Abstract
The halogen bond, a noncovalent interaction involving polarizable chlorine, bromine, or iodine molecular substituents, is now being exploited to control the assembly of small molecules in the design of supramolecular complexes and new materials. We demonstrate that a halogen bond formed between a brominated uracil and phosphate oxygen can be engineered to direct the conformation of a biological molecule, in this case to define the conformational isomer of a four-stranded DNA junction when placed in direct competition against a classic hydrogen bond. As a result, this bromine interaction is estimated to be ≈2–5 kcal/mol stronger than the analogous hydrogen bond in this environment, depending on the geometry of the halogen bond. This study helps to establish halogen bonding as a potential tool for the rational design and construction of molecular materials with DNA and other biological macromolecules.
Keywords: biomolecular engineering, DNA structure, molecular interactions
Halogen bonds have recently seen a resurgence of interest as a tool for “bottom-up” molecular design. Chlorines, bromines, and iodines in organic and inorganic compounds are known to polarize along their covalent bonds to generate an electropositive crown; the halogen thus acts as a Lewis acid to pair with Lewis bases, including oxygens and nitrogens. These electrostatic pairs, originally called charge-transfer bonds (1), are now known as halogen bonds (X-bonds), recognizing their similarities to hydrogen bonds (H-bonds) in their strength and directionality (2). In chemistry, X-bonds are being exploited in the design and engineering of supramolecular assemblies (3) and molecular crystals (for review, see ref. 4), with an iodine X-bond estimated to be ≈3.5 kcal/mol more stable than an O–H⋯O H-bond in organic crystals (5).
The X-bond, however, has not generally been a part of the biologist's lexicon. Although halogens are widely used in drug design and to probe molecular interactions, X-bonds have only recently been recognized as a distinct interaction in ligand recognition and molecular folding and in the assembly of proteins and nucleic acids (6, 7). With the growing application of biological molecules (biomolecule), particularly nucleic acids (for review, see ref. 8), in the design of nanomechanical devices, we ask here whether specific X-bonds can be engineered to direct conformational switching in a biomolecule.
To compare X- and H-bonds in the complex environment of a biomolecule, we have designed a crystallographic assay to determine whether an intramolecular X-bond could be engineered to direct the conformational isomerization of a DNA Holliday junction by competing an X-bond against a classic H-bond and, consequently, we are able to compare the stabilization energies afforded by these two types of interactions. The stacked-X form of the DNA Holliday junction (Fig. 1), seen in high-salt solutions (9) and in crystal structures (10–12), is a simple and well controlled biomolecular assay system that can isomerize between two nearly isoenergetic and structurally similar conformers (Fig. 2) (13–15). The presence of an X- or H-bond at the junction cross-over will be reflected, in this assay, by the preference for one of the two isomer forms.
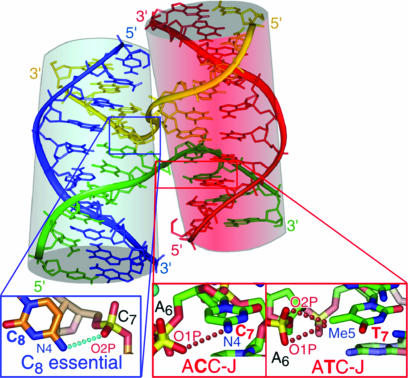
Fig. 1.
Structure of the stacked-X DNA Holliday junction. The structure of d(CCGGTACCGG) (ACC-J) as a four-stranded junction (11) is shown with the inside cross-over strands colored in yellow and green and the outside noncrossing strands in blue and red. The pairs of stacked duplex arms are highlighted with cylinders. Details of the molecular interactions that stabilize junctions are in crystals are shown, with the essential H-bond from the C8 cytosine to the phosphate of the cross-over C7 nucleotide in the blue box, and the weaker H-bond from C7 to A6 in the ACC-J or the weak electrostatic interaction from the methyl of T7 to A6 in d(CCGATATCGG) (ATC-J) in the red boxes (12, 16).
