Abstract
The ATM (ataxia telangiectasia mutated) protein plays a central role in sensing and responding to DNA double-strand breaks. Lymphoid cells are unique in undergoing physiologic double-strand breaks in the processes of Ig class switch recombination and T or B cell receptor V(D)J recombination, and a role for ATM in these processes has been suggested by clinical observations in ataxia telangiectasia patients as well as in engineered mice with mutations in the Atm gene. We demonstrate here a defect in thymocyte maturation in ATM-deficient mice that is associated with decreased efficiency in V-J rearrangement of the endogenous T cell receptor (TCR)α locus, accompanied by increased frequency of unresolved TCR Jα coding end breaks. We also demonstrate that a functionally rearranged TCRαβ transgene is sufficient to restore thymocyte maturation, whereas increased thymocyte survival by bcl-2 cannot improve TCRα recombination and T cell development. These data indicate a direct role for ATM in TCR gene recombination in vivo that is critical for surface TCR expression in CD4+CD8+ cells and for efficient thymocyte selection. We propose a unified model for the two major clinical characteristics of ATM deficiency, defective T cell maturation and increased genomic instability, frequently affecting the TCRα locus. In the absence of ATM, delayed TCRα coding joint formation results both in a reduction of αβ TCR-expressing immature cells, leading to inefficient thymocyte selection, and in accumulation of unstable open chromosomal DNA breaks, predisposing to TCRα locus-associated chromosomal abnormalities.
Keywords: ataxia telangiectasia mutated, recombination, T cell development
The ATM (ataxia telangiectasia mutated) protein, first identified in patients with ataxia telangiectasia syndrome, plays a critical role in sensing and responding to chromatin changes such as DNA double-strand breaks induced by ionizing irradiation (1, 2). ATM is a member of a family of phosphatidylinositol 3-kinase-related proteins that function in cell cycle regulation, monitoring of telomere length, meiotic recombination, and DNA repair (1, 2). After activation by as-yet-incompletely understood signals associated with changes in DNA structure, ATM mediates an early step in the damage response, by phosphorylating a variety of protein targets and activating multiple signal transduction pathways (3–5). The downstream outcomes of these events can include cell cycle arrest and apoptotic cell death.
In addition to the well characterized role of ATM in responses to DNA damage caused by environmental insults such as ionizing radiation, both clinical and laboratory observations have indicated that ATM also plays an important role in the unique physiological instances of DNA recombination that occur during development and differentiation of T and B lymphocytes (6–8). The generation of diversity in the antigen-specific receptors of T and B lymphocytes is mediated by induction of DNA breaks by recombination-activating gene recombinase followed by repair through the nonhomologous end-joining pathway. Recombination-activating-gene-dependent recombination targets double-strand DNA cleavage events at the boundaries of Ig or T cell receptor (TCR)-encoding (V, D, or J) gene segments and flanking recombination signal sequences. DNA cleavage at these sites initially generates coding end (CE) breaks, terminated in a hairpin structure, and blunt phosphorylated signal end (SE) breaks. Completion of the recombination process requires opening and processing of the hairpin-terminated CE followed by ligation of the two gene segments to generate the rearranged Ig or TCR gene.
Patients with ataxia telangiectasia have a high incidence of lymphomas marked by translocations preferentially involving Ig and TCR genes. Consistent with a causal role of ATM deficiency in these abnormal recombination events, ATM-deficient mice die in high proportion with thymic lymphomas that involve recombination-activating-gene-dependent translocations of the TCRα/δ locus (9–11). Translocations involving the TCRα/δ locus were also observed at high frequency in nonlymphoma peripheral T cells of ATM mice (9). Consistent with a role of the ATM protein in V(D)J recombination, chromatin immunoprecipitation has also indicated that ATM localizes to the Igκ locus in pre-B cell lines and to TCRα in mouse thymocytes (12). Most recently, elegant experiments have provided evidence that ATM can function in vitro in the resolution of DNA double-strand breaks generated during V(D)J recombination (13).
We have therefore systematically analyzed thymic development in mice deficient in ATM and demonstrate here a defect in recombination of the TCRα locus that results in reduced efficiency in production of mature T cells. This defect involves a significant decrease in Vα-Jα recombination accompanied by increased accumulation of unresolved CE in ATM-deficient CD4+CD8+ double-positive (DP) thymocytes. Our data argue that ATM plays a significant, direct role in the resolution of CE breaks during TCRα recombination.
Results
Impaired Development of Single-Positive (SP) Mature Thymocytes and Accumulation of TCRlo DP Cells in ATM-Deficient Mice.
