Abstract
SnRK2.8 is a member of the sucrose nonfermenting-related kinase family that is down-regulated when plants are deprived of nutrients and growth is reduced. When this kinase is over expressed in Arabidopsis, the plants grow larger. To understand how this kinase modulates growth, we identified some of the proteins that are phosphorylated by this kinase. A new phosphoproteomic method was used in which total protein from plants overexpressing the kinase was compared with total protein from plants in which the kinase was inactivated. Protein profiles were compared on two-dimensional gels following staining by a dye that recognizes phosphorylated amino acids. Candidate target proteins were confirmed with in vitro phosphorylation assays, using the kinase and target proteins that were purified from Escherichia coli. Seven target proteins were confirmed as being phosphorylated by SnRK2.8. Certain targets, such as 14-3-3 proteins, regulate as yet unidentified proteins, whereas other targets, such as glyoxalase I and ribose 5-phosphate isomerase, detoxify byproducts from glycolysis and catalyze one of the final steps in carbon fixation, respectively. Also, adenosine kinase and 60S ribosomal protein were confirmed as targets of SnRK2.8. Using mass spectrometry, we identified phosphorylated residues in the SnRK2.8, the 14-3-3κ, and the 14-3-3χ. These data show that the expression of SnRK2.8 is correlated with plant growth, which may in part be due to the phosphorylation of enzymes involved in metabolic processes.
Keywords: 14-3-3, nutrient deprivation, plant, potassium
The mammalian AMPK (AMP-activated protein kinase), the yeast SNF (sucrose nonfermenting) 1 protein kinase, and the plant SnRK (SNF1-related protein kinase) are highly conserved and play broad roles in growth and metabolic responses to cellular stress (1). Mammalian cells sense glucose levels through the mammalian target of rapamycin kinase, which is part of an AMPK complex. AMPK is implicated in the development and treatment of metabolic disorders, including obesity and type 2 diabetes (2), and mutations in AMPK causes cardiac abnormalities (3). In yeast, SNF1 is required for regulation of glucose-responsive genes necessary for pseudohyphal growth in response to nutrient limitations (4) and for controlling the onset of meiosis in yeast (5). SNF1 provides a mechanism for crosstalk between metabolic pathways and cell cycle signaling processes (6). Mammal AMPK and yeast SNF1 act as energy-level sensors that function to regulate metabolism during low-energy conditions.
In contrast to the single family of SNF kinases in mammals, the plant SnRKs have been grouped based on sequence similarity and domain structure into three subfamilies: SnRK1, SnRK2, and SnRK3 (1, 7). SnRK1 kinases in plants have been well characterized at molecular and biochemical levels (8). In potato, SnRK1 is necessary for the sucrose-induced expression of sucrose synthase (9). In barley, the absence of SnRK1 expression in pollen leads to abnormal development of pollen and a lack of starch accumulation in pollen (10). In wheat, an embryo-expressed SnRK1 has been suggested to regulate the expression of α-amylase that is involved in starch accumulation. In moss, SNF1 is required for growth under normal and low light conditions (11). Plant SnRK1s play roles in regulating energy metabolism, which is similar to the proposed roles of the yeast SNF1-kinase and mammalian AMPK (1).
The SnRK2 and SnRK3 gene subfamilies are unique to plants (6). To date, most studies on SnRK2 and SnRK3 kinases focus on their involvement in responses to different stresses. The SnRK2 family is activated by hyperosmotic stress and is also involved in abscisic acid (ABA) signaling in response to water stress (12, 13). Overexpression of SnRK2.8 increased the expression of stress-related genes that enhance drought tolerance (14). It has been suggested that the SnRK2 family members may play different functional roles in planta. For example, ABA activates only five of the ten members of the SnRK2 family (12). SnRK2.8 is strongly activated by salt and mannitol treatments and only slightly activated by ABA, whereas SnRK2.9 activation is unaffected by ABA, mannitol, or hyperosmotic stress (12). This highlights a possible role beyond abiotic stress responses for these kinases. Several members of the SnRK3 subfamily have been extensively characterized. One of best studied kinases in the SnRK3 family is SOS2, which is required for sodium and K+ homeostasis and abiotic stress tolerance (15). The activity of SnRK3 kinases are regulated in a Ca2+-dependent manner, and their activity is induced by interactions with a calcineurin-B like Ca2+ binding protein in response to abiotic stress (16–18).
AMPK, SNF1, and SnRK regulate assorted metabolic processes through the phosphorylation of target proteins. Enzymes regulated by AMPK include acetyl-CoA carboxylase (fatty acid synthesis), glucose-6-phosphatase (gluconeogensis), a glucose transporter (glucose uptake), tuberin (cell growth and protein synthesis), and 3-hydroxy-3-methylglutaryl CoA (HMG-CoA) reductase (isoprenoid and sterol synthesis) (19). In plants, SnRK1 is inactivated by dephosphorylation and reactivated by phosphorylation (20). The activated form of SnRK1 phosphorylates and inactivates HMG-CoA reductase, sucrose phosphate synthase, and nitrate reductase (NR) in vitro (21). Even though AMPK, SNF1, and SnRK1 share similar functions in animals, yeast, and plants, the SnRK2 and SnRK3 families may have plant-specific targets.
Identifying molecular targets of the SnRK2 kinases is important for understanding how these kinases modulate plant growth and development. Currently, most of the downstream components involved in these kinase-signaling pathways are not known. Studies in rice and Arabidopsis showed that certain transcription factors containing ABREs (ABA response element) are targets of SnRKs or other types of Ser/Thr kinases (22, 23). Mutant screening, microarrays, and yeast-two-hybrid screening have been performed to find kinase targets; however, only limited numbers of phosphorylated target proteins have been identified (24). In this study, we report that SnRK2.8 is down-regulated by nutrient deprivation, and that this down-regulation is accompanied by a decrease in growth. Overexpression of this kinase resulted in increased growth of Arabidopsis under normal and nutrient-limited conditions. To understand how SnRK2.8 increases growth, we used a phosphoproteomic method to identify potential targets in vivo.