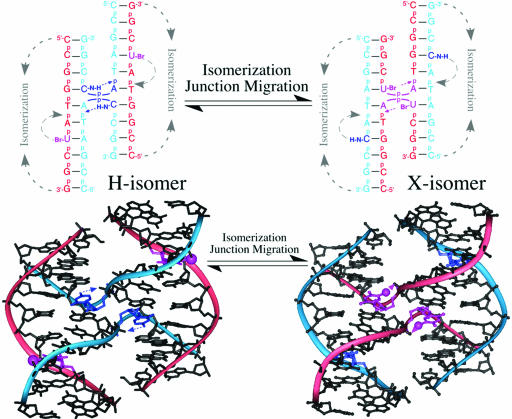
Fig. 2.
Assay for competing X- against H-bonds. The isomeric forms of the stacked-X junction, resulting from restacking of the arms and migration of the junction (top), place an H-bond (H-isomer) or X-bond (X-isomer) at the junction cross-over. The crystal structures of H2J in the H-isomer (with its H-bonding interaction) and Br2J in the X-isomer (with its X-bond) are shown below. Bromines have been modeled at the outside strand of the H2J structure to indicate their positions, if present, in the H-isomer of Br2J.
A cytosine to phosphate H-bond at the N7 nucleotide position has been shown to help stabilize the junction in the sequence d(CCGGTACCGG) (ACC-J) in crystals (11, 12, 16) and in solution (17). The stability of this junction, formed by a single inverted repeat sequence, depends entirely on this set of intramolecular interactions. The brominated junctions of the current competition assay can adopt a conformer (the H-isomer) that is stabilized by a similar cytosine to phosphate H-bond. Alternatively, they can adopt a conformer (the X-isomer) that is stabilized by an X-bond from a BrUra that replaces the cytosine (Fig. 2). Thus, the isomer form of the junction will be explicitly identified by the position of the bromine: if the BrUra sits at the inside crossing strands, then the junction is the X-isomer, but if it sits at the outside strand, it is the H-isomer. Furthermore, the distribution of X- and H-isomers observed in the crystal will reflect the differences in the overall energies and, by extension, the differences in energies of the intramolecular interactions that stabilize these isomers, assuming identical crystal lattice energies for the conformers.
Results
We have designed three DNA constructs (Table 1) in which bromine X-bonds compete against H-bonds in ratios of 2X:2H (Br2J), 1X:2H (Br1J), and 0X:2H (H2J) at the cross-over to drive the isomeric form of the resulting stacked-X junction. The single-crystal structure directly identifies the isomeric form and the stabilizing interactions for each conformer (Fig. 2). For the Br2J and Br1J constructs, a bromine at the inside cross-over strand would indicate formation of the X-isomer, whereas a bromine at the outside strand would be indicative of the H-isomer. Because the crystals of the three constructs are isomorphous (Table 2) and the sites of stabilizing interactions distant from any direct intermolecular DNA lattice contacts, we can assume that the isomer preference is not defined by the crystal lattice. Therefore, the ratio of H- to X-isomers in the crystals reflects the differences in energy between the H- and X-bonding interactions. The sequence-dependent formation of DNA junctions in solution has been directly correlated to the structure and molecular interactions seen in crystals (17), indicating that the properties of the DNA junction in crystals closely mirror those in solution and are not explicitly induced by the crystal; the crystal structure, therefore, is a means by which we can directly identify the type of stabilizing interaction for a particular conformer form of each DNA construct.
Table 1.
Constructs and sequences that compete halogen bonds against hydrogen bonds in DNA junctions
| Construct | Sequences* | X:H |
|---|---|---|
| Br2J | d(CCGGTAbrUCGG)2/d(CCGATACCGG)2 | 2:2 |
| Br1J | d(CCGGTAbrUCGG)/d(CCGGTAUCGG)/d(CCGATACCGG)2 | 1:2 |
| H2J | d(CCGGTAUCGG)2/d(CCGATACCGG)2 | 0:2 |
The ratios of halogen to hydrogen bonds that potentially compete in each construct are listed as X:H.
*BrU, 5-bromouracil.
Table 2.
