based approach was used to statistically resolve gene expression differences using the R/maanova package (34, 35). Log-transformed expression measures were expressed in fixed effects ANOVA models as the sum of different components that contributes to the overall intensity value of each gene on the array. For each tested difference between time, the model Yi = μ + TIME + Ei was used to fit the log-transformed gene expression measures Yi within a condition (RP, P, R, or N), where μ is the mean for each array, TIME is the effect for each of the time points (i.e. 2, 24, or 72 h) and Ei captures random error. All statistical tests for differences between treatment conditions were performed using Fs, a modified F-statistic incorporating shrinkage variance components (36). Critical P values were calculated through permutation analyses incorporating 1000 residual shuffles and pooled t statistics. The proportion of false positives generated in these probe set lists was estimated by implementing the false discovery rate adjustment of Storey (37).
Overrepresented classifications of genes were determined from the statistical outcomes by testing for association with gene ontology biological process terms and gene product relationships (i.e. canonical and high-level functions) available in an expert-curated database of biological networks (Ingenuity Pathways Analysis) (38). The Ingenuity Pathways Knowledge Base (IPKB) (www.ingenuity.com) consists of expert-curated information from over 400 journals with known biological relationships between genes and gene products. A right-tailed Fisher’s exact test for 2 × 2 contingency tables was used to determine the significance of overrepresentation of pathway members in IPKB gene lists. Enrichment of pathway members among top-ranking genes derived from statistical tests based on biological data available in the gene ontology terms and IPKB was assessed with the one-tailed Fished exact test for 2 × 2 contingency tables (IPA) or a conservative adjustment to the Fisher’s exact probability that weights significance according to the number of genes (EASE score) (39).
Other statistical methods
Statistically significant differences between treatment groups in the mouse studies were detected using one-way ANOVA followed by post hoc analysis by Student-Neuman-Keuls within the SigmaStat software (SPSS, Inc., Chicago, IL) after establishing the homogeneity of variances and normal distribution of data. In all cases, results are expressed as the mean value ± SEM; P < 0.05 and were considered significant. For analysis of the randomized human study, procedures of the statistical analysis system SAS (version 9.2; SAS Institute Inc., Cary, NC) were used. All statistical tests were two-tailed, and a 5% significance level was maintained throughout. All treatment evaluations were performed on the principle of intention to treat. A mixed-models approach to repeated measures was used to examine the time course of response in treatment and control arms at baseline and at 2, 4, 8, and 14 wk. The correct covariance structure was determined by likelihood ratio test (i.e. the first order auto regression matrix was compared against an unstructured covariance matrix). Maximum likelihood imputation was used to ensure all the randomized patients could be included in the mode.
Results
In vitro studies
Overview of microarray data
Comparing U-33/γ2 and U-33/c cells, we performed high-throughput analyses to determine the effects of PPARγ2 activation with Rosi on MSC gene expression. One of the objectives of the analysis was to characterize a sequence of events controlled by PPARγ2, which led to the conversion of cells from the osteoblastic lineage into fat-laden cells (19). This allowed us to speculate on possible clusters of IGF regulatory pathways, which would be regulated early and possibly in a direct manner by PPARγ2. Therefore, in addition to comparison of two cell lines and two treatments, we compared changes in gene expression at three different time points. Thus, cells with or without the transcription factor PPARγ2 (U-33/γ2 and U-33/c, respectively) were cultured in the presence or absence of Rosi for 2, 24, and 72 h followed by RNA isolation. Different experimental layouts and model formulations were considered before determining that the most useful framework for this experimental data set was the: 2 × 2 × 3 full factorial design.
Overall, F-test analyses revealed that differences in mRNA expression between different cell culture treatments were highly dependent upon time after addition of Rosi. There was a progressive increase in the total number of differentially expressed probe sets with time for a range of statistical thresholds. The number of genes whose expression was significantly affected in U-33/γ2 cells increased exponentially with a time of exposure to Rosi resulting in 35 genes after 2 h, 2510 after 24 h, and 6768 after 72 h (P < 0.01). None of the analyzed genes was affected by Rosi in U-33/c cells, indicating that effects seen in U-33/γ2 are specific for PPAR-γ2. A pathway-specific correlation analysis identified several gene clusters, whose function are essential for osteoblast and adipocyte differentiation, and bone turnover (see Table 1). Significant changes in expression of cell proliferation and fatty acid metabolism regulators were among the earliest observed. These changes were followed by a suppression of osteoblast-specific gene markers essential for their development and function, among them transcriptional regulators such as Dlx5, Runx2, and Osterix (see Table 1). Most importantly, activation of PPAR-γ2 had a pleiotropic effect on the expression of several critical proliferation and differentiation factors (see Table 1).
TABLE 1.
Fold change at 72 h for Rosi-treated U-33/γ2 cells compared with Rosi-treated U-33/c cells in expression for proliferation and differentiation genes in the adipocyte and osteoblast pathways
| Gene expressiona |
Fold change in U-33/γ2 cells treated with Rosi relative to nontreated U-33/γ2 cells | Downstream pathway | |
|---|---|---|---|
| Gene | Probe set | ||
| CD36 | [1423166_at] | +178.8 | Adipocyte differentiation |
| FABP4 | [1424155_at] | +69.6 | Adipocyte differentiation |
| Resistin | [1449182_at] | +66.0 | Adipocyte differentiation |
| FSP 27 (Cidec) | [1452260_at] | +454.9 | Adipocyte differentiation |
| Adipsin | [1417867_at] | +153.4 | Adipocyte differentiation |
| p21 | [1420979_at] | +4.2 | Cell cycle inhibitor |
| Cyclin D1 | [1417420_at] | −12.2 | Cell proliferation |
| Cyclin D2 | [148229_s_at] | −3.2b | Cell proliferation |
| Cyclin B2 | [1450920_at] | −5.3 | Cell proliferation |
| FGF 7 | [1438405_at] | −8.9 | Cell proliferation |
| FGF receptor2 | [1420847_a_at] | −4.4 | Cell proliferation |
| C-jun | [1417409_at] | −3.8 | Cell proliferation |
| Runx 2 | [1424704_at] | −7.7 | Bone differentiation factor |
| Dlx5 | [1449863_a_at] | −7.1 | Bone differentiation factor |
| Osterix (Sp7) | [1418425_at] | −5.5 | Bone differentiation factor |
| BMP-4 | [1422912_at] | −6.9 | Bone differentiation factor |
| PTHR (Pthr1) | [1417092_at] | −6.2 | Bone differentiation receptor |
All statistically significant at P < 0.001.
