Abstract
Plants rely on ribulose bisphosphate carboxylase/oxygenase (Rubisco) for carbon fixation. Higher plant Rubisco possesses an L8S8 structure, with the large subunit (LS) encoded in the chloroplast by rbcL and the small subunit encoded by the nuclear RBCS gene family. Because its components accumulate stoichiometrically but are encoded in two genetic compartments, rbcL and RBCS expression must be tightly coordinated. Although this coordination has been observed, the underlying mechanisms have not been defined. Here, we use tobacco to understand how LS translation is related to its assembly status. To do so, two transgenic lines deficient in LS biogenesis were created: a chloroplast transformant expressing a truncated and unstable LS polypeptide, and a line where a homolog of the maize Rubisco-specific chaperone, BSD2, was repressed by RNAi. We found that in both lines, LS translation is no longer regulated by the availability of small subunit (SS), indicating that LS translation is not activated by the presence of its assembly partner but, rather, undergoes an autoregulation of translation. Pulse labeling experiments indicate that LS is synthesized but not accumulated in the transgenic lines, suggesting that accumulation of a repressor motif is required for LS assembly-dependent translational regulation.
Keywords: autoregulation, gene expression, synthesis, plant
Macromolecular organellar energetic complexes are essential and require assembly in defined stoichiometric ratios. Their biogenesis is complicated by their dual genetic origins, with subunits encoded both in the organelle and the nucleus. Prior studies led to the notion of concerted accumulation: the absence of one core subunit is accompanied by the loss of its assembly partners, as exemplified by mitochondrial mutants in yeast (1) or chloroplast mutants in Chlamydomonas (2). Two main mechanisms responsible for unassembled subunits' fate have been identified. The most common is rapid proteolytic degradation, and, in certain cases, substrates for specific chloroplast proteases have been identified (3–7). Alternatively, some chloroplast-encoded proteins are subject to assembly-dependent translational regulation or control by epistasy of synthesis (CES) (8). The chloroplast uses CES in a hierarchical manner, whereas proteolysis is a relatively nonspecific mechanism for disposing of excess or incorrectly folded subunits.
Insights into CES were first described for Chlamydomonas cytf, which is encoded by petA. Unassembled cytf could be shown to repress petA translation initiation, acting through the 5′ untranslated region (9) and the cytf C-terminal domain (10). Analogous regulatory features were subsequently uncovered for the PS I subunits PsaA and PsaC (11) and the PS II subunits D1 and CP47 (12). Together, these results suggest that assembly-dependent CES is a general feature of chloroplast gene expression in Chlamydomonas. However, its prevalence in higher plants is unclear.
Here, we have used Rubisco assembly as a model to investigate whether CES occurs in higher plants. Rubisco is a complex consisting of only two subunits, the large subunit (LS) (53 kDa) being encoded by the chloroplast rbcL gene, and the small subunit (SS) (14 kDa) being encoded by the RBCS nuclear gene family. The holoenzyme in higher plants is a 550-kDa L8S8 form whose assembly requires the BSD2 chaperone, identified through study of the eponymous maize mutant, which fails to accumulate Rubisco (13). BSD2 includes a DNAJ-like domain (14), suggesting that it might interact with nascent LS. Additional chaperones mediate the remainder of the L8S8 assembly pathway (15).
Essential features of concerted Rubisco subunit accumulation were identified in earlier studies. Chloramphenicol-treated Chlamydomonas cells, impaired in chloroplast and thus LS synthesis, showed rapid degradation of unassembled SS (16). On the other hand, deletion of Chlamydomonas RBCS genes (17) or knocked-down expression using antisense technology in tobacco (18, 19), showed that LS translation declines in absence of its assembly partner. LS synthesis is also subject to repression under oxidative stress (20). Nevertheless, the mechanism(s) leading to translational inhibition have not been solved. Two previously evoked mechanisms (21) could account for reduced LS synthesis in the absence of SS: either LS could feedback-regulate its own synthesis, or SS could act as a positive factor for LS synthesis. Here, we provide evidence that the CES mechanism responding to Rubisco assembly state is exerted by LS feedback regulation in higher plants.
Results
Rubisco LS Is a CES Protein.
By using an antisense strategy directed against RBCS, tobacco transformants accumulating ≈30% of WT Rubisco were previously obtained (18). Because there was a concomitant reduction in RBCS mRNA accumulation, rbcL mRNA polysome association, and LS synthesis, this demonstrated coordination between rbcL and RBCS expression. In particular, the correlation between reduced LS synthesis and the absence of its assembly partner marks LS as a CES subunit.