Crystallographic and geometric parameters for the Holliday junction constructs Br2J, H2J, and Br1J (for sequences, see Table 1)
| Junction | Br2J | H2J | Br1J |
|---|---|---|---|
| Crystallographic parameters | |||
| Space group | C2 | C2 | C2 |
| Unit cell | |||
| a, Å | 65.78 | 65.61 | 65.89 |
| b, Å | 24.41 | 24.17 | 24.21 |
| c, Å | 37.32 | 36.96 | 37.29 |
| β-angle | 111.07° | 110.01° | 111.02° |
| Unique reflections (for refinement) | 3,362 (3,200) | 4,176 (3,992) | 4,350 (4,247) |
| Resolution, Å | 2.00 | 1.90 | 1.85 |
| Completeness, %* | 82.8 (49.2) | 89.6 (63.4) | 87.6 (61.7) |
| I/sig, I* | 13.70 (2.31) | 20.41 (3.76) | 21.18 (7.73) |
| Rmerge, %* | 4.9 (28.1) | 5.1 (25.2) | 3.2 (9.3) |
| Refinement statistics | |||
| Rcryst (Rfree), % | 21.8 (27.2) | 21.8 (26.8) | 22.3 (24.9) |
| No. of atoms: DNA (solvent) | 404 (106) | 403 (101) | 405 (102) |
| <B-factor > DNA (solvent) | 13.1 (15.6) | 16.3 (20.9) | 15.4 (17.5) |
| rmsd bond length, Å | 0.004 | 0.007 | 0.009 |
| rmsd bond angle | 0.8° | 1.1° | 1.4° |
| PDB ID codes | 2ORG | 2ORH | 2ORF |
| Junction geometry | |||
| Jtwist | 40.2° | 39.6° | 42.4° |
| Jroll | 142.5° | 137.9° | 138.6° |
| Jslide, Å | 0.21 | 1.13 | 1.10 |
| rmsd relative to Br2J, Å | 0.00 | 0.71 | 0.49 |
Listed are the crystallographic and refinement parameters, and the geometric parameters that describe the rotation of the stacked duplex arms across the junction (Jtwist), and the rotation (Jroll) and translation (Jslide) of each set of stacked arms along the respective helical axes (24, 26) for each junction construct.
*Values for the highest resolution shell are shown in parentheses.
The Br2J construct shows that two X-bonds compete effectively against two classic H-bonds in the DNA junction. This structure (Fig. 2) was initially refined without specifying the nucleotides at the N7 or complementary N4 positions of the strands. The strong positive difference density adjacent to the C5 carbon of the N7 pyrimidine showed that the bromines sit at the junction cross-overs, leaving the H-bonding cytosine at the outside strands (Fig. 3a). The short distance (2.87 Å) and approximately linear alignment between the bromines and the phosphate oxygens between T5 and A6 of the crossing strandsand the orientation of the halogens toward the nonbonding electrons of the oxygens are all hallmarks of a strong X-bond (4, 6, 7), indicating that Br2J adopts the X-isomer induced by an X-bond.
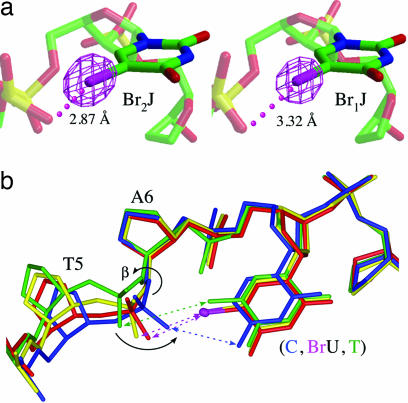
Fig. 3.
Geometries of X-bonds in Br2J and Br1J. (a) Omit electron density maps contoured at 5σ comparing geometries at the tight U-turns of the Br2J and Br1J junctions. Closest distances from the bromines to the X-bonded phosphate oxygens are labeled. (b) Overlay of all common DNA atoms for nucleotides N5, N6, and N7 at the core of the junctions of Br2J (red), Br1J (yellow), H2J (blue), and the previously published structure of ATC-J (green). Conformational rearrangements are seen at the N5 nucleotide to allow rotation of the phosphate to form a weak electrostatic interaction (green arrow) with the methyl group of T7 in ATC-J, halogen bonds (magenta arrows) to the bromines in Br2J and Br1J, and a hydrogen bond (blue arrow) to the amino group of C7 in H2J.
