Abstract
Current methods to analyze gene expression measure steady-state levels of mRNA. To specifically analyze mRNA transcription, we have developed a technique that can be applied in vivo in intact cells and animals. Our method makes use of the cellular pyrimidine salvage pathway and is based on affinity-chromatographic isolation of thiolated mRNA. When combined with data on mRNA steady-state levels, this method is able to assess the relative contributions of mRNA synthesis and degradation/stabilization. It overcomes limitations associated with currently available methods such as mechanistic intervention that disrupts cellular physiology, or the inability to apply the techniques in vivo. Our method was first tested in serum response of cultured fibroblast cells and then applied to the study of renal ischemia reperfusion injury, demonstrating its applicability for whole organs in vivo. Combined with data on mRNA steady-state levels, this method provided a detailed analysis of regulatory mechanisms of mRNA expression and the relative contributions of RNA synthesis and turnover within distinct pathways, and identification of genes expressed at low abundance at the transcriptional level.
Keywords: newly transcribed RNA, posttranscriptional regulation, RNA degradation/stabilization
In response to environmental changes, cells alter their gene expression program. Eukaryotic gene expression is a complex process that is fine-tuned at different levels, such as transcription, turnover, and translation of mRNAs. Microarray-based measurements of steady-state mRNA levels do not distinguish between different regulatory processes. Both transcriptional and posttranscriptional regulation, however, can profoundly influence steady-state mRNA levels (1–4). To evaluate de novo transcription and the relative contributions of mRNA transcription and turnover, nuclear transcription run-on and its modifications have long been the methods of choice (4–6). These methods, however, are either inherently cell-invasive and/or are not compatible with the assay formats currently used to analyze global gene expression. To avoid mechanistic intervention that disrupts cellular physiology for example by cell permeabilization, we modified a method in which we take advantage of the cellular pyrimidine salvage pathway. Pyrimidine ribonucleotides may be de novo synthesized from simple molecules by energetically expensive multistep pathways. Alternatively, ribonucleotide pools are sustained by recycling of the basic components derived from their catabolism during normal RNA turnover, the so-called salvage pathway, a mechanism that is energetically less expensive. By this more favorable pathway, it is possible to label and selectively enrich newly transcribed (nascent) RNA molecules (NT-RNA; refs. 7–9) in vivo by incorporation of exogenous 4-thiouridine (s4U) and subsequent RNA purification by affinity chromatography. The RNA can then be further processed by standard protocols for hybridization to oligonucleotide microarrays. We refer to our method with the acronym NIAC-NTR (noninvasive application and capture of newly transcribed RNA). We have established this protocol in a mouse embryonal fibroblast serum-response model. Fibroblasts are ubiquitous mesenchymal cells in the stroma of epithelial organs playing important roles in development, wound healing, inflammation, and fibrosis. The genomic response of fibroblasts to serum, the soluble fraction of coagulated blood, represents a coordinated “wound-healing program,” including regulation of hemostasis, cell cycle progression, epithelial cell migration, inflammation, and angiogenesis (10–11). We then applied NIAC-NTR to analyze mRNA synthesis in a murine renal ischemia reperfusion injury (IRI; refs. 12 and 13). The murine IRI is a model for the ischemia/reperfusion-induced acute renal failure, which is a common clinical problem associated with high morbidity and mortality. IRI is associated with a depletion of energy-rich phosphates in renal epithelial cells, thereby activating systems leading to (i) disruption of the cytoskeleton and cell polarity, and (ii) cell death, either in form of necrosis or apoptosis (14). Our studies have demonstrated the potential use of our method for the analysis of mRNA transcription in living organisms.
Results
Thiolated RNA Can Specifically Be Isolated by an Organomercurial Affinity Matrix.
Successful isolation of thiolated RNA can be achieved by exploiting the strong chemical interaction between thio groups and mercury. Therefore, an agarose-based organomercurial affinity matrix [Bio-Rad (Hercules, CA) Affi-Gel 10-modified p-aminophenylmercuric acetate agarose, mercury content 10 μmol/ml] was tested for its efficiency to bind thiolated RNA molecules synthesized in vitro. Synthetic mRNA molecules of different lengths were generated either in the absence or presence of 4-thiouridine-5′-triphosphate. Capillary gel-electrophoresis data demonstrated that thiolated RNA of ≥200 bases length was bound to the matrix and was elutable by addition of 20 mM 2-mercaptoethanol [supporting information (SI) Fig. 3 and SI Table 1]. Because the binding efficiency of thiolated molecules to the organomercurial matrix strongly depends on salt concentration and pH (7), optimal binding and wash buffer combinations were evaluated (SI Methods and Protocols).
s4U Is Efficiently Incorporated into Cellular RNA and Can Be Specifically Isolated.