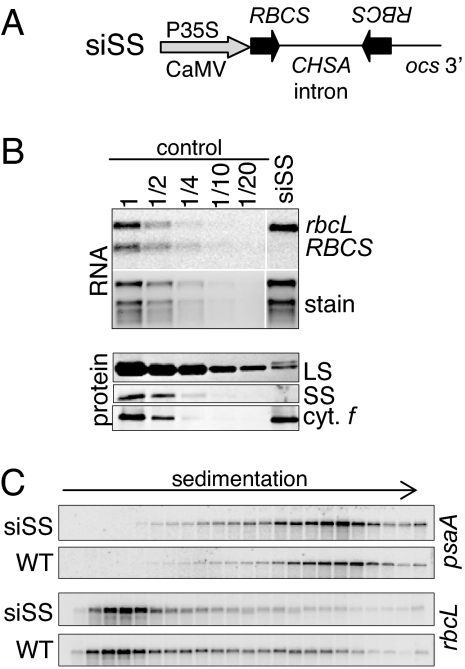
As a first step to obtain further insight into LS regulation, we established RNAi lines deficient in RBCS expression, using the vector siSS (Fig. 1A). Three transformants exhibiting strong Rubisco defects were further characterized, and results from a representative transformant are shown here. When mRNA accumulation was compared between siSS and a WT control, we found at least a 20-fold decrease for RBCS but no effect on rbcL (Fig. 1B). As expected from reduced RBCS expression, LS accumulation decreased to 5% or less of the control. We then investigated rbcL translational status using polysome analysis from sucrose density gradients. As shown in Fig. 1C, rbcL mRNA is less associated with polysomes in siSS vs. the control, because rbcL transcript is less prevalent in the heavier gradient fractions in contrast to the control chloroplast transcript psaA. These results agree with previous results (18, 19) and show that LS synthesis is subject to assembly state-dependent regulation that operates at the translational level. These results also validate the siSS line as a tool to study LS CES behavior.
Fig. 1.
Characterization of RBCS RNAi transformants. (A) Silencing construct targeting RBCS genes. An RBCS inverted repeat, separated by the chalcone synthase intron (CHSA), is under the control of the CaMV 35S promoter and octopine synthase (ocs) 3′ UTR. (B) RNA and protein analysis from a representative RBCS RNAi transformant (siSS). Total leaf RNA (5 μg or the indicated dilution of WT) was hybridized by using probes directed against rbcL or RBCS transcripts. The ethidium bromide stain is provided as a loading control. Total leaf proteins (30 μg or the indicated dilution of WT) were separated as described in Methods, and analyzed by using a Rubisco antibody. Cytf is provided as a loading control. The controls were a vector transformant for RNA analysis and the WT for protein analysis. (C) For polysome analysis, total leaf extract from siSS or WT were fractionated on sucrose gradients. An equal proportion of RNA from each fraction was analyzed by gel blot.
Generation of a Mutant Expressing Truncated LS and Reduced RBCS mRNA.
One alternative for LS CES regulation is that LS synthesis requires SS as a translation activator. In this case, the lack of SS (such as in siSS) would lead to reduced LS synthesis. A second mechanism envisions negative autoregulation of LS synthesis in the absence of assembly, which would also occur in siSS. We therefore undertook an in vivo approach to discriminate between these two possibilities, by creating a mutant affected in both LS and SS. If SS is an activator, the double mutant should exhibit the same phenotype as siSS, i.e., decreased rbcL polysomal association. Should the autoregulatory model prevail, however, down-regulation of LS translation should no longer occur in the double mutant.
To disrupt LS availability without affecting rbcL mRNA accumulation, a necessity because rbcL polysome association was to be measured, we introduced a premature stop codon into the rbcL gene using the vector pLS* (Fig. 2A), where a 4-bp insertion causes a frame shift leading to a 44-kDa polypeptide as well as a DNA polymorphism. We expected this mutation to dramatically destabilize LS, as did truncations in Oenothera (22) and Chlamydomonas (23), and a missense mutation in tobacco (24). An aadA selectable marker was included to allow transformant selection on the basis of spectinomycin and streptomycin resistance.
Fig. 2.
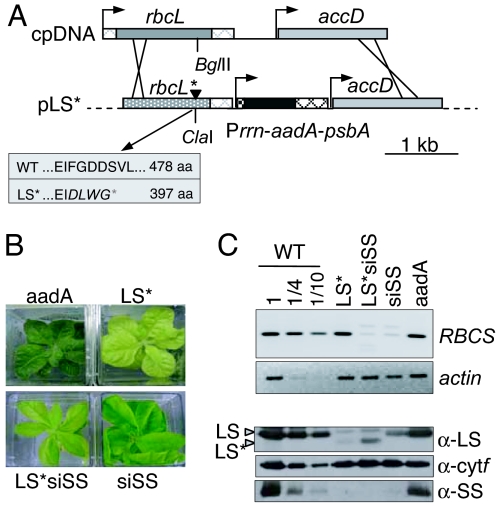
Creation of an LS/SS double mutant. (A) The relevant region of the chloroplast transformation vector is shown beneath the chloroplast rbcL–accD genomic region. The black arrowhead indicates the 4-bp insertion discussed in the text. In the gray box below the diagram, the mutant and WT LS sequences are compared, with altered amino acids italicized. (B) The four strains were photographed under similar lighting and magnification. (C) RT-PCR (Upper) and protein (Lower) analyses were performed, with RBCS transcript accumulation assessed by RT-PCR with actin as a control and immunoblotting with antisera shown at the right.
Two spectinomycin-resistant lines were obtained after biolistic transformation of WT tobacco with pLS*. RFLP analysis showed that one of them bore the rbcL* frame shift, whereas the second contained a WT rbcL gene. These lines were brought to homoplasmy by subcloning, generating the LS* and aadA transplastomic lines. The aadA line showed a WT phenotype (Fig. 2B), whereas LS* was chlorotic, more affected than siSS, and similar to a tobacco rbcL gene deletion (25). As expected, neither line was affected in RBCS transcript accumulation, as assessed by RT-PCR (Fig. 2C, lanes LS* and aadA).