The H2J structure (Fig. 2) shows that the H-isomer can be accommodated by this crystal lattice. A detailed analysis showed a guanine at the N4 position of the outside strand and an adenine at the inside strand [supporting information (SI) Fig. 5]. Thus, the N7 cytosines of this structure are located at the cross-over strands, with the uracil bases located exclusively at the outside strands, indicating that the junction adopts the H-isomer, opposite of Br2J. The uracils of this construct cannot form X- or H-bonds to the phosphate backbone; therefore, in the absence of an X-bond, the junction is stabilized by H-bonding interactions similar to those originally seen in the ACC-J junction (11).
The Br1J construct is midway between Br2J and H2J, competing a single X-bond against two H-bonds. The resulting structure showed two bromines (≈half-occupancy for each of the two symmetry-related strands, essentially accounting for the single bromine of the construct) at the junction cross-over (Fig. 3a). This structure can be interpreted as either a homogeneous population where half of the uracils are brominated and half are unbrominated in each junction, or a heterogeneous mixture of equal populations of the fully brominated Br2J and unbrominated H2J junctions. The Br1J structure is seen to be structurally unique, with no evidence for multiple conformations; therefore, the crystal represents a nearly homogeneous population of singly brominated four-stranded DNA complexes in the X-isomer form with insignificant amounts of Br2J or H2J.
The three junctions are nearly identical in structure, with significant structural perturbations localized at the core of the junction, consistent with the intramolecular interactions being the primary determinant of the stability (11, 12) and the isomeric form of the junction. Interestingly, the X- and H-bonding pyrimidine bases of N7 overlay almost exactly in all three structures (Fig. 3b). Compared with the analogous junction of d(CCGATATCGG) (ATC-J, with a thymine replacing BrUra), the bromines of Br2J and Br1J do not appear to affect the base pair geometry at N7 significantly, indicating that the substituents do not perturb the base pairing or stacking of the DNA arms in the junction. The most dramatic perturbations are seen as rotations about the β-angle of N6, which subsequently defines the interaction distance of the phosphate to the N7 nucleotide. We note, however, that these angles are not unusual for B–DNA duplexes. The Br⋯O distances for Br2J and Br1J are closer than the sums of their respective van der Waals radii, indicating the formation of X-bonds, and they are closer than the analogous methyl to phosphate distance in the ATC-J junction. ATC-J is amphimorphic (crystallizing as both junction and B–DNA duplex) (12), suggesting that the thymine to phosphate electrostatic interaction is significantly weaker than either the X- or H-bonds. In Br2J and Br1J, the phosphate oxygens are drawn toward the bromines to establish the respective X-bonds, similar to the H-bond in H2J.
The energies of the X- vs. H-bond could be estimated by quantifying the bromines at nucleotide N7 on the inside and outside strands of Br1J. The estimated occupancy of bromine was 0.41 Br per uracil (±0.02) (or 0.82 bromines for the two uracils) at the junction center and 0.05–0.15 Br per uracil at the outside strands (SI Fig. 6). These values translate to ≈1 kcal/mol (range from 0.6 to 1.3 kcal/mol) greater stability for the X- vs. H-isomer. Because Br1J has 0.5 X-bonds/H-bond, the normalized single bromine X-bond is extrapolated to be twice this energy, or ≈2 kcal/mol more stable than the N–H⋯O− H-bond, assuming that there is no entropy difference between the two conformers of this construct.
Discussion
In the current work, we have shown that bromine type X-bonds compete effectively against a classic N–H⋯O− type H-bond in defining the isomeric form of DNA junctions. This intramolecular assay system is thus an elegant method to compare directly the ability of X- and H-bonds to direct biomolecular conformation. Using the distributions of X- and H-isomer forms observed in the Br1J crystals, we estimate an energy difference between X- and H-bonds in this system to be ≈2 kcal/mol. One may ask whether thermodynamic quantities can be derived from crystallographic assays, where the conformers are not in equilibrium but locked into a defined lattice. We had previously applied a similar competitive crystallographic assay to estimate the relative energies of reverse base pairs in DNA, taking advantage of the contributions of such base pairs to the crystal lattice (18). In the current system, however, the distribution of isomer forms of the junction in crystals are defined by the intramolecular interactions that stabilize the conformers and not by any differences in the crystal lattice; therefore, we can consider the crystals as sampling a preestablished equilibrium in solution. Thus, this ≈2 kcal/mol is a valid estimate for the difference in energy of the bromine X-bond vs. N–H⋯O− H-bond in the Br1J crystals.