Probe set ends in _s_at.
Rosi suppresses IGF-I, IGF-II, IGFIR, and IGFBP-4 in U-33/γ2cells
Our a priori hypothesis, based on previous genetic studies, one preliminary in vivo experiment, and the microarray pattern for other genes (see Table 1), was that elements of the IGF regulatory system in pre-OBs were affected by Rosi activation of PPARγ2. At 2 and 24 h after Rosi exposure, we found no changes in any of the principal IGF components in U-33/γ2 or U-33/c cells. However, at 72 h, IGF-I, IGF-II, IGF1R, IGF2R, and IGFBP-4 mRNA were significantly down-regulated by Rosi in U-33/γ2 cells compared with nontreated cells (P < 0.05 for all genes; see Table 2). In addition to significant changes in both IGF ligands, IGFBP-4 and the type I and II IGF receptor, we found several key molecules in the IGF signaling pathway were also down-regulated, including two MAPKs, Src homology 2 domain, and paxillin (see Table 3). On the other hand, the GH receptor (GHR) was up-regulated more than 3-fold at 24 h, and more than 4-fold at 72 h (P < 0.00000), in U-33/γ2 cells treated with Rosi although GH binding protein and Stat 5b transcripts were unchanged over the course of these experiments (Table 2). IGFBP-1, -2, -3, -5, or -6 expression in U-33/γ2 or U-33/c cells did not change significantly at any time point. In the microarray, at 2, 24, and 72 h, Rosi had no statistically significant effect on any IGF pathway genes in the U-33/c cells.
TABLE 2.
Micro-array expression changes in components of the GH/IGF-I axis in U-33/γ2 cells treated with Rosi compared with nontreated cells
| Probe ID | IGF component | Time (h) | Fold change | P value |
|---|---|---|---|---|
| 1419519 | IGF-I | 72 | −1.75 | 0.00560 |
| 1437401 | IGF-I | 72 | −2.80 | 0.000002 |
| 1448152 | IGF-II | 72 | −3.32 | 0.0005 |
| 1426565 | IGF1R | 72 | −2.14 | 0.0018 |
| 1445660 | IGF1R | 72 | −1.44 | 0.0500 |
| 1424111 | IGF2R | 72 | −1.90 | 0.0030 |
| 1421991 | IGFBP4 | 72 | −3.00 | 0.000000 |
| 1423757 | IGFBP4 | 72 | −2.63 | 0.000020 |
| 1417962 | GHR | 72 | −3.79 | <0.00000 |
| 1451501 | GHR | 72 | −4.18 | <0.00000 |
TABLE 3.
Microarray expression changes for genes involved in the IGF-I signaling pathway in U-33/γ2 cells treated with Rosi vs. nontreated cells
| Probe ID | IGF component | Time (h) | Fold change | P value |
|---|---|---|---|---|
| 1434831 | FOXO3A | 72 | −2.15 | 0.0001 |
| 1417409 | c-Jun | 72 | −3.76 | 0.000005 |
| 1426565 | Kras | 72 | −2.57 | 0.00006 |
| 1453104 | MAPK1 | 72 | −1.63 | 0.0007 |
| 1437045 | MAPK8 | 72 | −2.00 | 0.007 |
| 1456135 | Paxillin | 72 | −3.01 | 0.000005 |
| 1422854 | SHC | 72 | −1.64 | <0.007 |
| 1418255 | SRF | 72 | −2.40 | <0.00006 |
To confirm the microarray findings, we performed real-time PCR for each component of the IGF-I regulatory system that was statistically different in the U-33/γ2 cells treated with Rosi compared with vehicle or to U-33/c cells treated in the same manner. Because of alternate promoter usage and splicing in the IGF-1 gene, we also recognized that there could be multiple transcripts differing in stability, as well as tissue-specific expression (40). In particular, high molecular weight transcripts, predominately expressed in extrahepatic tissues possess different lengths of exon 6. But because the cDNA probes present on the analyzed microarrays corresponded only to exon 6, we performed real-time PCR analysis using primers for both exon 6 and exon 4. As shown in Fig. 1, A and B, in U-33/γ2 cells, Rosi down-regulated by 5-fold the levels of transcripts which possessed exon 6 (P < 0.01) but did not change the level of exon 4 transcripts. Similarly, in comparison to U-33/c cells treated with Rosi, transcripts for IGF1R and IGFBP-4 were more than 50% suppressed in treated-U-33/γ2 cells (P < 0.01) (data not shown). Interestingly, in U-33/c cells that do not express the PPARγ2, isoform, Rosi treatment modestly increased IGF-I transcript expression for both exon 4 and exon 6 (P < 0.05) without changing IGF-IR or IGFBP-4 message (data not shown). A similar increase was noted in the microarray, but those changes were not statistically significant (P = 0.88).
Fig. 1.

IGF-I mRNA and protein expression in U-33/γ2 cells after treatment with Rosi. Cells were treated with 1 μM Rosi for 72 h followed by either RNA isolation for real-time PCR or media change to serum-free media and conditioned media were collected for detection of IGF-1 protein as described in Materials and Methods. A and B, Effect on mRNA expression assessed by real-time PCR. IGF-I mRNA expression was detected using primers specific to exon 6 (A) and exon 4 (B) and results were normalized to 1 ng 18S rRNA. C, IGF-I protein concentrations in conditioned media. Protein concentrations were normalized to the total protein content in the remaining cells. The graph represents the relative levels of IGF-I protein production (P < 0.01 for both cell types) during 48 h after media change in Rosi-treated cells vs. vehicle-treated control. Gray bars, Vehicle (DMSO); black bars, Rosi; *, P < 0.01 vs. vehicle.
Rosi down-regulates IGF-I protein in U-33/γ2 cells
Next, we asked whether changes in transcript expression were related to IGF-I protein production in the conditioned media of U-33/γ2 cells after 72 h of Rosi treatment. As shown in Fig. 1C, there was 50% reduction in IGF-I protein normalized to total protein in U-33/γ2 cells exposed to Rosi compared with no-Rosi (P < 0.01 vs. vehicle).