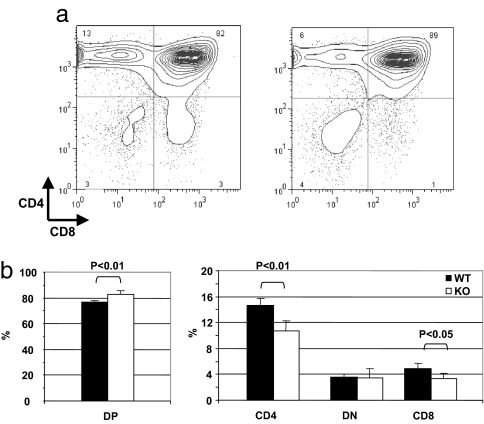
As previously reported, ATM-deficient mice exhibit generalized growth deficiency and have body weights that are ≈80% those of WT littermates (14). Total thymocyte recovery is comparably reduced to ≈75% of the WT controls (ref. 14 and data not shown). However, subpopulation analysis by flow cytometry revealed significant alterations in the composition of ATM-deficient thymocytes. Whereas the percentage of CD4−CD8− double-negative (DN) thymocytes is similar, the percentage of DP cells is increased [82.5 ± 1.3% in ATM knockout (KO) vs. 76.6 ± 1.1% in WT, P < 0.01], and the proportions of both CD4 SP (10.8 ± 1.1% in ATM KO vs. 14.7 ± 1.4% in WT) and CD8 SP cells (3.3 + 0.2% in KO vs. 4.9 ± 0.8% in WT) are significantly reduced in ATM-deficient thymi (Fig. 1b). ATM inactivation thus results in a decreased maturation of SP thymocytes from DP precursors. Despite this reduction in the proportion of SP cells, the T cell repertoire appears to maintain its normal heterogeneity because Vβ expression is similar to that in SP thymocytes from ATM WT control mice (data not shown).
Fig. 1.
Thymic subpopulations in ATM-deficient mice. (a) The proportion of DP cells is increased and the proportions of CD4 and CD8 SP cells are decreased in ATM-deficient mice (Right) compared with WT mice (Left). (b) Proportions of DP, DN, and SP thymocytes from WT and ATM KO thymi are summarized.
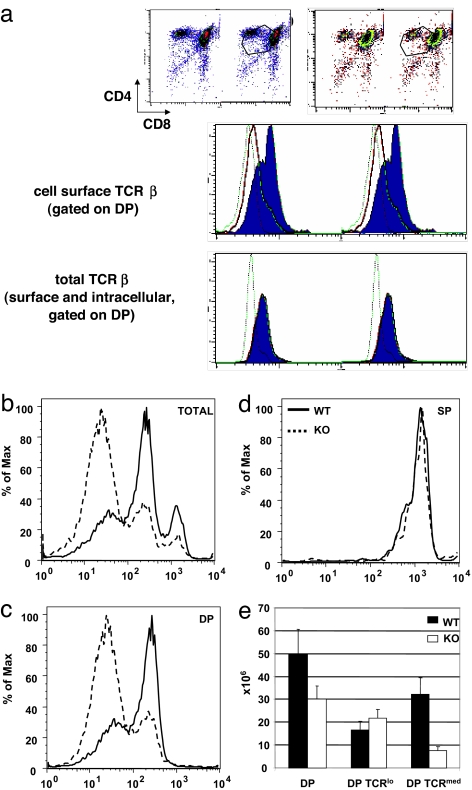
In addition to decreased percentages of SP thymocytes, analysis of cell surface TCR expression on DP thymocytes revealed a skewing toward lower expression on ATM-deficient cells (Fig. 2a, cell surface TCRβ), consistent with previous observations (15). Thymocytes from WT or ATM-deficient mice were cultured for 4 h as single-cell suspensions to allow up-regulation of TCR and, therefore, maximal discrimination between those DP cells expressing low surface levels of TCR (TCRlo) and those expressing a higher level of cell surface TCR (TCRmed) (16). Thymocytes displayed a trimodal distribution of cell surface TCR expression, defining TCRlo, TCRmed, and TCRhi subpopulations in both WT and ATM-deficient thymi (Fig. 2b). However, the relative proportions of these subpopulations differed strikingly in WT and ATM-deficient thymocytes. Thymi from ATM-deficient mice had a substantially higher proportion of TCRlo cells, and lower proportions of TCRmed and TCRhi cells. Gating on DP thymocytes demonstrated bimodal distributions of TCR cell surface density, with ATM-deficient cells showing a predominance of TCRlo cells, whereas TCRmed cells predominated in WT control DP thymocytes (Fig. 2c). The high levels of TCR found on the ATM-deficient SP population were virtually superimposable on the WT SP profiles (Fig. 2d). The number of TCRlo DP cells is slightly increased in ATM-deficient mice, whereas the number of TCRmed DP thymocytes is markedly lower than WT controls (Fig. 2e). These data indicate a substantial defect in thymocyte development in ATM-deficient mice at the transition from the DP TCRlo to DP TCRmed cells.
Fig. 2.
TCR expression in ATM-deficient mice. (a) (Top) Freshly explanted ATM WT (Left) and KO (Right) thymocytes were stained with anti-CD4, anti-CD8, and anti-TCRβ antibodies and gated to allow analysis of DP thymocytes. (Middle and Bottom) Anti-TCRβ staining was either cell-surface-specific or carried out after permeabilizing to measure total cellular TCRβ. The solid line indicates staining of ATM-deficient thymocytes, and the shaded area represents WT control thymocytes. The dotted line indicates staining of TCRβ-deficient thymocytes. (b–d) Thymocytes from ATM WT and KO mice were cultured for 4 h to allow the up-regulation of TCR expression and then stained with anti-CD4, anti-CD8, and anti-TCRβ antibodies and analyzed by flow cytometry. (b) Cell surface TCRβ expression is presented for the total number of thymocytes. The solid lines represent WT control thymocytes, and the dotted lines represent ATM KO thymocytes. (c) Thymocytes were gated to assess TCRβ expression on CD4+CD8+ DP thymocytes. ATM-deficient DP thymocytes have an increased proportion of TCRlo and a decreased proportion of TCRmed cells. (d) TCRβ expression is equivalent on TCRhi SP ATM KO and WT thymocytes. (e) Numbers of total DP cells, TCRlo DP cells, and TCRmed DP cells in ATM KO and WT thymi.