The X-bond in the Br1J junction is longer and, consequently, expected to be weaker than that of Br2J (Fig. 3), as might be predicted with only half the interactions present. Ab initio calculations on atomic models derived from the coordinates of the Br1J and Br2J crystal structures showed the interaction between the BrUra and phosphate groups to be ≈3 kcal/mol more favorable in the shorter Br2J geometry (SI Fig. 7). As with H-bonds, the stabilizing potential of an X-bond may also depend on competing solvent effects, but an analysis of the solvent accessible surfaces showed no significant differences in free energies of hydration for the X- and H-isomeric forms. Thus, each X-bond in Br2J can be estimated (through a simple thermodynamic cycle) to be ≈5 kcal/mol more stable than the H-bond of H2J (Fig. 4). This magnitude of difference in the energies is consistent with the lack of any observable quantity of the competing H-isomer in the Br2J crystals.
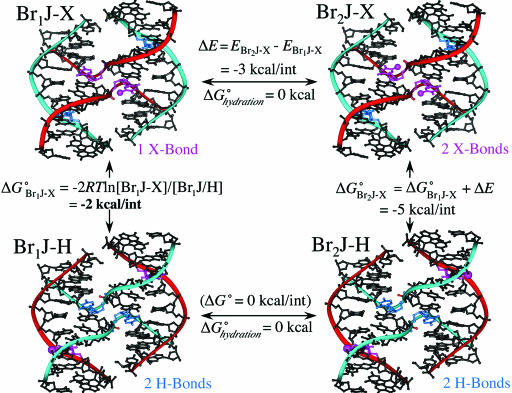
Fig. 4.
Thermodynamic cycle to estimate the free energies of the X- relative to H-bonds. A free energy difference of ≈1 kcal/mol was estimated between the X- and H-isomers from the occupancies of bromine at the inside and outside strands of Br1J (Br1J-X and Br1J-H) (Si Fig. 6) or ≈2 kcal/mol for 1 X-bond vs. 1 H-bond. Because there is very little contribution of hydration free energy to placing the bromine in the H- or X-isomeric forms (ΔGhydrationo ≈0), we can assume that the primary effect on the energy of the X-bonds is electrostatic. Finally, if we assume that the is no difference in the energies of the H-isomer for either the Br1J or Br2J constructs (Br1J-H and Br2J-H, bottom of cycle, with the bromines on the outside strands), then the energy of the X-bond in the Br2J construct (ΔG°Br2J-X) can be estimated as the sum the Br1J X-bond and the difference in electrostatic energy (ΔE) estimated from ab initio calculations (SI Fig. 7).
DNA junctions have been applied to a number of different biomolecule-based nanodevices, including molecular arrays and lattices, computers and translational devices or walkers (8). The ability to direct the isomer form extends the design aspects of junctions from simple sequence complementarity to true three-dimensional control of such devices. We can imagine, for example, that cycling between brominated and nonbrominated strands of a junction would allow a DNA junction to scissor between isomeric forms where the DNA arms pair and repair in a well controlled manner.
X-bonds as a general tool in biomolecular engineering, as opposed to nonhydrated organic systems (5), would typically see contributions from both electrostatic effects from polarization and hydrophobic effects from the sequestering of the hydrophobic halogen from water. Unlike H-bonds (19), however, both effects should help to stabilize the X-bond in a buried environment. By extension from the DNA junction described here, we would thus expect the stabilizing effects of X-bonds to be even greater in molecular systems where the halogen is buried, for example within protein folds or at protein–protein or protein–nucleic acid interfaces, making this general interaction attractive for controlling and manipulating biomolecule structures and complexes.