Results
Overexpression of SnRK2.8 Enhanced Plant Growth.
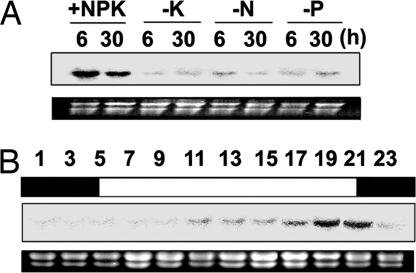
In a microarray study, we found that the gene encoding SnRK2.8 was down-regulated in roots under K+ deprivation. To confirm and extend the previous result, Northern blot analyses were performed on root RNA isolated from plants that had been deprived of K+, N, or P. The abundance of the SnRK2.8 transcript decreased in roots under N, P, and K+ deprivation (Fig. 1A). Because SnRK2.8 is related to the SNF kinases and may be regulated by diurnal factors that link them to carbon supply, we explored the expression of SnRK2.8 in leaves over a 24-h period. Fig. 1B shows that the expression of SnRK2.8 in leaves gradually increased following illumination. The accumulation of SnRK2.8 in leaves was highest from 1900 (7:00 p.m.) to 2100 (9:00 p.m.) and dropped off when the plants were in the dark (Fig. 1B).
Fig. 1.
Northern blot analysis of SnRK2.8 under nutrient-deprived conditions and during a 24-h light-dark cycle. (A) The expression of SnRK2.8 was down-regulated by nutrient deprivation. Shown are Arabidopsis Col-0 roots 6 h and 30 h after deprivation, grown in full nutrient conditions (+NPK) or without K+ (−K), N (−N), and P (−P). (B) Expression of SnRK2.8 in leaves from soil grown Col-0 plants. Each number indicates the time of harvest during one 24-h period. The lights went on at 500 and off at 2100. The rRNA bands in ethidium bromide-stained gels are shown as loading controls.
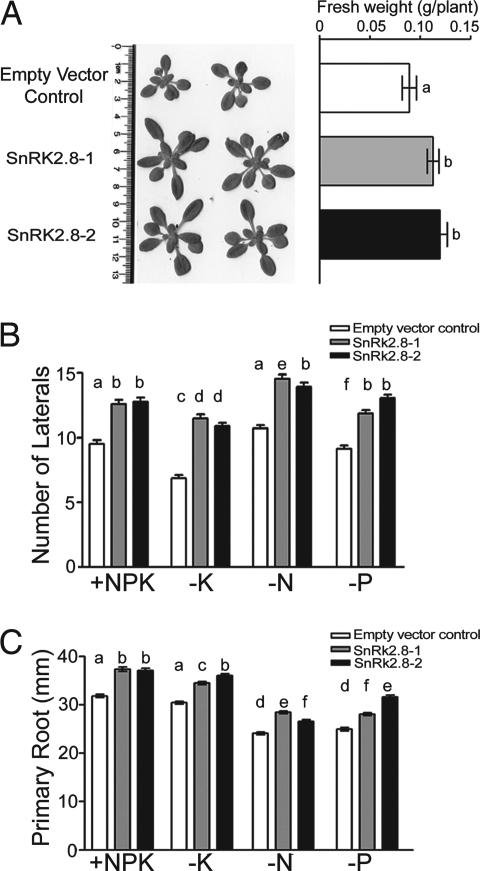
To further elucidate the function of SnRK2.8, homozygous T-DNA inactivation lines snrk2.8-1 (Salk_73395) and snrk2.8-2 (Salk_53423) were isolated from the SALK collection and eight lines overexpressing SnRK2.8 under the control of the figwort mosaic virus promoter were created. These lines were studied under a range of conditions and replicated numerous times. For more detailed analyses, two overexpression lines were chosen. When grown under control conditions, SnRK2.8-1 and SnRK2.8-2 had significantly higher biomass than the plants containing the empty vector (Fig. 2A). Under nutrient-deprived conditions primary root growth and lateral root number were also significantly higher in the plants overexpressing SnRK2.8 than plants containing the empty vector (Fig. 2 B and C). The increased growth of the SnRK2.8 overexpressing plants was most obvious under the nutrient-sufficient conditions. The growth of snrk2.8-1 was tested multiple times, and we found no significant differences in growth as compared with the wild type. These data on the overexpression lines suggest that SnRK2.8 is involved in regulating shoot and root growth.
Fig. 2.
Overexpression of SnRK2.8 enhances freshweight and root growth. (A) Picture and freshweight of shoots from plants containing the empty vector and two overexpressing lines, SnRK2.8-1 and SnRK2.8-2. (B) Lateral root numbers. (C) Primary root length. +NPK indicates full nutrients. −K contained 10 μM K+, −N contained 100 μM N, and −P contained 12 μM P. Bars indicate standard error. Different letters indicates a significant difference between means at P < 0.05 (Tukey honestly significantly different test) (n ≥ 70 plants).
Screening for Targets of SnRK2.8 in Planta.
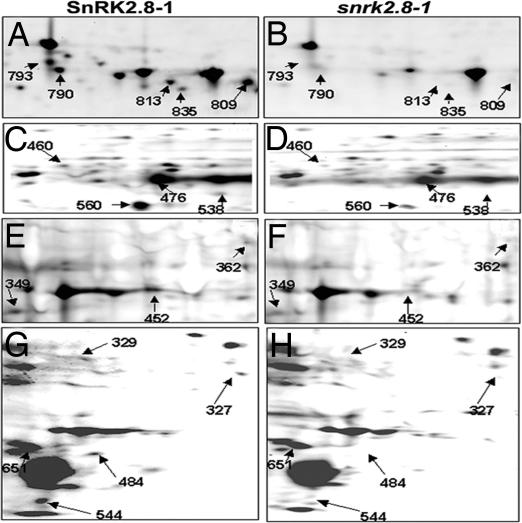
To gain insight into how SnRK2.8 modulates plant growth, a phosphoproteomics approach was used to identify phosphorylation targets [supporting information (SI) Fig. 7]. The study compared root protein from SnRK2.8 overexpressing to knockout lines. Total protein was extracted from the roots of three biological replicates and run on two-dimensional electrophoresis gels. The gels were stained with Pro-Q Diamond to detect phosphorylated amino acids (25). Image analysis was used to compare the intensity of spots from the replicate gels. When there were quantitative differences in spot volume, Student t tests were conducted to identify significant differences. The protein spots that were detected only from gels containing protein from the SnRK2.8 overexpressing lines or were significantly less abundant than on gels containing protein from the wild-type or knockout lines were identified by MS. More than 15 putative differentially phosphorylated spots were found from pI 3–10 broad range gels (SI Table 2); however, each spot contained multiple proteins. Because >70% of phosphorylated protein spots were located between pI values 3 and 6, and to reduce the number of proteins in each spot, narrow-range immobilized pH gradient strips (pI 3.9–5.1 and pI 4.7–5.9) were used to further analyze the phosphorylated proteins. Thirteen different spots from the 3.9–5.1 immobilized pH gradient strips and 11 different spots from 4.7–5.9 strips were identified by image analysis and sequenced (SI Table 2). A few examples of the differences in the abundance of phosphorylated proteins from SnRK2.8-1 and snrk2.8-1 are shown in Fig. 3.