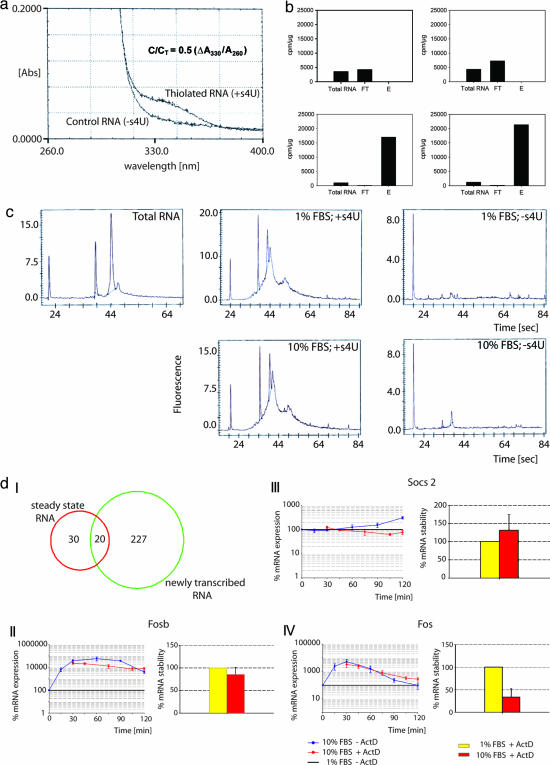
In the next experiment, we labeled serum-starved primary mouse embryonal fibroblast cells with s4U and 3H-cytidine (to follow s4U incorporation into newly transcribed cellular RNA) at the time serum was added to the medium. s4U (200 μM) for 2 h was chosen based on time-dose-response analyses (data not shown). Two hours after s4U addition, total RNA (T-RNA) was isolated, and s4U-incorporation efficiency was checked by OD measurement based on the specific absorption of s4U at ≈330 nm. Successful incorporation into total RNA leads to a characteristic pattern when compared with control RNA (Fig. 1a). Based on these data, the average approximate incorporation rate was calculated to be 4–5 s4U/1000b (9). T-RNA was then loaded onto the organomercurial affinity matrix for capture of thiolated RNA with total RNA from non-s4U-treated cells as control RNA. NT-RNA in serum-starved and -induced cells was determined by measuring 3H-cytidine incorporation. As demonstrated in Fig. 1b Upper, there was no binding of NT-RNA to the affinity matrix in the absence of s4U. In contrast, labeling with s4U leads to retention of NT-RNA (Fig. 1b Lower). Electrophoretic analysis of NT-RNA showed a broad size distribution, indicating an enrichment of heteronuclear RNA and mRNA compared with steady-state RNA (Fig. 1c Center and Left, respectively). This pattern may be regarded as a characteristic fibroblast “signature” for early (2-h) serum response, demonstrating the power of our method for enrichment of newly transcribed RNA. Background binding of RNA to the organomercurial matrix was minimal (Fig. 1c Right) and is most likely because of RNA thiolation in the absence of exogenous s4U rather than from nonspecific molecular interactions. s4U naturally occurs in prokaryotic cells, and 2-thiouridine, a chemically related base, is found in eukaryotic cells. Both bases are incorporated into tRNA of the respective cells. Thus, to minimize contamination by serum-containing thiolated nucleotides produced by nanobacteria (15), only dialyzed serum (cutoff 10,000) was used. In fact, standard qualitative HPLC analysis that was performed on RNase-treated samples showed the presence of two closely located peaks, one of them indicative for s4U (SI Fig. 4). Because s4U labeling depends on the length of the target RNA molecules, we analyzed the fibroblast RNA labeling efficiency of all expressed genes [Affymetrix (Santa Clara, CA) P-calls] for NT-RNA and steady-state RNA samples. Data showed that with the s4U conditions used, we were able to efficiently label and isolate RNA molecules independent of their lengths (SI Fig. 5). To test whether the presence of s4Us, using our labeling conditions, had any toxic influence on the fibroblasts, a cell viability assay was performed, and no impact on cell viability could be observed (SI Fig. 6). Similarly, we analyzed a potential s4U impact on the global fibroblast transcriptional profile and found only a moderate differential expression of eight genes attributable to s4U (SI Fig. 7 and SI Table 2). Importantly, a 2-h labeling period with s4U had no significant influence on decay rates of RNAs (SI Figs.8 and 9).