Materials and Methods
DNA sequences (Table 1) were synthesized with the trityl-protecting group attached and subsequently purified by HPLC followed by size exclusion chromatography on a Sephadex G-25 column after detritylation. Crystals were grown by the sitting-drop vapor diffusion method with solutions of 0.7 mM DNA and 25 mM sodium cacodylate (pH 7.0) buffer with 10–20 mM calcium chloride and 1.0–1.2 mM spermine, and equilibrated against reservoirs of 26–30% aqueous 2-methyl-2,4-dimethylpentanediol. Data for the crystals were collected at liquid nitrogen temperatures by using CuKα radiation from a Rigaku (Tokyo, Japan) RU-H3R rotating anode generator with an Raxis-IV image plate detector, and processed by using DENZO and SCALEPACK from the HKL suite of programs (20). Structures were solved by molecular replacement using EPMR (21) with d(CCAGTACbrUGG) [Protein Data Bank (PDB) ID code 1P54] as the starting model. Distinct solutions with correlation coefficients of ≈79% and Rcryst from 39.4% to 45.1% were obtained with two unique strands in the asymmetric unit; the other pairs of strands of the full four-stranded junctions were generated from the unique crossing and outside strands of the asymmetric units by the crystallographic 2-fold symmetry axis running through the center of the four-stranded DNA junction. Subsequent refinement of the initial junction structures in CNS (22) with rigid body refinement, simulated annealing, several rounds of positional and individual B-factor refinement, and addition of solvent resulted in the final models for subsequent analyses (Table 2). The brominated structures were initially refined in the absence of bromines, and the halogens were subsequently located from difference maps. The bromine occupancies on the inside crossing and outside strands of Br1J were initially refined in CNS, and the errors were estimated by monitoring the range of occupancy values that had little or no effect on the minimum Rfree value of the refinement (SI Fig. 6).
Ab initio calculations were performed with the program GAMESS (23) by using the WebMO interface (WebMO, version 6.0; www.webmo.net) for importing and constructing models. The models of the X-bonding interactions were constructed by using the BrUra base and the oxygen and phosphorus atoms of the interacting phosphate groups from the Br1J and Br2J crystal structures. Methyl groups were added to the equivalent O3′ and O5′ oxygens of the phosphates (with an overall charge of −1) and to the N1 carbon of the uracil base, and hydrogens were added to complete the valence states of the appropriate atoms. Ground-state energies for these models were calculated by density function theory applying the B3LYP function and the 6–31G(d) basis set (the highest set applicable to row 4 elements). The infinite interaction distance energy for each complex was approximated as the sum of the energies for the BrUra and phosphate dimethylester components.
To estimate the effects of solvation on the relative free energies of the X- and H-isomers, we first calculated the solvent-accessible surfaces (SAS) of the bromine of the X-bonding BrUra base and the H-bonding N2 amino group of the opposing cytosine base at the outside and inside strands of the respective conformers. The SAS were translated to free energies of hydration (ΔGhydrationo) by using an atomic solvation parameter (ASP) of 36.8 cal/mol/Å2 for a bromine atom (derived from the difference in the partition coefficient of bromobenzene versus benzene) and −63.0 cal/mol/Å2 for the extracyclic amino group according to the following relationship (25): ΔGhydrationo = SAS × ASP. The exposed surfaces for the bromine atoms were calculated to be 12.89 Å2 at the inside strand (X-isomer) and 21.46 Å2 on the outside strand (H-isomer), whereas the exposure of the amino group was 10.42 Å2 for the outside strand (X-isomer) and 14.67 Å2 at the inside strand (H-isomer) of the Br1J junction. This calculation translates to a difference in ΔGhydrationo ≈ −0.2 kcal/mol for the X- versus H-isomers, which is negligible compared with the electrostatic effects.
Supplementary Material
Acknowledgments
We thank P. A. Karplus and his group for helpful discussions. This work was supported by National Institutes of Health Grant R1GM62957A. The x-ray diffraction facilities are supported by the Proteins and Nucleic Acids Facility Core of the Environmental Health Sciences Center at OSU (National Institute on Environmental Health Sciences Grant ES00210) and a by grant from the Murdock Charitable Trust.
Abbreviations
- H-bond:
hydrogen bond
- SAS
solvent-accessible surfaces
- X-bond
halogen bond.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB codes 2ORG, 2ORH, and 2ORF).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610531104/DC1.