Rosi suppresses IGF-I expression and protein production in primary MSCs
To determine whether IGF-I was suppressed in primary bone cells by Rosi (1 μM), we harvested marrow stromal cells (MSCs) from adult (6 months) and old (24 months) B6 mice. In MSCs from adult B6 exposed to Rosi, there was a marked reduction in IGF-I mRNA (P < 0.01) containing both exon 6 and exon 4, compared with no Rosi (Fig. 2, A and B). Aged mice had markedly fewer IGF-I mRNA transcripts, compared with adults, and there was a nonsignificant reduction in those transcripts with Rosi treatment (Fig. 2A). We then measured IGF-I protein in the conditioned media collected from adult and old MSCs cultures at 24 and 48 h after treatment with Rosi. We found a similar suppression in IGF-I protein, as noted for the U-33/γ2 cells, particularly at 48 h in adult cells (P < 0.01), and a tendency for a decrease in aged mouse cells (Fig. 2C). In a manner analogous to the stable cell lines, Rosi exposure of primary MSCs from adult B6 mice resulted in significant down regulation of both IGF1R and IGFBP-4 transcripts (P < 0.01) (Fig. 3). Thus, activation of PPARγ down regulates components of the IGF regulatory system in primary MSCs.
Fig. 2.
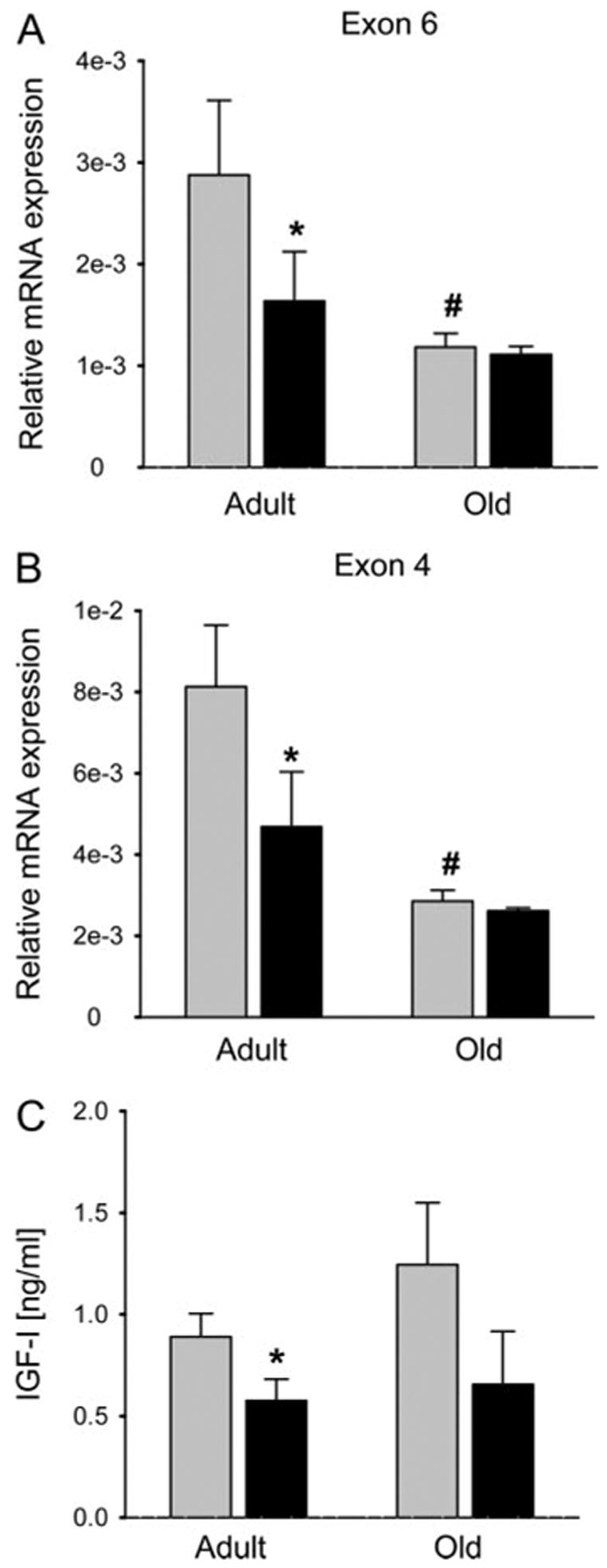
Effect of Rosi on IGF-I production in primary bone marrow cultures derived from adult (6 months) and old (24 months) C57BL/6 mice. A and B, Effect on mRNA expression assessed by real-time PCR. IGF-I mRNA expression was detected using primers specific to exon 6 (A) and exon 4 (B) and results were normalized to 1 ng 18S rRNA. B, IGF-I protein production. Primary bone marrow cultures were treated with Rosi for 72 h followed by media change to serum-free media. Conditioned media were collected after 48 h and the levels of IGF-I protein were detected as described in Materials and Methods and normalized to the protein content in remaining cells. Gray bars, Vehicle (DMSO); black bars, Rosi; *, P < 0.01 vs. vehicle; #, old vs. adult untreated; P < 0.01.
Fig. 3.