To make the transition from the DN to the DP stage, developing T cells must first successfully rearrange and express the TCRβ locus, allowing expression of the pre-TCR and transition from the DN3 to DN4 developmental stage (17). To further assess TCRβ expression in DP TCRlo thymocytes from ATM-deficient mice, thymocytes were permeabilized and stained with TCRβ-specific H57 Ab to measure total cellular TCRβ protein. Gating on DP thymocytes, total TCRβ protein expression was superimposable between the WT and ATM-deficient cells (Fig. 2a), in contrast to the substantially lower cell surface expression on ATM-deficient versus WT cells. Thus, TCRβ protein is present at normal levels in ATM-deficient DP thymocytes, but is expressed on the cell surface at greatly reduced levels. The accumulation of these TCRlo cells indicates that thymocyte development in the absence of ATM is severely impaired predominantly beyond the stage of TCRβ rearrangement and pre-TCR expression, but before thymocytes acquire the expression of intermediate levels of TCR.
Expression of Rearranged TCR Transgenes Overcomes the Defect in ATM-Deficient Thymic Differentiation.
Up-regulation of cell surface TCR expression on DP thymocytes, as well as subsequent differentiation and selection of SP thymocytes, requires expression of TCRα and pairing with TCRβ to generate the mature TCR. The observed defect in ATM-deficient thymic development therefore suggests a defect in TCRα expression. This defect could be due to defective gene rearrangement or reduced expression of functionally rearranged genes. To distinguish between these two possibilities, already rearranged TCRα and TCRβ transgenes were introduced into ATM-deficient mice. ATM WT or deficient mice were bred to mice expressing a transgenic TCR specific for the male antigen H-Y in association with the class I molecule Db (18). In female mice, expression of the transgenic TCR leads to strong positive selection of DP thymocytes to the CD8 SP lineage. Because this transgenic system is “leaky,” with many female DP thymocytes rearranging and expressing endogenous TCRα (19), it allows parallel analysis of the development and selection of those cells expressing the rearranged clonotypic TCRαβ receptor and those cells rearranging and expressing endogenous TCRα.
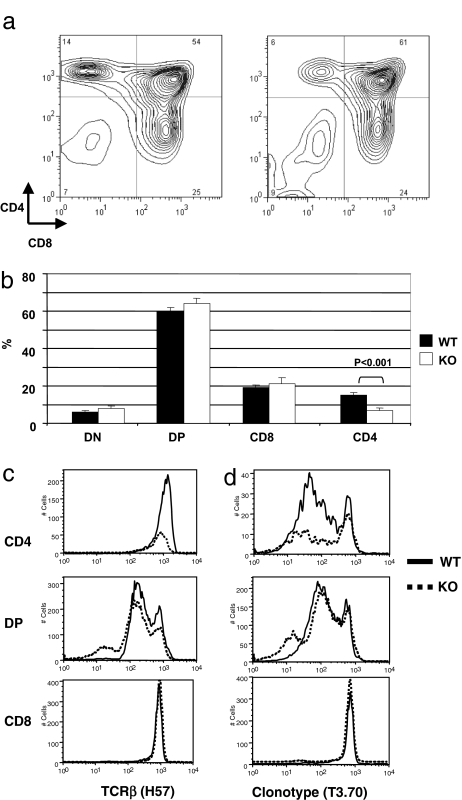
Analysis of female TCR transgenic mice revealed that, in contrast to the defects identified in non-TCR transgenic ATM KO thymocytes, there was no significant difference in the percentages of DP or DN thymocytes between ATM WT and ATM-deficient mice (Fig. 3a and b). In addition, there was no significant difference in the recovery of CD8 SP thymocytes between WT and ATM-deficient female TCR transgenic littermates. Thus, forced expression of rearranged TCR transgenes allows thymocyte-positive selection to proceed unhindered in the absence of ATM, suggesting that ATM does not play an integral role in thymocyte development independent of TCR rearrangement.
Fig. 3.
Thymic subpopulations and cell surface TCR expression in ATM-deficient TCR transgenic mice. Thymocytes from female ATM WT or KO mice expressing the H-Y TCR transgene were stained with anti-CD4, anti-CD8, and anti-TCRβ antibodies and analyzed by flow cytometry. (a) CD4 and CD8 expression by ATM WT (Left) and KO (Right) thymocytes. (b) Percentages of DN, DP, and SP thymocytes in female ATM WT or KO mice expressing the H-Y TCR transgene. (c) Cell surface expression of TCRβ on DP, CD4 SP, and CD8 SP thymocytes. Overall TCRβ expression is similar on ATM WT and KO DP and SP thymocytes. (d) Clonotype (T3.70)lo CD4 SP thymocytes, which express endogenous TCRα, are reduced in ATM KO mice.
CD4 SP Thymocytes That Express Endogenous TCRα Are Reduced in ATM-Deficient TCR Transgenic Mice.