To obtain an LS*–siSS double mutant, LS* was supertransformed with the siSS RNAi construct. One LS*siSS transformant was obtained on selective medium and exhibited an intermediate pale-green phenotype (Fig. 2B). As expected, RT-PCR revealed a >95% decrease in RBCS mRNA, comparable with siSS (Fig. 2C). Unexpectedly, LS*siSS could not be propagated on spectinomycin-containing medium, and, ultimately, DNA (data not shown) and RNA analysis (Fig. 3A) revealed that the aadA cassette had been lost in LS*siSS. However, RFLP analysis (data not shown) and detection of truncated LS (Fig. 2C Lower) indicated that the line retained the LS* mutation, whereas mRNA sequence analysis indicated that rbcL mRNA 3′ formation was not affected (data not shown). LS*siSS is, hence, the desired double mutant.
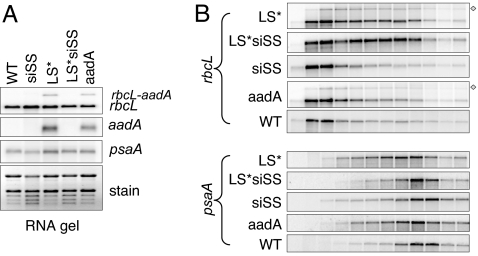
Fig. 3.
Transcript accumulation and polysome loading in transformed lines. (A) RNA gel blot for the lines shown across the top hybridized with rbcL, aadA, and psaA (top to bottom). rbcL-aadA is a dicistronic transcript present in LS* and aadA; that part of the gel has been cropped for the aadA probe. rRNA stained by ethidium bromide is shown as a loading control. (B) Polysomes were prepared and analyzed as described in the Fig. 1 legend for the lines shown at left. The discistronic rbcL–aadA mRNA is indicated by a diamond shape.
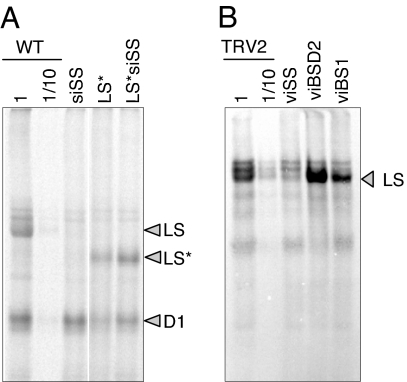
Immunoblot analysis (Fig. 2C) showed that the control line aadA normally accumulated LS and SS, whereas siSS underaccumulated Rubisco. The decrease in Rubisco accumulation in LS* and LS*siSS, where two faint bands were observed, the smaller of which is the size expected for truncated LS; the larger band, being an non-LS protein recognized by the antibody, is slightly more pronounced than in siSS. The difference in accumulation of the truncated LS* polypeptides in LS* and LS*siSS was not reproducible. As a result of concerted accumulation, SS was nearly undetectable. The more chlorotic phenotypes of LS* and LS*siSS compared with siSS probably result from the inability of LS*-expressing lines to form a functional Rubisco complex.
Truncated LS Is No Longer Regulated by Its Assembly State.
We next investigated rbcL polysomal association as a measure of LS translation. As a prerequisite, we confirmed that accumulation of monocistronic rbcL mRNA was similar in all of the lines (Fig. 3A). Dicistronic rbcL–aadA mRNA accumulated in LS* and aadA but was considered extraneous. Polysomal distributions are shown in Fig. 3B. As shown in Fig. 1, siSS has reduced rbcL polysome association. In contrast, rbcL polysomal association increases in LS* as compared with the controls, as it does for LS*siSS, whereas the psaA pattern was comparable between lines. We interpret these results to mean that SS does not promote LS translation, because in this experiment, LS* translation is insensitive to the expression of SS. A corollary conclusion is that, in LS* and LS*siSS, unassembled LS barely accumulates because of hyperinstability, preventing CES regulation.
Disruption of Tobacco BSD2 Prevents Rubisco Accumulation.
To substantiate our conclusions, we generated a second mutant affecting LS availability. Our strategy was to prevent LS accumulation by depleting the Rubisco chaperone BSD2, previously described in maize (14). To create transformants affected solely in BSD2, or in both BSD2 and RBCS, we used virus-induced gene silencing (VIGS) in Nicotiana benthamiana by the bipartite (TRV1 and TRV2) tobacco rattle RNA virus (26). For our experiments, either BSD2 or RBCS fragments were expressed in TRV2 and infiltrated with TRV1 into N. benthamiana seedlings at the 5-leaf stage. Infiltration with an empty TRV2 vector was used as a negative control, and TRV2 targeting phytoene desaturase (PDS), a carotenoid biosynthetic gene, was used as a positive control for leaf bleaching.