Fig. 3.
Sections of two-dimensional gels containing Arabidopsis root proteins from SnRK2.8-1 and the snrk2.8-1 stained with ProQ Diamond stain. Two sections are from the narrow-range gel 3.9–5.1 (A–D), one section from a narrow range gel 4.7–5.9 (E and F), and one section from the wide range gel 3–10 is shown (G and H). Spot numbers correspond to the spots identified in Table 1 and SI Table 2.
14-3-3, Adenosine Kinase, Glyoxalase I (glyI), Ribose 5-Phosphate Isomerase (R5PI), and Ribosomal Protein Are Phosphorylated by SnRK2.8.
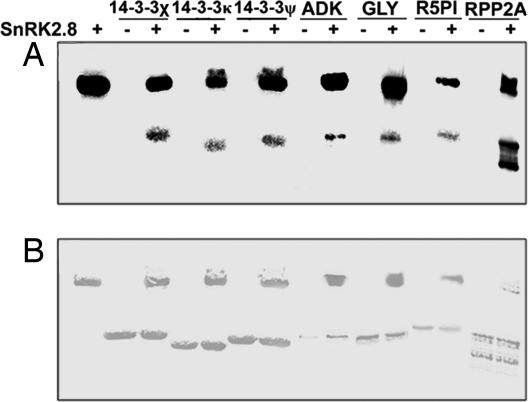
To reduce the number of candidates for further examination, we chose proteins that were identified several times during MS analysis of the ProQ Diamond stained spots. (SI Table 2). Adenosine kinase (ADK) 1, AALP protein, and R5PI were identified from both broad-range and narrow-range gels. Isoforms of tubulin, 14-3-3, 60S acidic ribosomal proteins (RPP2A), cysteine proteases, heat shock proteins, and glyI were also identified multiple times. Eleven putative protein targets were tested by using in vitro phosphorylation assays (Table 1). The in vitro assays were conducted with the SnRK2.8 kinase, which was purified as a fusion protein with GST. Target proteins were purified as hexahistidine-tagged proteins. Recombinant protein was obtained by using an Escherichia coli expression system for nine of the eleven proteins (Table 1). Of the nine proteins tested, phosphorylation by SnRK2.8 was confirmed for seven of the targets (Fig. 4). Fig. 4A shows the results of the in vitro phosphorylation assays. Autophosphorylation of SnRK2.8 is shown in the first lane, and subsequent lanes show that none of the proteins tested autophosphorylate (lanes marked with minus sign). The lanes marked with a plus sign show that three 14-3-3, glyI, ADK1, R5PI, and RPP2A are phosphorylated only in the presence of SnRK2.8. SnRK2.8 did not phosphorylate tubulin and the OTU-like cysteine protease (data not shown and Table 1). In each “plus lane,” the upper band represents the autophosphorylated kinase and the lower band the phosphorylated target. In most cases, we obtained purified protein for those being studied and those ran as a single band of the correct molecular weight (Fig. 4B). For RPP2A, two distinct bands were present, which may have been because of degradation or truncation when the protein was synthesized in E. coli. Attempts to purify the heat shock 70 protein and the protein phosphatase 2A were unsuccessful (Table 1). In summary, nine targets of the SnRK2.8 were identified by using proteomics approaches, and seven were confirmed as targets by using in vitro phosphorylation assays.
Table 1.
List of target proteins that were tested by using in vitro phosphorylation assays
| Name | Genome no. | Predicted/actual kDa | Fold change of spot vol(overexpressed/knockout) | Coverage, % | Spot, no. | Phosphorylated by SnRK2.8 |
|---|---|---|---|---|---|---|
| 14-3-3χ | At4g09000 | 30.0/33.8 | 52.6 | 27 | 503 | Yes |
| 14-3-3κ | At5g65430 | 27.8/29.8 | Present/absent | 33 | 538 | Yes |
| 14-3-3ψ | At5g38480 | 28.7/26.4 | 10.0 | 14 | 560 | Yes |
| Glyoxalase I | At1g11840 | 31.9/35.2 | Present/absent | 44 | 484 | Yes |
| Adenosine kinase I | At3g09820 | 38.2/36.8 | Present/absent | 15 | 452 | Yes |
| Ribose 5-phosphate isomerase | At3g04790 | 29.4/27.5 | Present /absent | 28 | 623 | Yes |
| 60S acidic ribosomal protein P2 | At2g27720 | 11.4/10.6 | Present/absent | 39 | 813 | Yes |
| OTU-like cysteine protease | At1g50670 | 23.6/24.5 | Present/absent | 13 | 544 | No |
| Tubulin α-2/α-4 chain | At1g04820 | 50.1/54.8 | 2.75 | 36 | 329 | No |
| PP2A-3 protein phosphatase | At2g42500 | 36.4/36.8 | Present/absent | 13 | 452 | Unable to produce protein |
| Heat-shock protein 70 | At3g09440 | 71.5/75.2 | Present/absent | 19 | 222 | Unable to produce protein |
Fig. 4.