Whereas there was no difference in the recovery of CD8 SP thymocytes, there was a striking decrease in the recovery of CD4 SP thymocytes in ATM-deficient female mice. TCR transgenic ATM WT females had 15.1 ± 1.4% CD4 SP thymocytes, whereas ATM-deficient TCR transgenic females had only 6.9 ± 1.2% CD4 SP cells (P < 0.001) (Fig. 3b). The CD4 SP population of thymocytes in WT TCR transgenic mice has been shown to express not only the transgenic TCRα, but also rearranged endogenous TCRα chains (19). Individual CD4 T cells thus express two TCRs, one being the clonotypic TCRαβ pair and the other consisting of the transgenic TCRβ chain in association with endogenous TCRα chain. Thymocyte subsets were therefore analyzed by flow cytometry for total TCR expression by using an antibody specific for TCRβ (clone H57-597) (Fig. 3c), or for expression of the transgenic TCR by using the clonotype-specific antibody T3.70 (Fig. 3d). Analysis with anti-TCRβ antibody reveals equivalent TCR surface expression on CD8 and CD4 SP thymocytes from ATM-deficient and WT mice. TCRβ expression on DP cells is also similar except for a small TCR-negative population detectable in the ATM-deficient but not the WT control thymocytes (Fig. 3c Middle). This is in marked contrast to the decreased TCRβ expression observed in ATM-deficient non-TCR transgenic DP thymocytes (Fig. 2a). When cell surface expression of the clonotypic transgenic TCR was examined, expression was found to be similar in CD8 and DP thymocytes from ATM-deficient and WT populations, consistent with the anti-TCRβ staining results. These data suggest that the defect in TCR expression observed in non-TCR transgenic ATM-deficient DP thymocytes is not due to defects in postrearrangement events. In contrast, ATM deficiency had a marked effect on expression of the clonotypic TCR on CD4 SP cells. TCR transgenic ATM WT females show a predominantly clonotype-dull population, consistent with a selective pressure favoring MHC class II-restricted CD4 cells that express TCR distinct from the clonotypic H-Y receptor, and requiring rearrangement and expression of endogenous TCRα (19). However, in contrast to ATM WT TCR transgenic female mice, the proportion of clonotype-dull CD4 cells is notably decreased in ATM-deficient TCR transgenic females (Fig. 3d), indicating a specific reduction in the number of thymocytes expressing an endogenous TCRα chain. When thymocytes were further analyzed for expression of endogenous TCRα chain by staining with a pool of Vα2- and Vα8-specific antibodies, substantial reductions in expression of these two Vα families were observed in DP and CD4 SP ATM subpopulations. Thus, in ATM-deficient thymi, surface expression of endogenous TCRα chains is markedly reduced relative to expression of the rearranged transgenic TCRα chain, indicating a role of ATM in the process of TCRα rearrangement that is necessary for expression of endogenous, but not transgenic TCRα.
In the Absence of ATM, DP Thymocytes Have Less Vα-Jα Rearrangement and More Unresolved, Open CE Breaks.
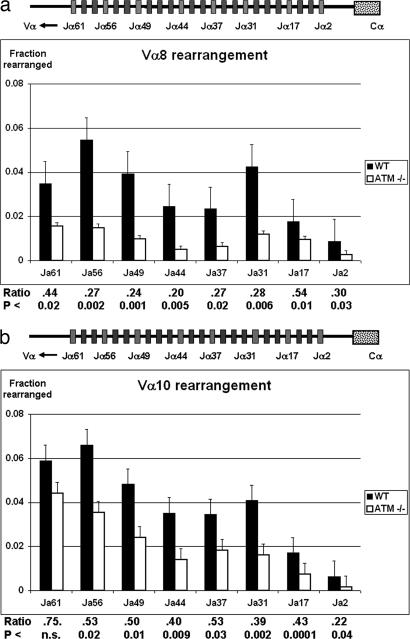
One mechanism by which ATM deficiency could result in decreased expression of endogenous, but not transgenic, TCRα chain is through a defect in TCRα rearrangement. To directly assess the role of ATM in TCRα recombination, we first compared the frequency of genomic TCR rearrangements in DP thymocytes from ATM-deficient and WT mice. TCR Vα-Jα rearrangements were assessed by quantitative, real-time PCR performed on genomic DNA purified from sorted DP thymocytes. We found a statistically significant decrease in the relative frequency of Vα8 rearrangement to all seven Jα loci analyzed (Fig. 4a) and of Vα10 rearrangement to six of the seven analyzed Jα loci (Fig. 4b). This reduction in Vα-Jα rearrangement was similarly observed in analysis of flow sorted TCRlo and TCRmed DP ATM thymocytes (data not shown). The reduction of Vα-Jα rearrangement across the entire Jα cluster indicates that loss of ATM results in a generalized defect in TCRα locus recombination.
Fig. 4.
Frequency of Vα-Jα rearrangements in DP thymocytes from ATM KO and WT mice. Genomic DNA was isolated from the sorted DP thymocytes of Atm−/− mice and control Atm+/+ littermates. Genomic rearrangements of Vα8 (a) or Vα10 (b) to multiple Jα genes were measured by quantitative PCR and normalized to the signal of the invariant Cα. Ratios of the amount of each Vα-Jα rearrangement in ATM KO DP thymocytes relative to the amount in WT control DP thymocytes and the significance of the differences between ATM-deficient and WT rearrangements for each Vα-Jα combinations are shown. Rearrangements in three to eight individual mice, which were analyzed for each data point, are shown. The organization of the TCR Jα cluster is displayed at the top.