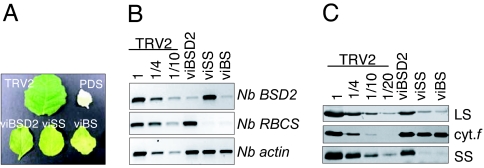
As shown in Fig. 4A, the controls worked as expected, because TRV itself confers only a mild growth phenotype. On the other hand, infiltration with TRV2 expressing either BSD2 (viBSD2) or RBCS (viSS) gene fragments resulted in slower growth and pale green leaves (the double mutant viBS is discussed below). RT-PCR analysis confirmed reduced mRNA accumulation for the targeted genes, with <10% of each transcript remaining (Fig. 4B). Immunoblot analysis showed that LS declined to ≈25% of the TRV2 control in viBSD2, and ≈10% in viSS (Fig. 4C), although rbcL mRNA levels were unchanged (data not shown). The difference in LS accumulation between viBSD2 and viSS reflects the relative efficiency of gene silencing rather than leakiness of the bsd2 mutation, because stable RNAi transformants targeting BSD2 exhibited a stronger LS deficiency (data not shown). The Rubisco deficiency in viBSD2 agrees with data from maize and supports a role of BSD2 in tobacco and perhaps all plants' Rubisco biogenesis. Thus, viBSD2 and viSS represent single mutants affected in LS or SS, respectively.
Fig. 4.
VIGS analysis of LS regulation. (A) Leaf phenotypes observed 3 weeks after inoculation for the lines shown. (B) RT-PCR was performed as described in Methods, with dilutions of cDNA from the control TRV2 line used to estimate the degree of silencing for the BSD2 and RBCS transcripts. Actin was used as a loading control. (C) Immunoblot analysis for the lines shown across the top and antibodies shown at right. Dilutions of protein from the control TRV2 were used to help estimate residual LS accumulation. Cytf was a loading control.
Reduced BSD2 Expression Leads to LS CES Deregulation.
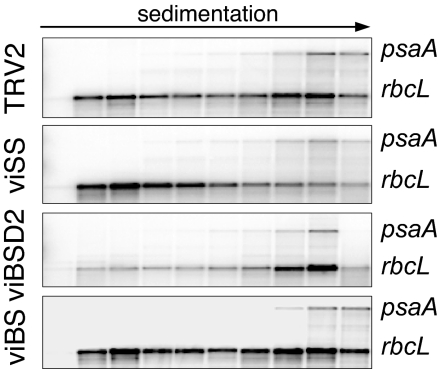
We then generated the viBS LS–SS double mutant by silencing simultaneously BSD2 and RBCS, after coinserting their gene fragments into the TRV2 vector. The double mutant exhibited pale-green leaves and accumulated ≈10% of the control LS protein level, and was shown by RT-PCR to have residual transcript accumulation comparable with the single mutants (Fig. 4), whereas it was unaffected in rbcL mRNA (data not shown). Next, we tested whether the CES autoregulatory model holds true in the VIGS context, by analyzing polysome gradients for each VIGS line (Fig. 5). psaA detection was used to show that the gradients were similarly prepared and, thus, that any variation observed for rbcL mRNA could be ascribed to gene silencing.
Fig. 5.
Polysome association of the rbcL transcript in VIGS-silenced lines. Polysomes were prepared as described in the Fig. 1 legend for the lines shown at left. RNA gel blots were probed simultaneously for rbcL and psaA.
Analysis of rbcL indeed confirmed that LS is a CES protein. In viSS, fewer rbcL transcripts are found in the heavier fractions of the gradient compared with the TRV2 control. viBSD2, in contrast, shows an almost complete disappearance of the free or monosome-associated rbcL transcripts, suggesting that, in this case, LS translation is increased. This is consistent with the assumption that LS is hyperunstable in viBSD2, rendering it unavailable for negative autoregulation. The expectation for the double mutant was that it would mimic viBSD2, because a hyperunstable LS should render the plant insensitive to the presence or absence of SS. We did not, however, obtain similar rbcL polysome profiles for viBSD2 and viBS, instead finding that viBS resembled most closely the TRV2 control. Although initially unexpected (see Discussion), these data still argue against a model where SS is required for LS translation.
LS Hyperinstability Prevents Accumulation of the LS CES Repressor Motif.
The above experiments revealed an increased proportion of polysome-associated rbcL mRNA in LS*, LS*siSS, viBSD2, and viBS, compared with cognizant lines silenced for RBCS alone. These observations strongly suggest that some amount of unassembled LS is required to down-regulate LS synthesis in the absence of its assembly. We hypothesized that either truncated LS or the absence of BSD2 would deplete this regulatory LS pool, for reasons discussed above. The absence of BSD2, however, has not yet been shown to destabilize LS and, in fact, has been argued to act on translation elongation (14). This raised the possibility that enhanced rbcL polysomal association in LS* and viBsd2 might arise through different mechanisms.
We subsequently investigated LS synthesis using pulse labeling. To do so, leaf discs of WT tobacco, siSS, LS*, and LS*siSS were labeled with [35S]Met/Cys in the presence of a cytosolic translation inhibitor, and equal quantities of radiolabeled products were separated by gel electrophoresis. As shown in Fig. 6A, siSS synthesized LS at <10% of the WT rate, in accordance with polysome data. In LS* and LS*siSS, synthesis of a shorter LS polypeptide was observed, as expected. Taking into account that LS* appears to be slightly underloaded, synthesis of truncated LS is comparable in LS* or LS*siSS, again in agreement with polysome data (Fig. 3). The polysome data also predicted an increased radiolabeling of the truncated polypeptide compared with the WT; however, this was not observed, suggesting that some of the newly synthesized LS* may undergo degradation during the 10-min pulse.