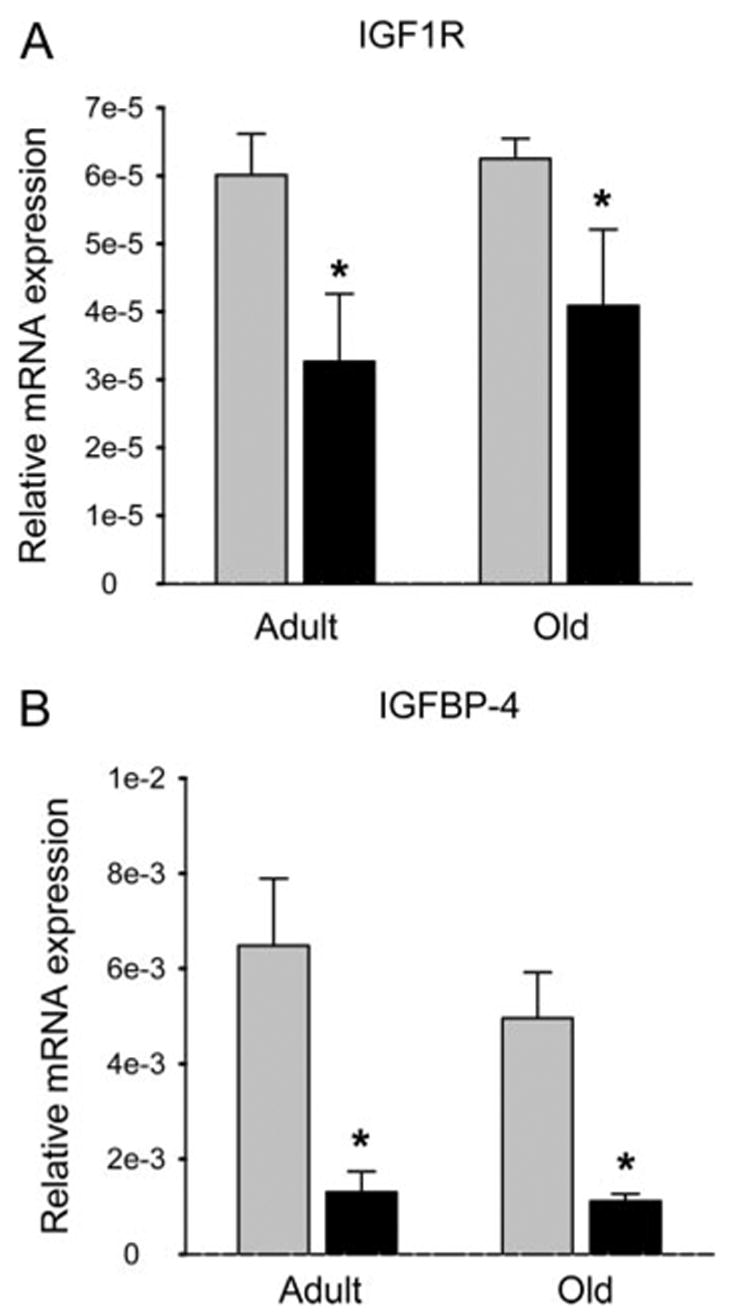
Effect of Rosi on IGF1R (A) and IGFBP-4 (B) expression in primary bone marrow cultures derived from adult (6 months) and old (24 months) C57BL/6 mice. Expression data were normalized to 1 ng 18S rRNA. Gray bars, Vehicle (DMSO); black bars, Rosi; *, P < 0.01 vs. vehicle.
Rosi suppresses IGF-I expression in mouse liver cells
Next, we asked whether Rosi suppressed IGF-I expression in a mouse liver cell line (AML). We treated AML cells at confluence with varying doses of Rosi for 48 h and measured IGF-I mRNA transcripts by RPA using 28S rRNA as a control. There was a dose-dependent decline in exon 4 IGF-I mRNA transcripts in the AML cells exposed to Rosi; i.e. there was no effect with 5 μM of Rosi, a nonsignificant 10% reduction with 10 μM of Rosi, and a 50% suppression of IGF-I mRNA (P < 0.00004) with 30 μM of Rosi (Fig. 4). There were no changes in the expression of IGFBP-3, IGFBP-2, or IGFBP-1 in response to Rosi (data not shown). Of note, the doses required for significant suppression of IGF-I were significantly greater in the AML cells than for U-33/γ2 cells or primary MSCs. Nonetheless, activation of PPARγ in vitro suppresses hepatic expression of IGF-I, but not the IGFBPs in a dose-dependent manner.
Fig. 4.
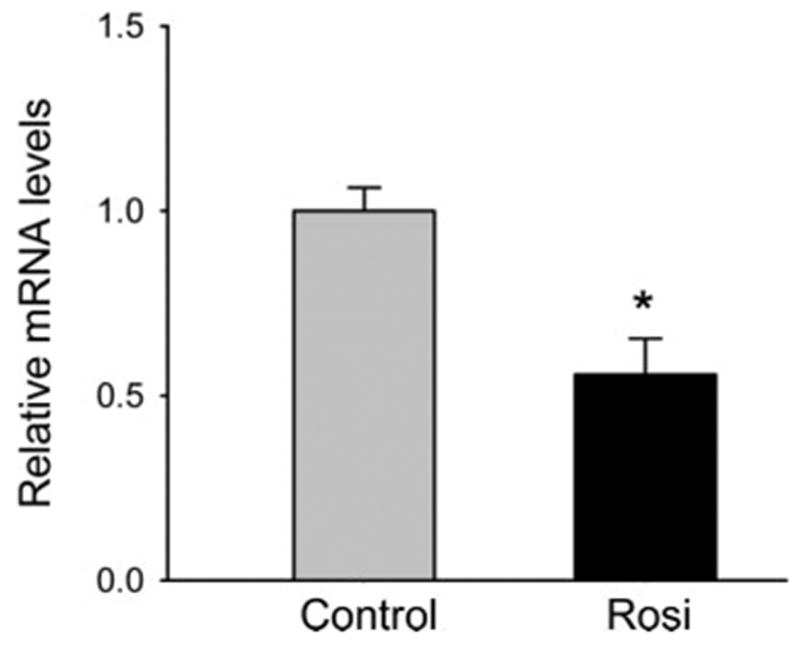
Effect of Rosi on IGF-I expression in hepatic AML12 cells. Cells were treated with 30 μM Rosi for 48 h and the level of exon 4 IGF-I mRNA expression was assessed using RPA as described (31). Gray bar, Nontreated control; black bar, cells treated with Rosi; *, P < 0.00004 vs. control. Five experiments were performed in this study. There were no significant changes in any of the predominant IGFBPs in liver including IGFBP-1,-2, or -3.
In vivo studies
Rosi treatment reduces circulating and hepatic IGF-I in intact and OVX B6 mice.
Short-term treatment
Because the majority of circulating IGF-I is derived from liver and there were significant changes in IGF-I expression in an hepatic cell line after exposure to Rosi, we then asked whether short-term or long term Rosi treatment would suppress circulating IGF-I and/or IGFBPs in mice (41). We administered Rosi (20 mg/kg·d) to female B6 mice (n = 14) at 9 wk of age for 4 d and compared them with control female B6 mice (n = 10) fed the same diet (NIH 31) but no active treatment for the same duration. After 4 d, serum levels of IGF-I in the Rosi-treated mice were 15% lower than controls (C, 380 ± 15 vs. Rosi, 330 ± 10 ng/ml; P < 0.05). At the time the mice were killed, livers from treated (Rosi) and nontreated mice (n = 4), revealed significantly lower IGF-I transcripts (i.e. exon 4) in the Rosi group (i.e. 2.4-fold reduction vs. controls; P = 0.04). On the other hand, IGFBP-1, -2, -3, or -5 mRNA levels by real-time PCR did not differ from untreated animals nor did Stat 5b, a transcription factor induced by GH. Like the liver, IGF-I transcripts in subcutaneous fat were also suppressed by nearly 90% (P < 0.15) in mice treated with Rosi compared with controls. We also noted lower IGR1R expression (i.e. 2.9-fold reduced vs. control) in the peripheral fat of Rosi-treated mice, but this change did not reach statistical significance (P = 0.25).