ATM deficiency could impair Vα-Jα rearrangement indirectly, by causing decreased survival of ATM-deficient thymocytes and therefore a shorter window in which to accomplish TCRα rearrangement. To address this possibility, ATM-deficient or TCR transgenic ATM-deficient mice were bred to express a bcl-2 transgene that prolongs thymocyte survival through its antiapoptotic activity. Introduction of the bcl-2 transgene had no effect on the frequency of Vα10 to Jα56 (proximal) or Jα31 (distal) gene segments despite decreasing the frequency of annexin V+ thymocytes from both ATM WT and ATM-deficient mice (data not shown). Moreover, increased expression of the bcl-2 transgene had no effect on the thymic subpopulation phenotype in ATM-deficient TCR transgenic or TCR nontransgenic mice compared with respective WT controls (data not shown). Decreased thymocyte survival thus does not appear to be a significant factor in the decreased frequency in TCRα rearrangement in ATM-deficient thymus.
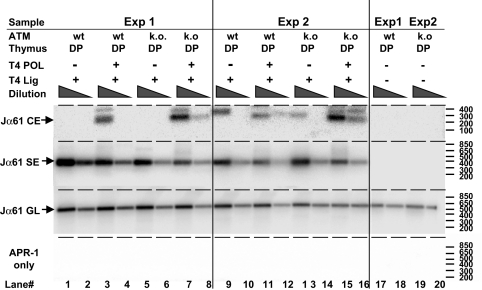
The decrease in TCR Vα-Jα rearrangement in ATM-deficient mice could be due to fewer attempts at rearrangement, or alternatively, to less efficient resolution of Vα and Jα CE breaks. To address these possibilities, thymocytes from ATM WT and deficient mice were assessed for the accumulation of CE and SE breaks in vivo (20). We found consistently increased amounts of Jα61-associated CE breaks in ATM-deficient DP thymocytes, whereas amounts of the corresponding SE breaks did not differ (Fig. 5). These results indicate that ATM deficiency does not affect cleavage, but rather compromises the joining phase of V-J recombination, resulting in decreased TCRα rearrangement and expression, and leading to a defect in generation of mature TCR-expressing T cells.
Fig. 5.
Detection of open CE breaks in WT and ATM-deficient DP thymocytes. Genomic DNA was isolated from sorted DP thymocytes of Atm−/− mice and control Atm+/+ littermates, as described in Fig. 5 and Materials and Methods, and ligation-mediated PCR was performed to measure open CE breaks. Results shown are PCR amplifications from 3-fold dilutions of ligation product.
Discussion
Our investigations were undertaken to define the role of ATM in T cell differentiation. We found that loss of ATM results in accumulation of a subset of DP thymocytes with a TCRlo phenotype, accompanied by a significant loss of DP thymocytes expressing a more mature TCRmed phenotype, as well as reduced numbers of mature TCRhi SP thymocytes. In studies assessing the basis for this developmental deficiency, we have identified a direct role for ATM in TCR gene rearrangement. Deficiency of ATM leads to decreased frequency of successful TCR Vα-Jα rearrangements in multiple Vα-Jα families studied. This decrease in TCR Vα-Jα rearrangement is not rescued by introduction of a bcl-2 transgene that would allow for prolonged DP survival, suggesting that ATM is not simply affecting DP thymocyte life span but rather is an integral part of the recombination process. Ligation-mediated PCR assay of Jα61 unresolved open CEs demonstrated that the frequency of CEs is higher in ATM-deficient DP thymocytes than in WT littermates, despite the observed decrease in Vα-Jα rearrangement in this population. These data imply a primary role for ATM in supporting efficient Vα-Jα CE joining in DP thymocytes. Deficiency of ATM thus leads to a reduction in TCRα locus recombination, which results in the accumulation of TCRlo DP cells and fewer TCRmed DP thymocytes, thereby generating a smaller pool from which SP cells can be selected.
Consistent with the above interpretation, we show that introduction of rearranged TCR transgenes rescues thymic development in ATM-deficient mice, consistent with the findings of Chao et al. (21). Our use of the leaky class I-restricted H-Y-specific TCR transgenic model system allowed analysis not only of the positively selected CD8 SP cells expressing the TCRαβ transgenes but also of the CD4 SP thymocytes expressing endogenous TCRα. This comparison most strongly indicates that ATM plays no role in development and selection of those thymocytes already expressing the rearranged TCR transgenes, but rather is important to TCRα gene rearrangement and expression.