Fig. 6.
Pulse labeling of chloroplast proteins. (A) Equal amounts of radiolabeled translation products for the transplastomic lines shown across the top were analyzed as described in Methods. LS and LS* indicate full-length and truncated LS, respectively, and a protein presumed to be D1 is indicated. (B) Labeled proteins from VIGS lines were analyzed in the same way. Two dilutions of control proteins were used to estimate residual translation levels in experimental lines.
Young leaves from the N. benthamiana TRV2 control or VIGS-silenced plants were then subjected to the same protocol. The viSS plants exhibited a strong decrease in LS synthesis, as expected. In viBSD2, enhanced synthesis of full-length LS was observed. This argues against a role of BSD2 in translational elongation, because impaired elongation would probably have resulted in labeling of shorter LS polypeptides and reduced synthesis of full-length LS. Rather, it confirms that LS is synthesized but unstable (when compared with immunoblot data), except for the fraction that is protected by residual BSD2. LS synthesis in viBS was slightly reduced compared with viBSD2, mirroring the results of the polysome experiments. In conclusion, mutations resulting in the expression of truncated LS, or the absence of its dedicated chaperone, result in the same outcome: hyperinstability of unassembled LS, leading to a failure to trigger regulatory feedback.
Discussion
In this work, we used Rubisco as a model to assess whether assembly-dependent regulation of translation, or CES, occurs in higher plants as it does in Chlamydomonas. Some candidate higher plant CES subunits had been identified, including LS and cytf in tobacco (18, 19, 27) and CP47 in the barley vir115 mutant, which is primarily affected in D1 synthesis (28). However, whether in any of these cases the unassembled CES subunit regulates translation had not been determined. As shown here, reduced polysomal association of rbcL transcripts and reduced synthesis of LS in SS-deficient plants, suggest that it is indeed translation initiation that is regulated. However, we cannot exclude that LS elongation may be blocked at an early stage, as was elegantly shown for LS in etiolated barley plastids (29).
We used in vivo approaches to test whether translational autoregulation is exerted by unassembled LS, which must normally accumulate in trace amounts. To do so, we introduced mutations altering LS stability, either in LS itself (LS*) or by reducing BSD2 expression. In these plants, LS CES regulation no longer occurred whether or not SS was present. Therefore, the two main features observed for CES in Chlamydomonas are conserved: assembly-dependence and autoregulation of translation.
Our data lead to the hypothesis that unassembled LS exposes a repressor motif, otherwise not accessible, which mediates translational repression. Because the truncated LS* polypeptide is no longer able to repress its own translation, one interpretation is that the repressor motif is located in the missing C-terminal part. However, the C-terminal deletion might also hinder a conformational change required for LS translational inhibition or, through misfolding, lead to rapid proteolysis, rendering it unable to persist sufficiently to regulate translation. Because LS* is synthesized at a WT rate, this unstable protein might exhibit a steady-state accumulation similar to full-length LS in the line siSS. Results obtained for the VIGS-silenced lines viSS and viBS supports this interpretation. Indeed, residual full-length LS is at comparable levels in the two lines, whereas it has a different outcome on LS synthesis.
The repressor motif within LS, whose location is still unknown, could occur in any structure other than the holoenzyme, such as free LS, a dimer, or a chaperone-complexed form. Two models can be envisioned for a repressor–rbcL mRNA interaction. One invokes a ternary effector such as a specific translational activator, as has been proposed for Chlamydomonas cytf (18, 19). In this model, the translational activator is trapped and inactivated by the unassembled repressor motif. Alternatively, repression may parallel bacterial ribosomal protein feedback regulation, where a direct interaction between the protein and its cognate RNA is observed (30). Interestingly, gel mobility shift assays reveal an LS N-terminal domain RNA-binding activity under oxidizing conditions, although this activity is nonspecific (20, 31). The regulatory significance of this domain remains to be tested in vivo.
Our results indicate that CES could allow a rapid readjustment of LS synthesis according to the pool of available SS. The differences observed between viBSD2 and viBS provide key information in this respect. The fact that viBSD2 accumulates ≈25% of Rubisco, but almost no free rbcL mRNA is found, could be explained by the imbalance between the production of SS and LS. If SS were in excess relative to LS in viBSD2 it would trap all available LS, leaving virtually no unassembled LS to down-regulate its translation, leading to increased polysomal association. On the other hand, depletion of the SS pool by gene silencing in viBS would lead to some accumulation of non-SS-associated LS, which would repress translation and lead to the appearance of some free rbcL mRNA. These hypotheses fit well with our observed result, were corroborated by pulse labeling, and lead to the interpretation that SS availability is limiting in the WT, allowing some unassembled LS to accumulate and repress translation. The system therefore is in a dynamic equilibrium, able to respond either positively or negatively to environmental or metabolic changes that may target RBCS gene expression.