Fig. 1.
NIAC-NTR allows for detailed analysis of RNA synthesis and turnover. (a) Efficiency of s4U incorporation into mouse embryonic fibroblast RNA. At 330 nm, thiolated RNA exhibited an absorption difference compared with control RNA. Approximate s4U incorporation rate can be calculated (9). C/CT = substitution level; ΔA330 = A330 (thiolated RNA) A330 (control RNA). (b) Graphs illustrating specific retention and enrichment of thiolated RNA. Specific activities (cpm/μg RNA) are shown for initial T-RNA, FT-RNA, and E-RNA. Ten percent FBS, serum-induced condition; 1% FBS, serum-starved condition; FT, flow-through RNA (not matrix-bound); E, pooled elution fractions. The specific activity of the E fraction indicates the fold enrichment of NT-RNA (12× on average). Upper Left, 1% FBS, control RNA; Upper Right, 10% FBS, control RNA; Lower Left, 1% FBS, thiolated RNA; Lower Right, 10% FBS, thiolated RNA. (c) Electrophoretic pattern of fibroblast NT-RNA (Center) compared with T-RNA (Left). Background binding (Right) is minimal. Note the different scaling of the y axis indicated by the assay-intrinsic leftmost constant marker peak. (d) I, Venn diagram illustrating the regulation of mRNA expression; II-IV, qRT-PCR analyses of the contributing actions of mRNA transcription, degradation, and stability for exemplary mRNAs. (Left) Time-resolved expression differences in serum-induced samples. (Right) Differences of stability of serum-induced samples (red bars) within an evaluated time frame of 90 min plotted as percentages relative to the stability of serum-starved controls (yellow bars). II, NT-RNA and T-RNA; III, NT-RNA; IV, posttranscriptionally regulated RNA. For more details, refer to the main text (Identification of Fibroblast NT-RNA in Response to Serum) and to SI Figs. 10 and 11 and SI Tables 3 and 4.
Identification of Fibroblast NT-RNA in Response to Serum.
A total of five biological replicates were analyzed by Affymetrix MG-U74Av2 microarrays, as described (16) (SI MIAME checklist). Data mining was performed with the SAS MicroArray Solution v1.0 software package (SAS Institute, Cary, NC) by using the loglinear mixed model strategy (17). Functional analysis of differentially expressed genes was performed by differential pathway analysis [based on Gene Set Enrichment Analysis (GSEA); ref. 18], coupled with functional group analysis by using the Ingenuity Pathways Analysis Program. Analysis of steady-state RNA (T-RNA) showed changes in gene expression of 50 genes, whereas analysis of NT-RNA identified 247 genes changing in expression after serum stimulation, with most of them (205) down-regulated. Detailed quantitative PCR (qPCR) analyses of exemplary genes indicated that changes in T-RNA levels resulted either predominantly from altered transcription (20 genes, Fig. 1 d-I and d-II; SI Fig. 10) or predominantly from mRNA turnover and altered stability, thereby masking transcriptional regulation [30 posttranscriptionally regulated genes, Fig. 1 d-I and d-IV; SI Fig. 10]. NT-RNA data also identified a total of 227 newly transcribed genes, the transcripts of which were not altered in steady-state RNA (Fig. 1 d-I and d-III; SI Fig. 10). Thus, our data indicated that (i) NT-RNA genes were predominantly regulated by de novo transcription with minimal contributions of mRNA degradation, (ii) genes detected in both NT-RNA and T-RNA were also predominantly regulated by de novo transcription with partial contribution of mRNA degradation/stabilization, and (iii) posttranscriptionally regulated genes were predominantly regulated by mRNA degradation, masking transcriptional regulation (SI Fig. 10 and SI Tables 3 and 4). These data and additional qPCR analyses thus were in good agreement with microarray data (SI Fig. 11). Our results indicated that the early transcriptional response to serum is characterized mainly by transcriptional down-regulation of the majority of genes involving pathways controlling (i) energy generation (Krebs cycle, fatty acid degradation), (ii) cell cycle and proliferation, and (iii) DNA damage and repair signaling. Genes which were found to be either predominantly transcriptionally up-regulated or predominantly regulated by mRNA degradation/ stabilization could be allocated to (i) immediate-early transcription factors, (ii) clusters involved in epithelialization and vascular development, and (iii) clusters involved in basic processes of fibroblasts including reprogramming of cytoskeletal properties and stem cell-related pathways such as wnt-signaling (SI Tables 5–7). These results fit with the current knowledge about the biology of fibroblasts (10) and their potential predictive role for cancer (11). But NIAC-NTR adds more detailed information, because it enriches for newly transcribed RNA and reveals information about the regulatory mechanism of each gene.