ATM could either indirectly affect gene rearrangement by influencing differentiation and/or survival of DP thymocytes or directly impacting the mechanism of V(D)J recombination. A transgenic Rag-reconstitution model allowed analysis of DP thymocytes that rapidly lose expression of Rag-1 and can only undergo an early, single round of TCRα recombination (22). In another model, a modified TCRα locus was generated by gene targeting that has a very limited capacity for revision of TCRα rearrangement (TCRαsJ) (23). In both of these models, the limited ability to rearrange the TCRα locus results in decreased SP maturation, as well as accumulation of the TCRlo DP thymocytes. In other models, prolongation of DP life span results in the utilization of more distal Jα gene segments, whereas increased apoptosis of DP cells leads to more proximal Jα usage bias (24). A common feature of all of these models is the profound alteration of Jα gene usage. In contrast, we did not observe a change in the pattern of Jα gene recombination in the thymus (Fig. 4) or in peripheral T cells (data not shown) of ATM-deficient mice compared with WT mice. Furthermore, we could not increase TCRα gene rearrangement in ATM-deficient cells by introducing the antiapoptotic bcl-2 transgene. These studies have thus failed to indicate a role for ATM in substantially affecting DP cell survival in response to endogenous Vα recombination, in accordance with earlier suggestions (25). We propose that ATM is critical for the rapid and efficient recruitment of the nonhomologous end-joining repair apparatus that is required for CE joining. Although ATM deficiency, unlike nonhomologous end-joining repair deficiency, does not completely abolish coding joint formation, it sufficiently delays TCRα rearrangement to severely reduce the size of the CD3med pool of DP thymocytes that are the subject of positive selection. Our findings thus directly link reduced Vα-Jα rearrangement, as the primary cause, to defective thymocyte maturation in ATM-deficient mice.
While this manuscript was in preparation, a report by Matei et al. (26) also presented evidence for defective thymocyte maturation in ATM-deficient mice. However, the conclusions of Matei et al. are significantly different from those reported here. They conclude that biallelic loss of distal Vα gene segments may underlie the defect in surface TCRαβ expression and SP thymocyte selection. In contrast, our model proposes that delayed coding joint formation results in reduced Vα-Jα coding joint formation and, consequently, defective thymocyte maturation. Matei et al. (26) found no reduction in TCRα locus rearrangement, as measured by a Southern blot assay of unfractionated thymocytes, whereas we show, by quantitative PCR, a statistically significant 25–75% reduction in Vα-Jα joining for multiple Vα and Jα combinations in purified ATM-deficient DP cells (Fig. 4), in good agreement with the magnitude of loss of CD3med DP thymocytes in ATM−/− mice. Notably, Matei et al. (26) also did not detect free CE in either ATM-deficient or control thymocytes, in contrast to our finding of increased CE (but not SE) in ATM-deficient DP thymocytes (Fig. 5). We therefore conclude that delayed TCRα coding joint formation and consequent reduction in functional Vα-Jα rearrangements is the major mechanism that results in a reduced level of surface TCRαβ expression and the accompanying defect in thymocyte maturation in ATM-deficient mice.
Our demonstration of a direct role for ATM in Vα-Jα rearrangement is consistent with the recent report from Bredemeyer et al. (13), who provided evidence that ATM can function in the in vitro resolution of DNA double-strand breaks that are generated during V(D)J recombination. By using reporter constructs expressed in v-Abl transformed pre-B cell lines, these authors observed that recombination-activating-gene-dependent inversion-mediated rearrangement was significantly reduced in ATM-deficient pre-B cell lines and that this was accompanied by a substantial increase in hybrid joints between CEs and SEs and a selective accumulation of unrepaired CE in ATM-deficient cells. It was noted that these effects of ATM inactivation were not attributable to defects in checkpoint control or apoptosis, again consistent with our characterization of the in vivo defect in TCRα rearrangement in ATM-deficient mice (13).
We have focused our analysis on the role of ATM in TCRα gene rearrangement in DP thymocytes. Rearrangement of other TCR loci was not directly investigated. However, our preliminary data reinforce previous suggestions that ATM affects recombination of all TCR, and possibly, all antigen receptor loci. We have found dramatically increased frequency of TCR Vδ5 and Vβ14 hybrid joint formation, as well as TCRγ–TCRβ transrearrangements in ATM-deficient thymocytes (data not shown), in accordance with earlier reports (13, 27). Formal demonstration of increased accumulation of CE breaks at these TCR loci will be required to establish the possible connection between these genetic aberrations and defective V(D)J recombination in the absence of ATM. Nevertheless, our model of ATM-dependent resolution of CE breaks offers a unified mechanism to explain two of the major characteristics of both human and engineered mouse ATM-deficient conditions. Accordingly, delayed coding joint formation would compromise progression through multiple rounds of TCRα locus recombination, resulting in defective thymocyte maturation. In addition, an increase in CE persistence may lead to increased genomic instability and elevate the risk of chromosome translocations, deletion, and other aberrations involving the TCRα locus.
In addition to its critical role in V(D)J recombination, site-specific recombination is involved in another lymphocyte-specific process, Ig class switch recombination in which specific donor and acceptor sequences allow recombination between two Ig constant regions to mediate the switching of an IgM molecule with a given antigen-binding specificity to a different class of Ig (IgG, IgA, or IgE) bearing the same antigen-specific variable region. Clinical observations in patients with ataxia telangiectasia have noted defects in expression of switched Ig isotypes (28). Subsequently, we and others have demonstrated in mouse models that Atm inactivation results in a substantial defect in class switch recombination, indicating a direct role of ATM in this process (7, 8). ATM thus appears to play an important role in both V(D)J recombination and class switch recombination, the two classes of physiologic DNA break/repair events that are critical to immune function.
Materials and Methods
Mice.