In vitro phosphorylation assays conducted by using purified SnRK2.8 protein with three 14-3-3, ADK, glyI (glyoxalase I), R5PI, and RPP2A. (A) Each band indicates a [γ-32P]dATP phosphorylated protein. Lane 1 on the left side contains SnRK2.8 alone. Subsequent lanes contain target proteins that have been incubated with SnRK2.8 (+) or without SnRK2.8 (−). (B) The Coomassie stained gel shows protein loading.
MS Identification of Phosphorylation Sites.
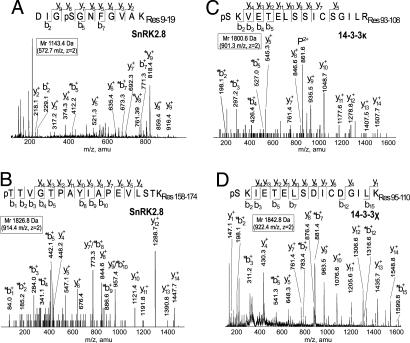
To determine the sites where the SnRK2.8 and target proteins were phosphorylated, we conducted in vitro phosphorylation assays followed by gel-purification and in-gel trypsin digestion. Using MS, we mapped two phosphorylation sites in the SnRK2.8 kinase, and mapped one phosphorylation site in two of the target proteins, 14-3-3κ and 14-3-3χ. The phosphopeptide candidates were identified by using a product ion scanning function in positive ion mode. The y and b sequence ions were searched with Mascot, using a database containing the full sequences of SnRK2.8, 14-3-3κ, and 14-3-3χ. MS data were matched to the specific phosphorylated peptides and the exact site of phosphorylation was identified. Fragmentation of the doubly charged ions 572.7 and 914.4 m/z from the SnRK2.8 peptide digests (Fig. 5A and B) resulted in the match of the sequence of two peptides from SnRK2.8 (1,143.4 and 1,826.8 Da) with a phosphate modification (80 Da). The phosphopeptides were identified as residues 9–19 with Ser-12 phosphorylated and residues 158–174 with Thr-158 phosphorylated (Fig. 5 A and B).
Fig. 5.
Phosphorylation site identification of SnRK2.8, 14-3-3κ, and 14-3-3χ by LC-MS/MS. (A and B) Fragmentation patterns of the doubly charged ions 572.7 (A) and 914.4 m/z from the SnRK2.8 peptide digests (B). Above each spectrum, the amino acid sequence of the peptide fragments 1143.4 Da and 1826.8 Da is shown with the phosphorylation site of Ser-12 and Thr-158 as pS and pT, respectively. (C) Fragmentation pattern of the doubly charged ions 901.3 from the 14-3-3κ. (D) Fragmentation pattern of the doubly charged ions 922.4 m/z from 14-3-3χ. The amino acid sequences of the peptide fragments 1800.6 and 1842.8 are shown with the phosphorylation site of Ser-93 and Ser-95 as pS. The mass of the b and y ions series visualized in the spectra marked with asterisks correspond to the mass of the phosphorylated fragment with the neutral loss of 98 Da of H3PO4.
The identification of phosphorylation sites in the 14-3-3κ and 14-3-3χ was performed in the same way. Fragmentation of the doubly charged ion 901.3 m/z gave a sequential b and y ion sequence that matched the sequence of one peptide from 14-3-3κ (1,800.6 Da) with a phosphate modification (Fig. 5C). Because the b sequence ions and the m/z of the b ions showed the loss of a phosphate, the phosphorylation site was deduced to be located at the Ser-93 position. Fragmentation of the doubly charged ion 922.4 m/z matched the sequence of the residues 95–110 of 14-3-3χ with a phosphate modification (Fig. 5D). The b ions showed that the phosphorylation site was located at the Ser-95 position.
Phosphorylation of glyI by SnRK2.8 Increases glyI Activity.
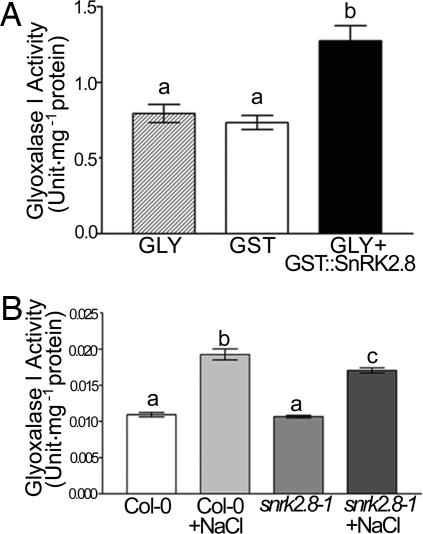
To demonstrate that phosphorylation by SnRK2.8 alters the in vitro and in vivo activity of a target identified in this study, we chose to examine one of the target proteins for which we were successful in developing an enzymatic assay to characterize activity in vitro and in crude plant extracts. Phosphorylation of the purified glyI by the purified SnRK2.8 GST fusion protein significantly increased the activity of this enzyme in vitro (Fig. 6A). This activity was reduced back to the levels measured without incubation with SnRK2.8 by the addition of alkaline phosphatase, indicating that the increased activity was due to phosphorylation (data now shown). The glyI activity was not affected by adding GST protein alone (Fig. 6A). The increased activity of glyI was also confirmed in the SnRK2.8 overexpression lines, which suggests that the activity of this enzyme is either directly or indirectly modified by SnRK2.8. In those lines, we observed 6–10% higher glyI activity than in the wild-type and 9–12% higher glyI activity than in the knockout line snrk2.8-1 under normal conditions (data not shown). Salinity stress has been shown to increase the activity of glyI in planta (26). Therefore, the activity of glyI in wild type and snrk2.8-1 knockout plants under control conditions and salt stress was tested. Salt treatment stimulated the activity of the enzyme in the wild type plant, but less stimulation was observed in the snrk2.8-1 (Fig. 6B). glyI activity was lower after salt treatment in the snrk2.8-1, but salt stress still increased the protein activity, indicating that the SnRK2.8 may not be the only kinase or factor modulating the gly activity.