Analysis of Newly Transcribed RNA in Vivo in Intact Animals.
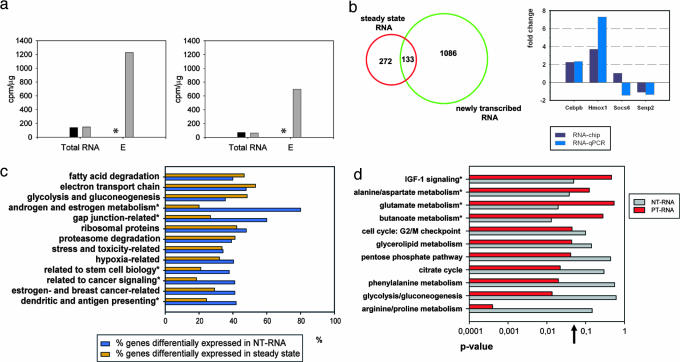
The previous results demonstrated that our method may potentially be useful for expression analysis of newly transcribed RNA in cell culture. We next applied our method in vivo in intact animals. A renal IRI model was chosen because the kidney has an active salvage pathway, and its catabolism of pyrimidine molecules is rather low (19–22). We have restricted analysis to early changes in RNA synthesis in the contralateral (noninjured) kidney of IRI-treated mice (23) compared with control mice. The contralateral kidney was chosen because the injured kidney may undergo substantial changes of internal nucleotide pools (24) early in the reperfusion phase, resulting in a massive incorporation of s4U and 3H-cytidine. Optimal doses of s4U were evaluated and injected i.p., mice were killed, and kidney RNA was isolated and analyzed (Fig. 2a). Labeling efficiency and potential s4U toxicity were analyzed (SI Fig. 12 and SI Tables 8 and 9) and revealed similar results as for the fibroblast study. T-RNA analysis showed 405 genes to be differentially expressed (100 up- and 305 down-regulated). NT-RNA analysis revealed 1,219 differentially expressed genes (642 up- and 577 down-regulated). Combinatorial analysis indicated that 272 genes were preferentially regulated by mRNA degradation/stabilization rather than transcription (Fig. 2b). qPCR validation data of genes found transcriptionally controlled (Cebpb and Hmox1) and unchanged (Socs6 and Senp2) were in good agreement with chip data (Fig. 2b). Gene Set Enrichment Analysis-based analysis (SI Tables 10–13) demonstrated that detection of many functional gene clusters involved in the disease response was possible only when NT-RNA data were analyzed (Fig. 2c). For example, steady-state data of differentially expressed genes in the contralateral kidney between IRI and control mice did not attribute any significance to the androgen and estrogen metabolism or to genes involved in mediating cell-to-cell information by gap junctions. Analysis of NT-RNA data, however, revealed the striking importance of these pathways because most of their genes were expressed differentially at the transcriptional level. Comparisons of genes predominantly regulated either by mRNA turnover or mRNA synthesis demonstrated their relative contribution to distinct pathways: some pathways most probably were preferentially regulated by mRNA turnover (e.g., arginine/proline metabolism), and others (glutamate and butanoate metabolisms, IGF-1 signaling) were preferentially regulated by mRNA synthesis (Fig. 2d).
Fig. 2.
NIAC-NTR can be applied in vivo. (a) Graphs show the specific activity (cpm/μg of RNA) of contralateral kidney control RNA (black bars) compared with thiolated RNA of the pooled elution-fractions (gray bars) for control (Left) and IRI-treated mice (Right) after 3 h. ∗, No cpm measured in the absence of s4U. (b) (Left) Venn diagram illustrating the regulation of mRNA expression. (Right) qPCR validation results of transcriptional regulation. Array and qPCR results are given back to back for each gene. Two independent qPCR validation experiments were performed with technical duplicates each. (c) Percentage of differentially expressed genes in a given functional group for NT-RNA (blue bars) and NT-RNA only (orange bars). ∗, Pathways significantly changed because of analysis of NT-RNA. (d) Relative contribution of mRNA synthesis and turnover of genes involved in specific pathways because of IRI-treatment. Genes regulated by mRNA turnover (posttranscriptionally regulated RNA) in steady-state analysis (see b; dark red bars) were compared with NT-RNA genes (gray bars). Pathways shown were significantly changed (P value on x axis). The order of pathways from top to down shows increasing contribution of mRNA turnover to the respective pathway. ∗, Pathways significantly changed because of mRNA transcription; ↑, statistical significance threshold (P = 0.05).