Atm−/− mice were generated by introducing a truncation mutation into the gene at nucleotide 5790 (14) and maintained as progeny of heterozygous matings (7, 10). Atm+/+ littermates were used as controls. H-Y-specific TCR transgenic mice were generated and used as described previously (18, 29). All mice used in this study were maintained at Bioqual (Rockville, MD) and analyzed at 7–12 weeks of age. All animal experiments were approved by the National Cancer Institute Animal Care and Use Committee.
Flow Cytometry.
Cells were prepared for flow-cytometric analysis as described previously (30). The antibody specific for the clonotypic H-Y-specific TCR, T3.70, was a generous gift from Dr. Elizabeth Shores (Food and Drug Administration, Bethesda, MD). Antibodies specific for CD4, CD8, TCRβ (H57-597), TCR Vα, and TCR Vβ specificities were purchased from BD PharMingen (San Diego, CA). Detection of intracellular TCRβ was performed as described previously (31). Four-hour culture of thymocytes for up-regulation of TCR expression was carried out as described previously (16).
Genomic Real-Time Quantitative PCR.
Genomic DNA was isolated from flow-cytometrically sorted thymocyte populations of Atm−/− mice and control Atm+/+ littermates. Twenty-five to 50 ng of DNA was PCR amplified with a combination of Vα gene and Jα gene-specific primers (synthesized by IDT, Coralville, IA) by using the Power Sybr Green Master Mix kit (Applied Biosystems, Foster City, CA) for 40 cycles in 96-well plates. Forward (F) and reverse (R) primers used were as follows: Vα2(F), TGGAGACTCAGCCACCTACT; Vα6(F), CTCTGACAGAAAGTCAAGCAC; Vα8(F), GCAGCAGCTCCTTCCATC; Vα10(F), AAGAACGTCGCAGCTCTTTG; Jα2(R), TACCGGGTTGCAAATGGTG; Jα17(R), TGATGGCTAGGCTCCTTTTC; Jα31(R), CATATTAAATTGCATCTCTGCT; Jα37(R), AAATGAGCATAAAGCGACAG; Jα44(R), TACAGGAATACCCAGTCAAG; Jα49(R), GATAGCTCCTTCTGCAGGTT; Jα56(R), GACTTGTTCCTCACTGAAAC; Jα61(R), TCCAGCAATTTTCAGGTAGA; Cα(F), GTGTGAGCATGGGAGACAGA; Cα(R), TGGAGAGAGACAGGGGAAAG; Jα61 SE, GGTCCACGTCCAGATGCCAACT; Jα61 CE, TCGTCCCAAATTGTAGGTTGT (32).
Data were collected on an ABI 9700 Sequence Analyzer and analyzed by using the SDS 2.0 software (Applied Biosystems). For each assay, DNA sample was also analyzed for the control, nonrearranging Cα gene. The relative amount of V-Jα rearrangements in each sample was calculated by the following formula: 1.9−CtVxJy/1.9−CtCα. The relative ratios of each Vα-Jα rearrangements were averaged from all calculations and plotted together with the standard error of the mean. Statistical significance was calculated by using Student's t test. Every quantitative PCR was inspected for single product formation by performing melting curve analysis, and the specificity of each quantitative PCR was verified by running the amplified products on agarose gels to determine the size and specificity of products.
Ligation-Mediated PCR.
A total of ≈1 μg of genomic DNA, isolated as described above, was used per each sample by modification of the previously described procedure (20). DNA was treated with T4 polymerase (New England Biolabs, Boston, MA) at a ratio of 1.5 units per microgram of DNA in the presence of 200 μM dNTP at 15°C for 5 min. The DNA was phenol-chloroform extracted, ethanol-precipitated, and ligated to a double-strand, blunt, anchor oligonucleotide (20) by using T4 DNA ligase (New England Biolabs) at 15°C for overnight incubation. This procedure was shown to blunt 3′ overhang CE breaks and leave blunt SE breaks intact. Semiquantitative PCRs were performed on serial dilutions of ligation products by using anchor-specific and gene-specific primers to detect CE and SE breaks by using Platinum Taq polymerase (Invitrogen, Carlsbad, CA). Positive control reactions with the gene-specific primers and negative control reactions by using only the anchor-specific (APR-1) primer were also performed. All PCRs included a 5-cycle touchdown incubation followed by 25–35 cycles of amplification at 60°C in a 96-well plate on a thermocycler (MJ Research, Watertown, MA). PCR products were separated on composite agarose-sieving agarose gels (Cambrex Bioscience, Rockland, ME), transferred to charged nylon membranes (PerkinElmer, Boston, MA), and hybridized to specific, radioactive-labeled probes internal to the PCR amplification primer sites. Images were acquired and analyzed by using a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Acknowledgments
We thank Martin Gellert, Karen Hathcock, Joy Williams, Andre Nussenzweig, and Alfred Singer for their constructive comments during the course of this work and review of this manuscript; Jayne Danska and Cynthia Guidos for discussion of our work and for sharing their results before publication; Genevieve Sanchez-Howard and the staff at Bioqual for excellent animal care and husbandry; and Susan Sharrow, Tony Adams, and Larry Granger for flow cytometry and flow cytometric cell sorting. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Abbreviations