Next, to determine whether estrogen depletion affected the circulating IGF-I response to Rosi, we performed a second experiment, in which we took four groups of 6-month-old B6 female mice (n = 6 each), created two OVX groups, and treated two groups (sham and OVX) with Rosi 20 mg/kg·d × 4 d. We found, as expected, that serum IGF-I rose in the OVX group not treated with Rosi from 280 ± 10 ng/ml to 400 ± 25 ng/ml (P < 0.01). However, OVX mice treated with Rosi had serum levels of IGF-I similar to sham operated B6 mice and these were suppressed compared with OVX mice without Rosi (i.e. Rosi-treated OVX, 300 ± 12 ng/ml vs. OVX without treatment, 400 ± 25 ng/ml; P < 0.01). Finally, we performed a similar study that was longitudinal in nature. Ten female B6 mice at 8 wk of age were treated with Rosi 20 mg/kg·d for 4 d; eyebleeds were performed at the beginning and at the end of the study, specifically for IGF-I measurements. Serum IGF-I in the treated mice fell 25% from 303.5 ± 6.9 ng/ml to 225 ± 6.95 ng/ml (P < 0.0001) after only 4 d. Thus, Rosi reduces hepatic IGF-I expression and circulating IGF-I with short-term exposure in both intact and OVX B6 mice.
Long-term treatment
To test whether chronic activation of PPARγ would influence circulating IGF-I, post weaning 3-wk-old female B6 mice (n = 7) were treated with or without Rosi (20 mg/kg·d) on an NIH31 diet for 9 wk. Serum IGF-I at the end of the study was significantly lower in B6 Rosi-treated mice vs. controls (Rosi: 195 ± 12 vs. C: 260 ± 13 ng/ml, P < 0.01). Thus, we established that young (i.e. age 3–12 wk) B6 mice show suppression of circulating IGF-I in response to activation of PPARγ by Rosi.
Rosi treatment of postmenopausal women reduces circulating IGF-I but not IGFBP-3 nor IGFBP-2
To test the clinical relevance of our findings in mice, we measured IGF-I in women enrolled in a recently completed randomized, placebo-controlled trial examining the effects of Rosi on bone turnover markers. Fifty women who were more than 5 yr after menopause were randomized to 8 mg of Rosi (n = 25) or placebo (n = 25) for 16 wk. Forty-four women completed the study. At baseline, serum IGF-I did not differ between groups (C, 112 ± 13 ng/ml vs. Rosi, 107 ± 10 ng/ml; P = 0.90). Sixteen weeks later, serum IGF-I in the control group remained steady (C:104.0 ± 10.0 ng/ml, P = 0.90 vs. baseline), but the Rosi-treated women showed a significant decline (i.e. −25%) in serum IGF-I (Rosi, 82.5 ± 8.1 ng/ml vs. baseline: 107 ± 10 ng/ml, P < 0.05; and vs. control at 8 wk: 103.9 ± 10 ng/ml; P < 0.07). At the completion of the study, the relative difference in serum IGF-I between placebo and Rosi-treated women was −14.1% (P < 0.15). During the course of this study, neither serum IGFBP-3 nor serum IGFBP-2 changed in the placebo or Rosi-treated women. In sum, the effects of Rosi on circulating IGF-I in humans for a short course are comparable to the in vivo effects of Rosi in intact and OVX B6 mice.
Discussion
In the current study, we found that activation of PPARγ by Rosi, in primary and transformed MSCs, and in mouse liver cells, suppressed expression of IGF-I. Moreover, Rosi reduced expression of the IGF1R and IGFBP-4 in primary adult MSCs. Similarly, in mice and in postmenopausal women, we noted that Rosi treatment reduced circulating IGF-I concentrations by as much as 25%. The IGF-I circulatory changes occurred as early as 4 d after initiation of treatment, and were associated with reduced IGF-I transcripts in liver and peripheral fat. As such, these data raise a number of provocative questions, not only in respect to the effects of PPARγ activation on bone turnover, but also in regards to the mechanism of action of the PPARγ agonists.
This series of investigations started after we identified a major QTL for BMD and serum IGF-I between two inbred strains of mice, B6 and C3H (C3H/HeJ) (26, 42). This QTL was located on mouse chromosome 6 in the region of the Pparγ gene (26). To test the biological effect of this QTL, we generated a congenic mouse (i.e. 6T) that carried C3H genes from this locus in a pure B6 background (43). The resultant skeletal phenotype of 6T included low trabecular bone mass, increased marrow adiposity but enhanced insulin sensitivity, and low serum IGF-I (43). Subsequent expression profiling of the liver and marrow revealed a pattern consistent with endogenous PPARγ activation (24, 44). To pursue this further, we then asked whether exogenous activation of PPARγ could affect the IGF regulatory system in bone and liver.
PPARγ activation could have a deleterious effect on bone through a number of possible mechanisms. First, Rosi can directly suppress OB differentiation genes such as Runx2, Dlx5, and osterix, as nicely demonstrated in our microarray experiments (Table 1) (9, 11, 19). Second, work by our group and other investigators suggest there is an inverse relationship between the Wnt/β-catenin signaling pathway and PPARγ activation (12, 13, 18, 45–47). Third, we now show that IGF-I is also down regulated in marrow stromal cells by Rosi, as is the other important IGF ligand in bone, IGF-II. Changes in the bone IGF regulatory system in response to TZDs could affect the ultimate fate of pre-OBs, probably in combination with other key differentiation factors such as Runx2, osterix, and BMP4 (see Table 1). For example, Zhang et al. (48) demonstrated that targeted deletion of the IGF1R in OBs, using the Cre-loxP system with an osteocalcin promoter, markedly impaired two processes in late OB differentiation, matrix formation and mineralization. On the other hand, Jiang et al. (49) demonstrated that targeted over-expression of IGF-I using a 3.6 Col1A1 promoter, resulted in greatly enhanced bone turnover, implying that IGF-I was important in pre-OB recruitment and may be a determinant of lineage allocation. Interestingly, aged B6 mice expressed significantly less IGF-I mRNA than adult B6 mice, and the suppressive effect of Rosi on these cells was also dampened (see Fig. 2). Because “old” MSCs have enhanced PPARγ2 expression, it is interesting to speculate whether aging may affect IGF-I expression through this nuclear receptor activation (22).