References
- 1.Hassel O. Science. 1970;170:497–502. doi: 10.1126/science.170.3957.497. [DOI] [PubMed] [Google Scholar]
- 2.Metrangolo P, Resnati G. Chemistry. 2001;7:2511–2519. doi: 10.1002/1521-3765(20010618)7:12<2511::aid-chem25110>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 3.Brisdon A. Annu Rep Prog Chem Sect A. 2002;98:107–114. [Google Scholar]
- 4.Metrangolo P, Neukirch H, Pilati T, Resnati G. Acct Chem Res. 2005;38:386–395. doi: 10.1021/ar0400995. [DOI] [PubMed] [Google Scholar]
- 5.Corradi E, Meille SV, Messina MT, Metrangolo P, Resnati G. Angew Chem Int Ed. 2000;112:1852–1856. doi: 10.1002/(sici)1521-3773(20000515)39:10<1782::aid-anie1782>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Muzet N, Guillot B, Jelsch C, Howard E, Lecomte C. Proc Natl Acad Sci USA. 2003;100:8742–8747. doi: 10.1073/pnas.1432955100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auffinger P, Hays FA, Westhof E, Ho PS. Proc Natl Acad Sci USA. 2004;101:16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seeman NC. Trends Biochem Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duckett DR, Murchie AIH, Diekmann S, von Kitzing E, Kemper B, Lilley DMJ. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 10.Ortiz-Lombardía M, González A, Eritja R, Aymamí J, Azorín F, Coll M. Nat Struct Biol. 1999;6:913–917. doi: 10.1038/13277. [DOI] [PubMed] [Google Scholar]
- 11.Eichman BF, Vargason JM, Mooers BHM, Ho PS. Proc Natl Acad Sci USA. 2000;97:3971–3976. doi: 10.1073/pnas.97.8.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hays FA, Teegarden A, Jones ZJ, Harms M, Raup D, Watson J, Cavaliere E, Ho PS. Proc Natl Acad Sci USA. 2005;102:7157–7162. doi: 10.1073/pnas.0409455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grainger RJ, Murchie AIH, Lilley DMJ. Biochemistry. 1998;37:23–32. doi: 10.1021/bi9721492. [DOI] [PubMed] [Google Scholar]
- 14.Miick SM, Fee RS, Millar DP, Chazin WJ. Proc Natl Acad Sci USA. 1997;94:9080–9084. doi: 10.1073/pnas.94.17.9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKinney SA, Declais AC, Lilley DM, Ha T. Nat Struct Biol. 2003;10:93–97. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- 16.Hays FA, Watson J, Ho PS. J Biol Chem. 2003;278:49663–49666. doi: 10.1074/jbc.R300033200. [DOI] [PubMed] [Google Scholar]
- 17.Hays FA, Schirf V, Ho PS, Demeler B. Biochemistry. 2006;45:2467–2471. doi: 10.1021/bi052129x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mooers BH, Eichman BF, Ho PS. J Mol Biol. 1997;269:796–810. doi: 10.1006/jmbi.1997.1100. [DOI] [PubMed] [Google Scholar]
- 19.Baldwin RL. J Biol Chem. 2003;278:17581–17588. doi: 10.1074/jbc.X200009200. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Kissinger CR, Gehlhaar DK, Fogel DB. Acta Crystallogr D. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 22.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt MWK, Baldridge K, Boatz JA, Elbert ST, Gordon MS, Jensen JJ, Koseki S, Matsunaga N, Nguyen KA, Su S, et al. J Comput Chem. 1993;14:1347–1363. [Google Scholar]
- 24.Watson J, Hays FA, Ho PS. Nucleic Acids Res. 2004;32:3017–3027. doi: 10.1093/nar/gkh631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagawa TF, Stoddard D, Zhou GW, Ho PS. Biochemistry. 1989;28:6642–6651. doi: 10.1021/bi00442a017. [DOI] [PubMed] [Google Scholar]
- 26.Vargason JM, Ho PS. J Biol Chem. 2002;277:21041–21049. doi: 10.1074/jbc.M201357200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.