Fig. 6.
Glyoxylase I activity assay. (A) Incubation of purified glyI with SnRK2.8 increased the activity of the protein. GST is the GST protein purified from E. coli. GST::SnRK2.8 is the GST-tagged SnRK2.8 protein purified from E. coli. (B) Analysis of crude plant extracts show salt treatment increases glyI activity more in the wild type than in the snrk2.8-1 knockout. Experiments were repeated four times. Bars indicate standard error. Different letters indicate a significant difference at P < 0.001 (Tukey honestly significantly different test) (n ≥ 6) (A) and P < 0.05 (Tukey honestly significantly different test) (n ≥ 10) (B).
Discussion
Protein phosphorylation plays important roles in the regulation of cell signaling and growth by modifying enzyme activity, protein localization, protein structure, and protein-protein interaction (2, 7). Despite the importance of phosphorylation, the identification of phosphoproteins and phosphorylation sites has proceeded slowly in plants with relatively few of each being determined (24). To gain a more comprehensive understanding of the function of SnRK2.8, we devised a phosphoproteomics strategy for detecting targets, using a florescent dye that detects phosphoproteins combined with MS confirmation of the target identity. With this approach, potential target proteins of SnRK2.8 were identified to provide a better understanding of how this kinase might modulate growth in planta.
In this study, we identified three regulatory proteins in the 14-3-3 family that are phosphorylated by SnRK2.8. Homologues of the plant 14-3-3 are expressed in all eukaryotic cells and are highly conserved in structure and regulatory function in organisms from yeast to mammalian cells (27–29). By interacting with other proteins, 14-3-3s appear to have diverse regulatory roles (30). The 14-3-3σ positively regulates the activity of p53 and can suppress tumor growth (31). Plant 14-3-3s interact with enzymes involved in primary biosynthetic and energy metabolism (32) and are comprised of a family containing 13 genes. A key enzyme in plant N metabolism, NR, is a well known target of a 14-3-3 (33). The activity of plant NR is controlled by phosphorylation at a specific Ser residue. Phosphorylation per se does not alter NR activity, but it allows for the binding of a 14-3-3. Once a 14-3-3 binds to NR, the phosphoserine-NR is inactive (34). Many enzymes in C metabolism, such as sucrose phosphate synthase, sucrose synthase, and 3-hydroxy-3-methylglutaryl CoA reductase (8, 32, 35), are also controlled by phosphorylation and the binding of 14-3-3. The targets of most of the plant 14-3-3 have not been identified, and many of the plant 14-3-3 are poorly characterized (36). Our results suggest that the 14-3-3s phosphorylated by SnRK2.8 may regulate C or N metabolism through interaction with unknown targets.
Most studies of the role of 14-3-3 in cellular regulation have focused on changes in target protein phosphorylation as the initial regulatory event; however, 14-3-3 proteins have been shown to be phosphorylated in animal systems (37, 38) and in plants (39–41). Phosphorylation of 14-3-3 is another way in which these proteins are regulated (29), and in animal systems, a few kinases reportedly phosphorylate 14-3-3. The sphingosine-dependent kinase phosphorylates the 14-3-3ζ at Ser-58, the 14-3-3β at Ser-60, and the 14-3-3η at Ser-59 but does not phosphorylate 14-3-3σ or 14-3-3τ (42). The phosphorylation sites of 14-3-3 in mammals have not been found to be highly conserved (28). The non-ε family of 14-3-3s may be found only in plants (43), and little is known about where they are phosphorylated. Lu et al. (41) found that the 14-3-3ω was phosphorylated at a Ser residue by plant extracts. The maize GF14–6 (14-3-3) was phosphorylated at Tyr-137 by insulin-like growth factor receptor 1 in vitro (40). The specific kinases that phosphorylate plant non-ε 14-3-3 have not been identified. To our knowledge, SnRK2.8, which phosphorylates 14-3-3κ, χ, and ψ, is the first plant kinase shown to phosphorylate non-ε 14-3-3. Although each isoform of 14-3-3 may have a distinct function (44), the three 14-3-3 identified in this study are phosphorylated by SnRK2.8. Because they are regulated in a similar way in plants overexpressing SnRK2.8, these proteins may have similar functions related to plant growth and metabolism in these overexpression lines.
The phosphorylation of 14-3-3 can inhibit the interaction between 14-3-3 and their targets (38, 40). The phosphorylation of 14-3-3ζ in vivo by the casein kinase I inhibits binding to c-Raf. In maize, the phosphorylation of the 14-3-3-named GF14–6 on Tyr-137 inhibits the binding to the H+-ATPase in maize and reduces H+-ATPase activity (40). The phosphorylation of 14-3-3 by SnRK2.8 also may inhibit the interaction between unknown target proteins, and this may change the activity of target proteins. We speculate that the 14-3-3 targeted by SnRK2.8 may be involved in energy metabolism in coordination with photosynthesis, ATP production, or the light-dark transition, as shown for NR (43). When light is limited, NR is phosphorylated and then bound by 14-3-3 to inhibit NR activity (43, 45). The expression of SnRK2.8 is regulated diurnally, and therefore, its activity may be enhanced in leaves late in the day, which may lead to more phosphorylation of 14-3-3, which could inhibit the interactions with their targets. In plants overexpressing SnRK2.8, the phosphorylation and dissociation of 14-3-3 from their targets could occur more than wild-type plants and result in enhanced activity of the 14-3-3 target proteins.
Other targets that are phosphorylated by SnRK2.8 are primarily related to energy metabolism such as glycolysis and carbon fixation. glyI is involved in the detoxification of methylglyoxal, a byproduct of glycolysis (43). Increased activity of glyI is correlated with cell proliferation in pea calli and in Datura (46). The phosphorylation of glyI affects the activity of this enzyme in vivo. In animals, the phosphorylation of glyI by protein kinase A plays a key role in TNF-induced cell death (47). Increased glyI activity during salt stress may be indicative of the increased metabolic activity required to cope with the stress (48). SnRK2 kinases have been shown to be activated by abiotic stress, and they plays roles in response to osmotic stress and drought tolerance in Arabidopsis (12–15, 23, 49). The enhanced growth of plants overexpressing SnRK2.8 might require increased activity of glyI through phosphorylation due to an increased rate of glycolysis.