One major metabolic cue for LS regulation in plants may be sucrose allocation. It is well known that sucrose inhibits photosynthetic gene expression (32), with the RBCS gene being a typical example (33). If CES were involved, one would expect a concomitant inhibition of LS translation. Indeed, rbcL polysome association was reduced in WT tobacco grown on sucrose-supplemented medium (Fig. 3), as compared with the VIGS TRV2 control, which was grown in soil (Fig. 5), albeit at different light intensity. On the other hand, rbcL polysomal association increased in both LS* and LS*siSS relative to controls, although all these plants were grown on sucrose-supplemented medium. Thus, LS translational repression triggered by sucrose does not occur in CES-deregulated strains.
LS CES regulation may also respond to light and oxidative stress. Light stress generates reactive oxygen species, which can reversibly or irreversibly inactivate Rubisco. It was recently shown that methyl viologen-induced oxidative stress leads to inhibition of LS synthesis in tobacco and the purple bacterium Rhodospirillum rubrum (20), as previously shown for Chlamydomonas (34). Because Rhodospirillum lacks the SS, LS translational regulation must be SS-independent, as we have concluded is the case in tobacco. The regulatory mechanisms involved, however, may differ. In addition, in vivo data from Chlamydomonas (35) and cucumber (36), and in vitro data from wheat and barley (37, 38) suggest that LS undergoes fragmentation upon light and oxidative stress, followed by proteolysis. One may speculate that CES is involved, and might be enhanced if these cleavages expose the LS repressor motif.
We conclude that in vivo, LS translational behavior in tobacco obeys the classical CES paradigm developed from studies in Chlamydomonas, thus describing the first example in higher plants. We suggest that this regulation may be physiologically relevant, as it could be involved in the chloroplast response to metabolic, environmental, and also developmental cues, such as bundle sheath cell-specific accumulation of Rubisco in C4 plants.
Materials and Methods
Growth Conditions.
N. tabacum (cv. petit Havana) and N. benthamiana were grown at 25°C with a 16-h light/8-h dark period under 30 μE·m−1·s−2 and 300 μE·m−1·s−2, respectively. Chloroplast transplastomic lines were propagated on MSO medium (containing 30 g·liter−1 sucrose) supplemented with 500 μg·ml−1 spectinomycin. The nuclear transformants siSS and LS*siSS were maintained on MSO plus 2 mg·liter−1 phosphinotricin.
Nucleic Acid Manipulations.
To generate the plasmid psiSS, 405 bp of RBCS sequence were amplified from N. tabacum DNA by using primers NtsiSSl_L1 and NtiSS_R1, whereas introducing XbaI and AscI restriction sites and BamHI and SwaI sites respectively, with sequences based on the genomic clone NtSS23 (39). SwaI-AscI and XbaI-BamHI fragments were sequentially cloned into the vector pFGC5941 (accession no. AY310901) digested with the same enzymes, yielding the psiSS RNAi plasmid, which was ultimately transformed into Agrobacterium strain LBA4404. These and other primer sequences are available upon request. To generate pLS*, the tobacco chloroplast rbcL–accD intergenic region (58440–60449) was amplified and cloned into pGemT (Promega, Madison, WI). The resulting plasmid pRAIR was digested with BglII, blunted, and self-ligated, generating pRAIRCla4. A NdeI-NruI fragment was excised and inserted into the chloroplast transformation vector pNT1 (40) digested with the same enzymes, generating pLS*.
To generate TRV derivatives, 396 bp of the tobacco BSD2 homolog (TIGR accession nos. CV019724 and CV018551) were amplified from cDNA by using the primers NtBsd2 Nco cod2 and NtBsd2 Kpn rev2 and inserted into pGemT, yielding pNtbsd2. An NcoI-SacI fragment was subcloned into TRV2 (accession no. AF406991), creating pviBSD2. To create pviSS, psiSS was digested with XbaI and BamHI, and the 428-bp SS fragment was inserted into TRV2. To create pviBS, BSD2 sequence was reamplified by using primers Ntbsd2 BamHI cod3 and Ntbsd2 Kpn rev2. The BamHI-KpnI fragment was inserted into pviSS, generating pviBS. All TRV2 derivatives were mobilized in Agrobacterium strain GV2260.
Plant Transformation.
Tobacco nuclear transformation was performed as described (41), and selective media contained 2 mg·liter−1 phosphinotricin. Biolistic chloroplast transformation was performed as described (42), with transformants obtained on RMOP-spectinomycin (500 mg·liter−1) subjected to three rounds of regeneration on RMOP-spectinomycin/streptomycin (500 mg·liter−1 each), then rooted on MSO-spectinomycin (500 mg·liter−1). VIGS transformation used GV2260 cultures grown overnight at 28°C in LB medium containing 50 μg·ml−1 kanamycin and 100 μg·ml−1 rifampicin. Bacteria were pelleted, washed in induction medium (50 mM Mes/28 mM glucose/1.7 mM NaH2PO4/750 mM NH4Cl/50 mM MgSO4.7H2O/80 mM KCl/3.6 mM CaCl2/0.36 mM FeSO4.7H2O/20 mM acetosyringone) for 3 h at room temperature and then pelleted and resuspended in the infiltration medium (10 mM Mes/20 mM acetosyringone). N. benthamiana plants were inoculated by infiltrating the cotyledons and the lower leaves ≈2–3 weeks after sowing.