Another provocative aspect of these data is the acute suppression of IGF-I transcripts from the liver by 4 d of Rosi administration. These findings were confirmed in vitro by demonstrating that Rosi reduced IGF-I mRNA by 50% in AML cells and in vivo by demonstrating lower levels of circulating IGF-I without changes in the IGFBPs. It is conceivable that the suppressive effects of TZDs on serum IGF-I could not only contribute to the deleterious skeletal changes, but also to the antiproliferative and antineoplastic properties of these agents. In fact, we found several cell cycle genes in MSCs suppressed by activation of PPARγ, some as early as 24 h after exposure. These findings are consistent with other reports which have noted the importance of cell cycle arrest in the antiproliferative response to PPARγ agonists (3, 5–8, 50). The mechanisms responsible for these changes are likely to be complex but may be partially mediated through suppression of IGF1R expression and its downstream signaling cascade (see Table 2). However, more studies are needed to delineate the exact role of PPARγ agonists in slowing cell proliferation and to define the proportion of change in cell proliferation that can be related to alterations in the IGF-I signaling network.
Several lines of evidence from our studies suggest that Rosi directly suppresses liver IGF-I gene expression, rather than acting through the GH/IGF-I axis. First, we found reduced hepatic IGF-I transcripts in AML cells treated with Rosi, independent of GH. Similarly, we found markedly suppressed IGF-I liver transcripts (i.e. ~60%) in B6 mice exposed to Rosi for as little as 4 d, without changes in Stat5b, a major transcription factor induced by GH (50). Second, serum IGFBP-3 did not change over the 16 wk of Rosi treatment despite a drop of nearly 25% in IGF-I at 8 wk. And, there was no effect of Rosi on IGFBP-3 expression in MSCs in vitro. IGFBP-3 is induced by GH, hence any significant change in its secretion should be reflected in ambient IGFBP-3 concentrations or tissue expression. Our results are similar to those of Bell et al. (51), who found that women with polycystic ovary syndrome treated with Rosi had significant suppression of serum IGF-I but not IGFBP-3. Third, we found in the microarray experiments that the GHR was actually up-regulated by exposure of U-33/γ2 cells to Rosi. As such, these changes may have clinical significance. A drop in circulating IGF-I levels could negatively impact the skeleton, particularly in younger individuals who may be insulin resistant yet are still acquiring peak BMD (41). On the other hand, low serum IGF-I concentrations could limit cell proliferation and enhance apoptosis in specific target tissues. It will be interesting to see whether changes in serum IGF-I can predict the effects of Rosi on a host of proliferative disorders.
There are several limitations to this study. First, we only examined one inbred strain, B6, to determine the effects of Rosi on IGF-I. It is conceivable that there are strain-related differences in the IGF-I response to activation of PPARγ, particularly in light of our previous genetic studies in B6 and C3H mice (43, 44, 52, 53). Another limitation is that we chose MSCs rather than OBs to study the effects of Rosi. Hence, we do not know whether TZDs can impact the differentiative function of mature OBs, particularly matrix production and mineralization. In addition, we have not explored the mechanism responsible for enhanced IGF-I expression in MSCs that do not have endogenous PPARγ2 activity (i.e. U-33/c cells), although unlike U-33/γ2 cells, these cells are biologically unique in that they cannot differentiate into adipocytes. Also, there is emerging evidence that not all TZDs work on adipocytes and OBs in the same manner (21). Studies with other ligand agonists should provide further insight as to whether the IGF suppressive effects are a class property or unique to Rosi. A limitation to the human study was the small number of subjects and the duration of the trial was short (i.e. 16 wk). It is uncertain whether suppression of IGF-I is persistent with chronic Rosi therapy, or whether GH secretion is enhanced by the drop in IGF-I, resulting in restoration of serum IGF-I levels. That scenario is plausible, particularly because we showed that Rosi enhances expression of the GHR in MSCs at 24 and 72 h. However, suppression of the IGF1R by Rosi means that even with restoration of circulating levels, the bioactivity of IGF-I could be compromised.
In conclusion, we established that activation of PPARγ suppresses elements of the IGF regulatory system in pre-OBs and in liver. This effect is evident within 72 h and occurs at the transcriptional and/or posttranscriptional levels, affecting both IGF-I mRNA and protein production. Similar changes occur in vivo as early as 96 h after exposure. The downstream consequences of reduced expression of IGF-I, its receptor, and several signaling factors, may contribute to the unique properties of the TZDs. Further studies should provide greater insight into the role IGF-I plays in mediating the antiosteoblastic actions of agents such as Rosi.
Abbreviations
- AML12
α Mouse liver 12
- BMD
bone mineral density
- BV/TV
trabecular bone volume
- DMSO
dimethylsulfoxide
- GHR
GH receptor
- IGFBP
IGF binding protein
- IGF1R and IGF2R
type I and II IGF receptor
- IPKB
Ingenuity Pathways Knowledge Base
- MSCs
pluripotent bone marrow stromal/stem cells
- OB
osteoblast
- OVX
ovariectomized
- PPAR
peroxisome proliferator-activated receptor
- QTL
quantitative trait locus
- RPA
ribonuclease protection assay
- Rosi
rosiglitazone
- TZDs
thiazolidinediones
Footnotes
Part of this work was presented at the 88th Annual Meeting of The Endocrine Society, June 2005.
Disclosure Summary: The authors have nothing to declare.
This work was supported by National Institutes of Health: National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR45433; National Institute on Aging Grant AG17482, and American Diabetes Association Grant 1-03-RA-46 and a Fulbright Fellowship for V.M.