The R5PI isoform that we studied is involved in the final step of the Calvin cycle, which is an important source of reducing power and metabolic intermediates, and is regulated in a light-dependent manner (50). The phosphorylation of R5PI through SnRK2.8 may increase flux through the Calvin cycle, which feeds into glycolysis and could be either a cause or the result of increased growth in plants overexpressing SnRK2.8. One component of the translation mechanism is the acidic RPP2A. These proteins have been found in phosphorylated forms in many plants species (51). The phosphorylated form of these acidic ribosomal proteins exhibit higher rates of translation (52), which may also affect the growth rates of plants overexpressing SnRK2.8. ADK plays a key metabolic role in the synthesis of adenylates, methyl recycling, and cytokinin interconversion (53). The decreased activity of ADK resulted in reduced and abnormal development. It is not clear how ADK plays roles in plants overexpressing SnRK2.8 at this stage; however, activating ADK in plants overexpressing SnRK2.8 may provide more input into secondary metabolism and it may help to increase overall metabolic activity.
The identification of phosphorylation sites is essential for understanding the regulation of kinases as well as target proteins. The phosphorylation sites of another SnRK2, OST1, were recently identified (54). OST1 is autophosphorylated at Ser-175 and this residue is essential for its activation and function and is located in the T-loop region (54). Phosphorylation sites of SnRK1 (AKIN10) and SnRK3 (SOS2) have also been found in the T-loop (20, 55, 56). SnRK2.8 is autophosphorylated at Thr-158, which corresponds to the Ser-175 in OST1. Therefore, phosphorylation of a Ser or Thr residue in the T-loop is a conserved feature in the SnRK2 and SnRK1 and SnRK3 families in plants.
Furihata et al. (22) found that members of SnRK2 can phosphorylate the bZIP transcription factor, AREB1 protein, in an ABA-dependent manner and that the SnRK2 target phosphorylation sequence motif was R-x-x-S/T (22, 23). In our study, we found that SnRK2.8 phosphorylates a conserved residue on both 14-3-3 proteins studied. The sequence DYRpSKVE on the 14-3-3κ and the DYRpSKIE on 14-3-3χ differ from the motif found in the AREB1 proteins. It is possible that we missed additional phosphorylated residues on the 14-3-3 proteins, because we obtained only 60% sequence coverage from the MS analysis. Alternatively, phosphorylation of more than one distinct target motifs by the same kinase family may explain this difference (57). Therefore, SnRK2.8 may recognize multiple target sequences.
In conclusion, we found that the expression of SnRK2.8 is down-regulated by potassium deprivation. This down-regulation is associated with a strong reduction in the growth of Arabidopsis under nutrient-deprived conditions. We also showed that overexpression of SnRK2.8 increases the overall growth of Arabidopsis. Using a proteomic approach, we identified and confirmed the phosphorylation of seven targets of SnRK2.8. The targets appear to be proteins involved in metabolism and translation or the regulation of metabolism, which may be related to overall growth. Therefore, the reduced growth observed in nutrient-deprived plants may in part be controlled by the SnRK2.8 kinase cascade, which appears to be linked to the activities of multiple proteins involved in metabolic processes.
Materials and Methods
Plant Material and Growth Conditions.
For constitutive expression of SnRK2.8, we used pCambia2300 with figwort mosaic virus promoter and nopaline synthase terminator. Plants were grown in nutrient solutions (58) for 6 weeks for proteomic analysis. For the nutrient deficiency assays, plants were grown on plates containing nutrient solution, 2% of sucrose, and 0.6% SeaKem agarose (Cambrex, Rockland, ME). For K+ deficiency growth assays, 10 μM KCl instead of 1.75 mM KCl was used, 12 μM phosphate was used for P deficient growth assay, and 100 μM Ca(NO3)2 was used for N deficient growth assay. The length of primary roots and numbers of lateral roots were measured 10 days after transfer to nutrient-deficient medium. For diurnal expression analysis, the leaves of soil-grown 3-week-old plants were collected, RNA was extracted, and Northern blot analysis was performed (58) by using 300 bp of the 3′ end of the SnRK2.8 cDNA.
Total Plant Protein Purification and Two-Dimensional Gel Analysis.
Total protein from wild-type, SnRK2.8-1, and snrk2.8-1 plants was isolated by trichloroacetic acid method (59). One hundred micrograms of protein was applied to immobilized pH gradient strips (110 mm, pH 3–10 nonlinear; pH 3.9–5.1, pH 4.7–5.9) (BioRad, Hercules, CA) and passively rehydrated for 16 h. Isoelectric focusing and two-dimensional electrophoresis was performed (59). Specific phosphorylated protein staining was performed by using ProQ Diamond phosphoprotein gel stain (Invitrogen, Carlsbad, CA) (60).
Protein Digestion, Peptides Separation, and Phosphorylation Site Determination, Using LC/MS/MS.
The in-gel protein digestion and protein identification were performed as described in ref. 60, except that a linear gradient from 5 to 60% in 25 min was used for online separation. Peptide sequencing and determination of the phosphorylated sites were performed with Mascot by searching the enhanced product ion scan spectra against a database containing the protein sequences of SnRK2.8 and the targets. Ser, Thr, and Tyr phosphorylation were also used as variable modifications, along with a charge state from 2 to 3. Each enhanced product ion scan spectrum was manually inspected to ensure acceptable ion coverage and phosphorylation site identification.
Protein Purification in E. coli and in Vitro Phosphorylation Assay.
SnRK2.8 cDNA was cloned into the pET41 vector (Novagen, San Diego, CA). The cDNAs of other targets were cloned into the pET28 vector (Novagen). All proteins were expressed in E. coli Rossetta (DE3) (Novagen). The SnRK2.8 protein was purified by using glutathione Sepharose 4B, and other target proteins were purified by using HIS-Select nickel affinity gel.