Gene Expression Analysis.
Total leaf RNA was extracted by using Tri-reagent (Molecular Research Center, Cincinatti, OH) and 3–5 μg were analyzed as in ref. 43. Gene-specific rbcL, psaA, and RBCS probes were generated by PCR using the following primer pairs: NtLSm cod1/NtLSm rev1, psaA-C5/-C3, and NtsiSSL1/NtsiSSR1. RT-PCR was performed by using cDNA generated by using SuperScript III (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions for 27 cycles by using the Ntactin cod1 and Ntactin rev1 primers to detect actin, and for 25 cycles by using NtsiSSl_L1 and NtsiSS_R1 for NtRBCS. The N. benthamiana NbBSD2 transcript was amplified by using the primers NbBSD2 cod1 and NbBSD2 rev1 for 23 cycles. NbRBCS cDNA was amplified by using the primers NbRBCS cod1 and NbRBCS rev1 for 23 cycles, whereas Nb actin cod1 and Nb actin rev1 were used to amplify actin cDNA by using 25 cycles.
Polysomes were analyzed from an extract prepared by grinding 150 mg of tissue in 1 ml of polysome extraction buffer as described in ref. 44, except that centrifugation was performed at 40,000 rpm for 90 min at 4°C in a SW-50Ti rotor.
Protein Analysis.
Leaves were ground in liquid nitrogen and total proteins extracted in 20 mM Hepes −0.1 M Na2CO3 supplemented with protease inhibitors. The slurry was filtered through miracloth and 20–30 μg of protein were separated in 12% SDS-polyacrylamide gels, which were analyzed by transfer to nitrocellulose membranes and enhanced chemiluminescence (Amersham, Piscataway, NJ).
For pulse labeling, young N. benthamiana leaves or 1-cm2 N. tabacum leaf discs were preincubated for 10 min in 100 μl of 1 mM KH2PO4 (pH 6.3), 0.1% Tween-20, and 40 μg·ml−1 cycloheximide at room temperature in the presence of light (20 μmol·m−2·s−1). Two hundred fifty microcuries (1 Ci = 37 GBq) of [35S]methionine/cysteine (>1,000 Ci mmol−1; ExpreSS protein labeling mix, PerkinElmer) was then added. After 10 min, the radioactive buffer was removed, and the samples were washed three times with 1 ml of ice-cold buffer, where cycloheximide was omitted. Proteins were extracted, and incorporation assayed from a 2-μl TCA-precipitated aliquot by using a liquid scintillation counter. Ten percent SDS-polyacrylamide–6 M urea gels were used for analysis.
Acknowledgments
We thank the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH) for distributing the plasmid CD3–447 (pFGC5941) described in www.chromdb.org/; Joyce Van Eck and Greg Martin for providing growth chamber space, the VIGS vectors, and PDS control strain and for helpful discussions on the use of VIGS; Bruce Cahoon (Middle Tennessee State University, Murfreesboro, TN) for the plasmid pRAIR; and Olivier Vallon (Institute de Biologie Physico-Chimique, Paris, France) and Amit Dhingra (Washington State University, Vancouver, WA) for providing us with LS and Rubisco antisera, respectively. This work was supported by National Science Foundation Award DBI-0211935.
Abbreviations
- LS
large subunit
- SS
small subunit
- CES
control by epistasy of synthesis
- VIGS
virus-induced gene silencing.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Fox TD. Hershey JWB, Matthews MB, Sonenberg N. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1996. pp. 733–758. [Google Scholar]
- 2.Wollman FA, Minai L, Nechushtai R. Biochem Biophys Acta. 1999;1411:21–85. doi: 10.1016/s0005-2728(99)00043-2. [DOI] [PubMed] [Google Scholar]
- 3.Adam Z, Clarke AK. Trends Plants Sci. 2002;7:451–456. doi: 10.1016/s1360-1385(02)02326-9. [DOI] [PubMed] [Google Scholar]
- 4.Majeran W, Wollman FA, Vallon O. Plant Cell. 2000;12:137–150. doi: 10.1105/tpc.12.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majeran W, Olive J, Drapier D, Vallon O, Wollman FA. Plant Physiol. 2001;126:421–433. doi: 10.1104/pp.126.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostersetzer O, Adam Z. Plant Cell. 1997;9:957–965. doi: 10.1105/tpc.9.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakamoto W, Zaltsman A, Adam Z, Takahashi Y. Plant Cell. 2003;15:2843–2855. doi: 10.1105/tpc.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choquet Y, Vallon O. Biochimie. 2000;82:615–634. doi: 10.1016/s0300-9084(00)00609-x. [DOI] [PubMed] [Google Scholar]