References
- 1.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 2.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 3.Theocharis S, Margeli A, Vielh P, Kouraklis G. Peroxisome proliferator-activated receptor-γ ligands as cell-cycle modulators. Cancer Treat Rev. 2004;30:545–554. doi: 10.1016/j.ctrv.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Lehrke M, Lazar MA. The many faces of PPARγ. Cell. 2005;123:993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 5.Freudlsperger C, Moll I, Schumacher U, Thies A. Anti-proliferative effect of peroxisome proliferator-activated receptor γ agonists on human malignant melanoma cells in vitro. Anticancer Drugs. 2006;17:325–332. doi: 10.1097/00001813-200603000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Aiello A, Pandini G, Frasca F, Conte E, Murabito A, Sacco A, Genua M, Vigneri R, Belfiore A. Peroxisomal proliferator-activated receptor-γ agonists induce partial reversion of epithelial-mesenchymal transition in anaplastic thyroid cancer cells. Endocrinology. 2006;147:4463–4475. doi: 10.1210/en.2005-1610. [DOI] [PubMed] [Google Scholar]
- 7.He G, Sung YM, Digiovanni J, Fischer SM. Thiazolidinediones inhibit insulin-like growth factor-I-induced activation of p70S6 kinase and suppress insulin-like growth factor-I tumor-promoting activity. Cancer Res. 2006;66:1873–1878. doi: 10.1158/0008-5472.CAN-05-3111. [DOI] [PubMed] [Google Scholar]
- 8.Kim KY, Kim SS, Cheon HG. Differential anti-proliferative actions of peroxisome proliferator-activated receptor-γ agonists in MCF-7 breast cancer cells. Biochem Pharmacol. 2006;72:530–540. doi: 10.1016/j.bcp.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Rzonca SO, Suva LJ, Gaddy D, Montague DC, Lecka-Czernik B. Bone is a target for the antidiabetic compound rosiglitazone. Endocrinology. 2004;145:401–406. doi: 10.1210/en.2003-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soroceanu MA, Miao D, Bai XY, Su H, Goltzman D, Karaplis AC. Rosiglitazone impacts negatively on bone by promoting osteoblast/osteocyte apoptosis. J Endocrinol. 2004;183:203–216. doi: 10.1677/joe.1.05723. [DOI] [PubMed] [Google Scholar]
- 11.Ali AA, Weinstein RS, Stewart SA, Parfitt AM, Manolagas SC, Jilka RL. Rosiglitazone causes bone loss in mice by suppressing osteoblast differentiation and bone formation. Endocrinology. 2005;146:1226–1235. doi: 10.1210/en.2004-0735. [DOI] [PubMed] [Google Scholar]
- 12.Akune T, Ohba S, Kamekura S, Yamaguchi M, Chung UI, Kubota N, Terauchi Y, Harada Y, Azuma Y, Nakamura K, Kadowaki T, Kawaguchi H. PPARγ insufficiency enhances osteogenesis through osteoblast formation from bone marrow progenitors. J Clin Invest. 2004;113:846–855. doi: 10.1172/JCI19900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD, MacDougald OA. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci USA. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz AV, Sellmeyer DE, Vittinghoff E, Palermo L, Lecka-Czernik B, Feingold KR, Strotmeyer ES, Resnick HE, Carbone L, Beamer BA, Park SW, Lane NE, Harris TB, Cummings SR. Thiazolidinedione use and bone loss in older diabetic adults. J Clin Endocrinol Metab. 2006;91:3349–3354. doi: 10.1210/jc.2005-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 16.Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, Hopkins N, Yaffe MB. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 17.Gimble JM, Robinson CE, Wu X, Kelly KA. The function of adipocytes in the bone marrow stroma: an update. Bone. 1996;19:421–428. doi: 10.1016/s8756-3282(96)00258-x. [DOI] [PubMed] [Google Scholar]
- 18.Mulholland DJ, Dedhar S, Coetzee GA, Nelson CC. Interaction of nuclear receptors with the Wnt/β-catenin/Tcf signaling axis: Wnt you like to know? Endocr Rev. 2005;26:898–915. doi: 10.1210/er.2003-0034. [DOI] [PubMed] [Google Scholar]
- 19.Lecka-Czernik B, Gubrij I, Moerman EJ, Kajkenova O, Lipschitz DA, Manolagas SC, Jilka RL. Inhibition of Osf2/Cbfa1 expression and terminal osteoblast differentiation by PPARγ2. J Cell Biochem. 1999;74:357–371. [PubMed] [Google Scholar]
- 20.Lecka-Czernik B, Moerman EJ, Grant DF, Lehmann JM, Manolagas SC, Jilka RL. Divergent effects of selective peroxisome proliferator-activated receptor-γ 2 ligands on adipocyte versus osteoblast differentiation. Endocrinology. 2002;143:2376–2384. doi: 10.1210/endo.143.6.8834. [DOI] [PubMed] [Google Scholar]
- 21.Lazarenko OP, Rzonca SO, Suva LJ, Lecka-Czernik B. Netoglitazone is a PPAR-γ ligand with selective effects on bone and fat. Bone. 2006;38:74–84. doi: 10.1016/j.bone.2005.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moerman EJ, Teng K, Lipschitz DA, Lecka-Czernik B. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-γ2 transcription factor and TGF-β/BMP signaling pathways. Aging Cell. 2004;3:379–389. doi: 10.1111/j.1474-9728.2004.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Botolin S, Faugere MC, Malluche H, Orth M, Meyer R, McCabe LR. Increased bone adiposity and peroxisomal proliferator-activated receptor-γ2 expression in type I diabetic mice. Endocrinology. 2005;146:3622–3631. doi: 10.1210/en.2004-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouxsein ML, Uchiyama T, Rosen CJ, Shultz KL, Donahue LR, Turner CH, Sen S, Churchill GA, Muller R, Beamer WG. Mapping quantitative trait loci for vertebral trabecular bone volume fraction and microarchitecture in mice. J Bone Miner Res. 2004;19:587–599. doi: 10.1359/JBMR.0301255. [DOI] [PubMed] [Google Scholar]
- 25.Beamer WG, Shultz KL, Donahue LR, Churchill GA, Sen S, Wergedal JR, Baylink DJ, Rosen CJ. Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res. 2001;16:1195–1206. doi: 10.1359/jbmr.2001.16.7.1195. [DOI] [PubMed] [Google Scholar]
- 26.Rosen CJ, Churchill GA, Donahue LR, Shultz KL, Burgess JK, Powell DR, Ackert C, Beamer WG. Mapping quantitative trait loci for serum insulin-like growth factor-1 levels in mice. Bone. 2000;27:521–528. doi: 10.1016/s8756-3282(00)00354-9. [DOI] [PubMed] [Google Scholar]