For the in vitro phosphorylation assay, SnRK2.8 was incubated in phosphorylation buffer (20 mM Hepes, pH 7.4/10 mM MgCl2/2 mM MnCl2/1 mM DTT/0.5 mM CaCl2/0.5 mM ATP) for 5 min. The target protein and preactivated SnRK2.8 were incubated in phosphorylation buffer with γ-32P-dATP for 30 min. Target protein without SnRK2.8 was used for a negative control. Each reaction was separated on 12% SDS/PAGE gel and detected by using the TYPOON 9410 system (Amersham Bioscience, Piscataway, NJ).
glyI Activity Assay.
50 mg of leaves from wild type and snrk2.8-1 ± NaCl treatment for 3 days were homogenized in 1 ml of 0.1 M sodium phosphate buffer (pH 7.0) containing 0.811 mM MgSO4 and 20% glycerol and centrifuged at 50,000 × g for 20 min. For assays of glyI activity in plant extracts, supernatant was diluted with potassium phosphate buffer (pH 6.6) and 0.2% (vol/vol) methylglyoxyal solution, and reduced glutathione was added as substrates. The ΔA240·min−1 value was obtained by using a spectrophotometer. glyI activity was calculated according to the following equation: activity (units·mg of protein−1) = [(ΔA240·min−1) × (dilution factor)]/[3.37 × volume (ml) × (mg of protein)]. Change in absorbance was corrected for background changes. One unit is defined as formation of 1.0 μmol of S-lactoylglutathione from methylglyoxal and reduced glutathione per minute at pH 6.6 at 25°C. For assays of purified recombinant Arabidopsis gly, the enzyme was incubated with SnRK2.8 or GST in phosphorylation buffer (see Protein Parification in E. Coli and in Vitro Phosphorylation Assay) for 30 min with and without 10 units of alkaline phosphatase then analyzed as described above.
Supplementary Material
Acknowledgments
We thank John Walker (University of Missouri) for his advice at key stages of the project and Sixue Chen (University of Florida) for critical comments on the manuscript and for allowing some of the phosphorylation analysis to be completed in his lab. This work was supported by Monsanto and startup funds from the Danforth Center.
Abbreviations
- ADK
adenosine kinase
- AMPK
AMP-activated protein kinase
- glyI
glyoxalase I
- NR
nitrate reductase
- R5PI
ribose 5-phosphate isomerase
- RPP2A
60S ribosomal proteins
- SNF
sucrose nonfermenting
- SnRK
SNF1-related protein kinase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610208104/DC1.
References
- 1.Hardie DG, Carling D, Carlson M. Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 2.Kemp BE, Stapleton D, Campbell DJ, Chen Z-P, Murthy S, Walter M, Gupta A, Adams JJ, Katsis F, van Denderen B, et al. Biochem Soc Trans. 2001;31:162–168. doi: 10.1042/bst0310162. [DOI] [PubMed] [Google Scholar]
- 3.Arad M, Benson WD, Perez-Atayde AR, McKenna WJ, Sparks EA, Kante RJ, McGarry K, Seidman JG, Seidman CE. J Clin Invest. 2002;109:357–362. doi: 10.1172/JCI14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen PJ, Sprague GF., Jr Proc Natl Acad Sci USA. 2000;97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honigberg SM, Lee RH. Mol Cell Biol. 1998;18:4548–4555. doi: 10.1128/mcb.18.8.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halford NG, Hey S, Jhurreea D, Laurie S, Mckibbin RS, Paul M, Zhang Y. J Exp Bot. 2003;54:467–475. doi: 10.1093/jxb/erg038. [DOI] [PubMed] [Google Scholar]
- 7.Hrabak EM, Chan CWM, Gribskov M, Harper JF, Choi JH, Halford NG, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker JHA, Slocombe SP, Ball KL, Hardie DG, Shewry PR, Halford NG. Plant Physiol. 1996;112:1141–1149. doi: 10.1104/pp.112.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell PC, Smith AM, Halford NG. Plant J. 1998;14:195–202. [Google Scholar]
- 10.Zhang Y, Shewry PR, Jones H, Barcelo P, Lazzeri PA, Halford NG. Plant J. 2001;28:431–441. doi: 10.1046/j.1365-313x.2001.01167.x. [DOI] [PubMed] [Google Scholar]
- 11.Thelander M, Olsson T, Ronne H. EMBO J. 2004;23:1900–1910. doi: 10.1038/sj.emboj.7600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boudsocq M, Hélène B-B, Lauriere C. J Biol Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 14.Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. Proc Natl Acad Sci USA. 2004;101:17306–17311. doi: 10.1073/pnas.0407758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelner A, Pekala I, Kaczanowski S, Muszynska G, Hardie DG, Dobrowolska G. Plant Physiol. 2004;136:3255–3265. doi: 10.1104/pp.104.046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halfter U, Ishitani M, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KN, Cheong YH, Gupta R, Luan S. Plant Physiol. 2000;124:1844–1853. doi: 10.1104/pp.124.4.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nozawa A, Koizumi N, Sano H. Plant Cell Physiol. 2004;42:976–982. doi: 10.1093/pcp/pce126. [DOI] [PubMed] [Google Scholar]
- 19.Hardie DG. Endocrinology. 2003;144:5179–5183. doi: 10.1210/en.2003-0982. [DOI] [PubMed] [Google Scholar]
- 20.Sugden C, Crawford RM, Halford NG, Hardie DG. Plant J. 1999;19:433–439. doi: 10.1046/j.1365-313x.1999.00532.x. [DOI] [PubMed] [Google Scholar]