- 9.Choquet Y, Stern DB, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman F. Proc Natl Acad Sci USA. 1998;95:4380–4385. doi: 10.1073/pnas.95.8.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choquet Y, Zito F, Wostrikoff K, Wollman FA. Plant Cell. 2003;15:1443–1454. doi: 10.1105/tpc.011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wostrikoff K, Girard-Bascou J, Wollman FA, Choquet Y. EMBO J. 2004;23:2696–2705. doi: 10.1038/sj.emboj.7600266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minai L, Wostrikoff K, Wollman FA, Choquet Y. Plant Cell. 2006;18:159–175. doi: 10.1105/tpc.105.037705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth R, Hall LN, Brutnell TP, Langdale JA. Plant Cell. 1996;8:915–927. doi: 10.1105/tpc.8.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brutnell TP, Sawers RJ, Mant A, Langdale JA. Plant Cell. 1999;11:849–864. doi: 10.1105/tpc.11.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutteridge S, Gatenby AA. Plant Cell. 1995;7:809–819. doi: 10.1105/tpc.7.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt GW, Mishkind ML. Proc Natl Acad Sci USA. 1983;80:2632–2636. doi: 10.1073/pnas.80.9.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khrebtukova I, Spreitzer RJ. Proc Natl Acad Sci USA. 1996;93:13689–13693. doi: 10.1073/pnas.93.24.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodermel S, Haley J, Jiang CZ, Tsai CH, Bogorad L. Proc Natl Acad Sci USA. 1996;93:3881–3885. doi: 10.1073/pnas.93.9.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodermel SR, Abbott MS, Bogorad L. Cell. 1988;55:673–681. doi: 10.1016/0092-8674(88)90226-7. [DOI] [PubMed] [Google Scholar]
- 20.Cohen I, Sapir Y, Shapira M. Plant Physiol. 2006;141:1089–1097. doi: 10.1104/pp.106.079046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis RJ. Biochem Biophys Acta. 1977;463:185–215. [Google Scholar]
- 22.Hildebrandt J, Bottomley W, Moser J, Herrmann RG. Biochem Biophys Acta. 1984;783:67–73. [Google Scholar]
- 23.Spreitzer RJ, Goldschmidt-Clermont M, Rahire M, Rochaix J-D. Proc Natl Acad Sci USA. 1985;82:5460–5464. doi: 10.1073/pnas.82.16.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avni A, Edelman M, Rachailovich I, Aviv D, Fluhr R. EMBO J. 1989;8:1915–1918. doi: 10.1002/j.1460-2075.1989.tb03594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanevski I, Maliga P. Proc Natl Acad Sci USA. 1994;91:1969–1973. doi: 10.1073/pnas.91.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. Plant J. 2004;39:734–746. doi: 10.1111/j.1365-313X.2004.02158.x. [DOI] [PubMed] [Google Scholar]
- 27.Monde RA, Zito F, Olive J, Wollman FA, Stern DB. Plant J. 2000;21:61–72. doi: 10.1046/j.1365-313x.2000.00653.x. [DOI] [PubMed] [Google Scholar]
- 28.Gamble PE, Mullet JE. J Biol Chem. 1989;264:7236–7243. [PubMed] [Google Scholar]
- 29.Kim J, Mullet JE. Plant Cell Physiol. 2003;44:491–499. doi: 10.1093/pcp/pcg061. [DOI] [PubMed] [Google Scholar]
- 30.Boni IV, Artamonova VS, Dreyfus M. J Bacteriol. 2000;182:5872–5879. doi: 10.1128/jb.182.20.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yosef I, Irihimovitch V, Knopf JA, Cohen I, Orr-Dahan I, Nahum E, Keasar C, Shapira M. J Biol Chem. 2004;279:10148–10156. doi: 10.1074/jbc.M308602200. [DOI] [PubMed] [Google Scholar]
- 32.Koch KE. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- 33.Sheen J. Plant Cell. 1990;2:1027–1038. doi: 10.1105/tpc.2.10.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irihimovitch V, Shapira M. J Biol Chem. 2000;275:16289–16295. doi: 10.1074/jbc.275.21.16289. [DOI] [PubMed] [Google Scholar]
- 35.Marin-Navarro J, Moreno J. Biochemistry. 2003;42:14930–14938. doi: 10.1021/bi035713j. [DOI] [PubMed] [Google Scholar]
- 36.Nakano R, Ishida H, Makino A, Mae T. Plant Cell Physiol. 2006;47:270–276. doi: 10.1093/pcp/pci245. [DOI] [PubMed] [Google Scholar]
- 37.Desimone M, Henke A, Wagner E. Plant Physiol. 1996;111:789–796. doi: 10.1104/pp.111.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishida H, Makino A, Mae T. J Biol Chem. 1999;274:5222–5226. doi: 10.1074/jbc.274.8.5222. [DOI] [PubMed] [Google Scholar]
- 39.Mazur BJ, Chui CF. Nucleic Acids Res. 1985;13:2373–2386. doi: 10.1093/nar/13.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monde RA, Greene JC, Stern DB. Plant Mol Biol. 2000;44:529–542. doi: 10.1023/a:1026540310934. [DOI] [PubMed] [Google Scholar]
- 41.Bollenbach TJ, Tatman DA, Stern DB. Plant J. 2003;36:842–852. doi: 10.1046/j.1365-313x.2003.01935.x. [DOI] [PubMed] [Google Scholar]
- 42.Svab Z, Maliga P. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cahoon AB, Harris FM, Stern DB. EMBO Rep. 2004;5:801–806. doi: 10.1038/sj.embor.7400202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barkan A. Methods Enzymol. 1998;297:38–57. [Google Scholar]