- 27.Lecka-Czernik B, Beamer W, Ackert-Bicknell C, Schwartz A, Rosen CJ. Activation of PPAR-y2 down regulates hepatic and skeletal IGF-I in vivo and in viro: a potential pathogenic mechanism in age-related osteoporosis. Endocrinology (Suppl) 2005;107 [Google Scholar]
- 28.Akilesh S, Petkova S, Sproule TJ, Shaffer DJ, Christianson GJ, Roopenian D. The MHC class I-like Fc receptor promotes humorally mediated autoimmune disease. J Clin Invest. 2004;113:1328–1333. doi: 10.1172/JCI18838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akilesh S, Shaffer DJ, Roopenian D. Customized molecular phenotyping by quantitative gene expression and pattern recognition analysis. Genome Res. 2003;13:1719–1727. doi: 10.1101/gr.533003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- 31.Delahunty KM, Shultz KL, Gronowicz GA, Koczon-Jaremko B, Adamo ML, Horton LG, Lorenzo J, Donahue LR, ckert-Bicknell C, Kream BE, Beamer WG, Rosen CJ. Congenic mice provide in vivo evidence for a genetic locus that modulates serum insulin-like growth factor-I and bone acquisition. Endocrinology. 2006;147:3915–3923. doi: 10.1210/en.2006-0277. [DOI] [PubMed] [Google Scholar]
- 32.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 33.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Churchill GA. Using ANOVA to analyze microarray data. Biotechniques. 2004;37:173–175. doi: 10.2144/04372TE01. [DOI] [PubMed] [Google Scholar]
- 35.Wu H, Kerr K, Churchill GA. MAANOVA: a software package for the analysis of spotted cDNA Microarray Experiements. In: Parmigiani G, Garrett ES, Irizarry RA, Zeger SL, editors. The analysis of gene expression data: an overview of methods and software. New York: Springer; 2003. pp. 313–431. [Google Scholar]
- 36.Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- 37.Storey JD. A direct approach to false discovery rates. J R Statistical Soc B. 2002;64:479–498. [Google Scholar]
- 38.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeRoith D, Werner H, Faria TN, Kato H, Adamo M, Roberts CT., Jr Insulin-like growth factor receptors. Implications for nervous system function. Ann NY Acad Sci. 1993;692:22–32. doi: 10.1111/j.1749-6632.1993.tb26202.x. [DOI] [PubMed] [Google Scholar]
- 41.Yakar S, Rosen CJ, Beamer WG, Ackert-Bicknell CL, Wu Y, Liu JL, Ooi GT, Setser J, Frystyk J, Boisclair YR, LeRoith D. Circulating levels of IGF-1 directly regulate bone growth and density. J Clin Invest. 2002;110:771–781. doi: 10.1172/JCI15463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosen CJ, Dimai HP, Vereault D, Donahue LR, Beamer WG, Farley J, Linkhart S, Linkhart T, Mohan S, Baylink DJ. Circulating and skeletal insulin-like growth factor-I (IGF-I) concentrations in two inbred strains of mice with different bone mineral densities. Bone. 1997;21:217–223. doi: 10.1016/s8756-3282(97)00143-9. [DOI] [PubMed] [Google Scholar]
- 43.Rosen CJ, Ackert-Bicknell CL, Adamo ML, Shultz KL, Rubin J, Donahue LR, Horton LG, Delahunty KM, Beamer WG, Sipos J, Clemmons D, Nelson T, Bouxsein ML, Horowitz M. Congenic mice with low serum IGF-I have increased body fat, reduced bone mineral density, and an altered osteoblast differentiation program. Bone. 2004;35:1046–1058. doi: 10.1016/j.bone.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 44.Rosen CJ, Ackert-Bicknell C, Beamer WG, Nelson T, Adamo M, Cohen P, Bouxsein ML, Horowitz MC. Allelic differences in a quantitative trait locus affecting insulin-like growth factor-I impact skeletal acquisition and body composition. Pediatr Nephrol. 2005;20:255–260. doi: 10.1007/s00467-004-1612-z. [DOI] [PubMed] [Google Scholar]
- 45.Jansson EA, Are A, Greicius G, Kuo IC, Kelly D, Arulampalam V, Pettersson S. The Wnt/β-catenin signaling pathway targets PPARγ activity in colon cancer cells. Proc Natl Acad Sci USA. 2005;102:1460–1465. doi: 10.1073/pnas.0405928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moldes M, Zuo Y, Morrison RF, Silva D, Park BH, Liu J, Farmer SR. Peroxisome-proliferator-activated receptor γ suppresses Wnt/β-catenin signalling during adipogenesis. Biochem J. 2003;376:607–613. doi: 10.1042/BJ20030426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lecka-Czernik B, Lazarenko OP, Czernik P, Rosen C, Shockley K, Churchill G. Osteoblastic activity of Wnt signaling pathway is controlled by PPAR-y2 nuclear receptor. J Bone Miner Res. 2006;21:S382. [Google Scholar]
- 48.Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, Malluche H, Zhao G, Rosen CJ, Efstratiadis A, Clemens TL. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 49.Jiang J, Lichtler AC, Gronowicz GA, Adams DJ, Clark SH, Rosen CJ, Kream BE. Transgenic mice with osteoblast-targeted insulin-like growth factor-I show increased bone remodeling. Bone. 2006;39:494–504. doi: 10.1016/j.bone.2006.02.068. [DOI] [PubMed] [Google Scholar]
- 50.Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine Growth Factor Rev. 1999;10:131–157. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 51.Belli SH, Graffigna MN, Oneto A, Otero P, Schurman L, Levalle OA. Effect of rosiglitazone on insulin resistance, growth factors, and reproductive disturbances in women with polycystic ovary syndrome. Fertil Steril. 2004;81:624–629. doi: 10.1016/j.fertnstert.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 52.Beamer WG, Donahue LR, Rosen CJ, Baylink DJ. Genetic variability in adult bone density among inbred strains of mice. Bone. 1996;18:397–403. doi: 10.1016/8756-3282(96)00047-6. [DOI] [PubMed] [Google Scholar]
- 53.Drake TA, Schadt E, Hannani K, Kabo JM, Krass K, Colinayo V, Greaser LE3, Goldin J, Lusis AJ. Genetic loci determining bone density in mice with diet-induced atherosclerosis. Physiol Genom. 2001;5:205–215. doi: 10.1152/physiolgenomics.2001.5.4.205. [DOI] [PubMed] [Google Scholar]