- ATM
ataxia telangiectasia mutated
- CE
coding end
- DN
double negative
- DP
double positive
- F
forward
- KO
knockout
- R
reverse
- SE
signal end
- SP
single positive
- TCR
T cell receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Kastan MB, Lim DS. Nat Rev Mol Cell Biol. 2000;1:179–186. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- 2.Shiloh Y. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 3.Bakkenist CJ, Kastan MB. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 4.Kitagawa R, Kastan MB. Cold Spring Harb Symp Quant Biol. 2005;70:99–109. doi: 10.1101/sqb.2005.70.002. [DOI] [PubMed] [Google Scholar]
- 5.Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A. Nature. 2006;443:222–225. doi: 10.1038/nature05112. [DOI] [PubMed] [Google Scholar]
- 6.Matei IR, Guidos CJ, Danska JS. Immunol Rev. 2006;209:142–158. doi: 10.1111/j.0105-2896.2006.00361.x. [DOI] [PubMed] [Google Scholar]
- 7.Lumsden JM, McCarty T, Petiniot LK, Shen R, Barlow C, Wynn TA, Morse HC, III, Gearhart PJ, Wynshaw-Boris A, Max EE, et al. J Exp Med. 2004;200:1111–1121. doi: 10.1084/jem.20041074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liyanage M, Weaver Z, Barlow C, Coleman A, Pankratz DG, Anderson S, Wynshaw-Boris A, Ried T. Blood. 2000;96:1940–1946. [PubMed] [Google Scholar]
- 10.Petiniot LK, Weaver Z, Barlow C, Shen R, Eckhaus M, Steinberg SM, Ried T, Wynshaw-Boris A, Hodes RJ. Proc Natl Acad Sci USA. 2000;97:6664–6669. doi: 10.1073/pnas.97.12.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petiniot LK, Weaver Z, Vacchio M, Shen R, Wangsa D, Barlow C, Eckhaus M, Steinberg SM, Wynshaw-Boris A, Ried T, et al. Mol Cell Biol. 2002;22:3174–3177. doi: 10.1128/MCB.22.9.3174-3177.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perkins EJ, Nair A, Cowley DO, Van Dyke T, Chang Y, Ramsden DA. Genes Dev. 2002;16:159–164. doi: 10.1101/gad.956902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 14.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, et al. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 15.Borghesani PR, Alt FW, Bottaro A, Davidson L, Aksoy S, Rathbun GA, Roberts TM, Swat W, Segal RA, Gu Y. Proc Natl Acad Sci USA. 2000;97:3336–3341. doi: 10.1073/pnas.050584897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama T, June CH, Munitz TI, Sheard M, McCarthy SA, Sharrow SO, Samelson LE, Singer A. Science. 1990;249:1558–1561. doi: 10.1126/science.2120773. [DOI] [PubMed] [Google Scholar]
- 17.von Boehmer H, Fehling HJ. Ann Rev Immunol. 1997;15:433–452. doi: 10.1146/annurev.immunol.15.1.433. [DOI] [PubMed] [Google Scholar]
- 18.Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Nature. 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 19.Borgulya P, Kishi H, Uematsu Y, von Boehmer H. Cell. 1992;69:529–537. doi: 10.1016/0092-8674(92)90453-j. [DOI] [PubMed] [Google Scholar]
- 20.Livak F, Schatz DG. J Mol Biol. 1997;267:1–9. doi: 10.1006/jmbi.1996.0834. [DOI] [PubMed] [Google Scholar]
- 21.Chao C, Yang EM, Xu Y. J Immunol. 2000;164:345–349. doi: 10.4049/jimmunol.164.1.345. [DOI] [PubMed] [Google Scholar]
- 22.Yannoutsos N, Wilson P, Yu W, Chen HT, Nussenzweig A, Petrie H, Nussenzweig MC. J Exp Med. 2001;194:471–480. doi: 10.1084/jem.194.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang CY, Sleckman BP, Kanagawa O. Proc Natl Acad Sci USA. 2005;102:14356–14361. doi: 10.1073/pnas.0505564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW. Nat Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 25.Bhandoola A, Dolnick B, Fayad N, Nussenzweig A, Singer A. J Exp Med. 2000;192:891–897. doi: 10.1084/jem.192.6.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matei IR, Gladdy RA, Nutter LM, Canty A, Guidos CJ, Danska JS. Blood. 2007;109:1887–1896. doi: 10.1182/blood-2006-05-020917. [DOI] [PubMed] [Google Scholar]
- 27.Lipkowitz S, Stern MH, Kirsch IR. J Exp Med. 1990;172:409–418. doi: 10.1084/jem.172.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavin MF, Shiloh Y. Annu Rev Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 29.Vacchio MS, Hodes RJ. J Exp Med. 2003;197:19–26. doi: 10.1084/jem.20021429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vacchio MS, Williams JA, Hodes RJ. Eur J Immunol. 2005;35:418–427. doi: 10.1002/eji.200424918. [DOI] [PubMed] [Google Scholar]
- 31.Williams JA, Hathcock KS, Klug D, Harada Y, Choudhury B, Allison JP, Abe R, Hodes RJ. J Immunol. 2005;175:4199–4207. doi: 10.4049/jimmunol.175.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak F, Schatz DG. Mol Cell Biol. 1996;16:609–618. doi: 10.1128/mcb.16.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]