- 21.Sugden C, Donaghy PG, Halford NG, Hardie DG. Plant Physiol. 1999;120:257–274. doi: 10.1104/pp.120.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Proc Natl Acad Sci USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T. Plant J. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 24.Peck SC. J Exp Bot. 2006;57:1523–1527. doi: 10.1093/jxb/erj126. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veena, Reddy VS, Sopory SK. Plant J. 1999;17:385–395. doi: 10.1046/j.1365-313x.1999.00390.x. [DOI] [PubMed] [Google Scholar]
- 27.Fu H, Subramanian RR, Masters SC. Annu Rev Phamacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 28.Roberts MR. Trend Plant Sci. 2003;8:218–223. doi: 10.1016/S1360-1385(03)00056-6. [DOI] [PubMed] [Google Scholar]
- 29.Tzivion G, Avruch J. J Biol Chem. 2002;277:3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 30.Thomas D, Guthridge M, Woodcock J, Lopez A. Curr Topics Dev Biol. 2005;67:285–303. doi: 10.1016/S0070-2153(05)67009-3. [DOI] [PubMed] [Google Scholar]
- 31.Yang H-Y, Wen Y-Y, Chen C-H, Lozano G. Mol Cell Biol. 2003;23:7096–7107. doi: 10.1128/MCB.23.20.7096-7107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber SC, Mackintosh C, Werner MK. Plant Mol Biol. 2002;50:1053–1063. doi: 10.1023/a:1021284002779. [DOI] [PubMed] [Google Scholar]
- 33.Moorhead G, Douglas P, Morrice N, Scarabel M, Aitken A, MacKintosh C. Curr Biol. 1996;6:1104–1113. doi: 10.1016/s0960-9822(02)70677-5. [DOI] [PubMed] [Google Scholar]
- 34.Kanamaru K, Wang R, Su W, Crawford NM. J Biol Chem. 1999;274:4160–4165. doi: 10.1074/jbc.274.7.4160. [DOI] [PubMed] [Google Scholar]
- 35.Ikeda Y, Koizumi N, Kusano T, Sano H. J Biol Chem. 2000;275:31695–31700. doi: 10.1074/jbc.M004892200. [DOI] [PubMed] [Google Scholar]
- 36.Sehnke PC, DeLille JM, Ferl RJ. Plant Cell. 2002;14(Suppl):S339–S354. doi: 10.1105/tpc.010430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Hoeven PC, van der Wal JC, Ruurs P, van Dijk MC, van Blitterswijk J. Biochem J. 2000;345:297–306. doi: 10.1042/0264-6021:3450297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubois T, Rommel C, Howell S, Steinhussen U, Soneji Y, Morrice N, Moelling K, Aitken A. J Biol Chem. 1997;272:28882–28888. doi: 10.1074/jbc.272.46.28882. [DOI] [PubMed] [Google Scholar]
- 39.Agrawal GK, Thelen JJ. Mol Cell Proteomics. 2006;5:2044–2059. doi: 10.1074/mcp.M600084-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Giacometti S, Camoni L, Albumi C, Visconti S, Michelis MID, Aducci P. Plant Biol. 2004;6:422–431. doi: 10.1055/s-2004-820933. [DOI] [PubMed] [Google Scholar]
- 41.Lu G, Rooney MF, Wu K, Ferl RJ. Plant Physiol. 1994;105:1459–1460. doi: 10.1104/pp.105.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Megidish T, Cooper J, Zhang L, Fu H, Hakomori S. J Biol Chem. 1998;273:21834–21845. doi: 10.1074/jbc.273.34.21834. [DOI] [PubMed] [Google Scholar]
- 43.Mackintosh C. Biochem J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paul AL, Sehnke PC, Ferl RJ. Mol Biol Cell. 2005;16:1735–1743. doi: 10.1091/mbc.E04-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulma A, Villadsen D, Campbell DG, Meek SEM, Harthill JE, Nielsen TH, Mackintosh C. Plant J. 2004;37:654–667. doi: 10.1111/j.1365-313x.2003.01992.x. [DOI] [PubMed] [Google Scholar]
- 46.Paulus C, Köllner B, Jacobsen H-J. Planta. 1993;159:561–566. doi: 10.1007/BF00198220. [DOI] [PubMed] [Google Scholar]
- 47.Herreweghe FV, Mao J, Chaplen FWR, Grooten J, Gevaert K, Vandekerckhove J, Vancompernolle K. Proc Natl Acad Sci USA. 2002;99:949–954. doi: 10.1073/pnas.012432399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain M, Choudhary D, Kale RK, Bhalla-Sarin N. Physiol Plant. 2002;114:499–505. doi: 10.1034/j.1399-3054.2002.1140401.x. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. J Biol Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- 50.Kruger NJ, von Schaewen A. Curr Opin Plant Biol. 2003;6:236–246. doi: 10.1016/s1369-5266(03)00039-6. [DOI] [PubMed] [Google Scholar]
- 51.Dresselhaus T, Cordts S, Heuer S, Sauter M, Lörz H, Kranz E. Mol Gen Genet. 1999;261:416–427. doi: 10.1007/s004380050983. [DOI] [PubMed] [Google Scholar]
- 52.Aguilar R, Montoya L, de Jiménez ES. Plant Physiol. 1998;116:379–385. [Google Scholar]
- 53.Moffatt BA, Wang L, Allen MS, Stevens YY, Qin W, Snider J, Schwa K. Plant Physiol. 2000;124:1775–1785. doi: 10.1104/pp.124.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S. Plant Physiol. 2006;141:1316–1327. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gong D, Guo Y, Jagendorf AT, Zhu J-K. Plant Physiol. 2002;130:256–264. doi: 10.1104/pp.004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Y, Halfter U, Ishitani M, Zhu J-K. Plant Cell. 2001;13:1383–1400. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernández SC, Hardin SC, Clouse SD, Kieber JJ, Huber SC. Arch Biochem Biophys. 2004;428:81–91. doi: 10.1016/j.abb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 58.Shin R, Berg RH, Schachtman DP. Plant Cell Physiol. 2005;46:1350–1357. doi: 10.1093/pcp/pci145. [DOI] [PubMed] [Google Scholar]
- 59.Zhu J, Chen S, Alvarez S, Asirvatham VS, Schachtman DP, Wu Y, Sharp RE. Plant Physiol. 2006;140:311–325. doi: 10.1104/pp.105.070219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alvarez S, Goodger JQD, Marsh EL, Chen S, Asirvatham VS, Schachtman DP. J Proteome Res. 2006;5:963–972. doi: 10.1021/pr050471q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.