Discussion
Cellular RNA metabolism is tightly regulated at different levels. For a detailed analysis of these processes, however, it is necessary to find methods to dissect and measure them separately. mRNA synthesis can be analyzed by nuclear run-on strategies (4–6); however, because of the nature of these techniques, they cannot be applied in vivo and are not suitable for fluorescence-based microarray techniques. Here we present a strategy, NIAC-NTR, that is compatible with fluorescence-based microarray analysis and that can be applied in vivo in intact animals, as evidenced by analysis in the reperfused mouse kidney. Our method is based on the incorporation of exogenous s4U into cellular RNA by using the pyrimidine salvage pathway. We showed that s4U is efficiently incorporated into nascent RNA. Albeit the potential toxicity of s4U (8, 9), we evaluated that an exposition to s4U for 2 h did not significantly change the RNA expression profile compared with nontreated cells. We demonstrated that s4U-labeled RNA can efficiently be isolated and enriched by an organomercurial affinity matrix. Furthermore, we presented evidence that the observed background binding depends on thiolated molecules rather than on unspecific molecular interactions. Background binding may be mediated mainly by the presence of cross-contaminating thiolated bases such as 2-thiouridine, which may be misincorporated into hnRNA. Exogenous s4U is processed intracellularly by the pyrimidine salvage pathway. The in vivo application of our method is therefore favored by organs with an active salvage pathway, such as the kidney. Based on comparative analyses, the relative contribution of mRNA synthesis and turnover to gene expression regulation could be demonstrated in the contralateral kidney between IRI and control mice. This enabled us to categorize canonical pathways and functionally related gene clusters according to preferentially synthesis-regulated or turnover-regulated. At present, very little is known about the role of posttranscriptional control, although mounting evidence supports its importance, e.g., under stress conditions (25–27). Based on this and our own data, it may be hypothesized that under certain conditions, posttranscriptional control may be favored to keep pace with specific needs such as timely and energetically economic control of (specifically localized) gene expression (28). It seems plausible that members of distinct pathways or functional groups may therefore be controlled by a similar mechanism. Strikingly, the enrichment of NT-RNA by our method allowed for the detection of a plethora of genes predominantly regulated at the transcriptional level, which would normally have escaped detection by an analysis of steady-state mRNA alone, because of dilution in the pool of total RNA.
The brain may serve as well as a target for NIAC-NTR, because exogenous uridine passes the blood–brain barrier and may be sufficiently incorporated into brain RNA, in specific brain areas such as cerebellum and cerebral cortex (29–32). Organs exhibiting a limited use of the pyrimidine salvage pathway such as liver and intestine (19–22, 33) may be less suited for s4U-based NIAC-NTR application. The analysis of organs with a low level salvage pathway, however, may be performed with thiolated orotic acid rather than with s4U because orotic acid is the precursor molecule of uridine in the de novo biosynthesis pathway. Such a “bypass” could further increase the potential of NIAC-NTR applications in vivo. Our technique may represent a potentially useful approach to specifically analyze RNA de novo synthesis, a goal also aimed at by nuclear run-on techniques (4–6) and a technique that has recently been published describing the use of animal cells, which have been infected with the parasite Toxoplasma gondii expressing uracil phosphoribosyltransferase (34). This method, however, is useful only to distinguish between parasite RNA from animal RNA. In contrast to the nuclear run-on, our method can potentially be applied in vivo and to current microarray formats and enables specific isolation of predominantly newly transcribed animal RNA without the need for transgene experiments.
In summary, our method provides a potentially useful tool to discriminate between the relative contributions of mRNA synthesis and turnover. It may therefore allow a more detailed understanding of basic molecular mechanisms in gene expression in vitro and in whole organs in vivo. Analysis of NT-RNA in whole organs may be especially useful for testing (side) effects of chemicals intended for medical disease treatments.
Materials and Methods
Labeling and Preparation of RNA.
RNAs of different lengths were synthesized in vitro either in the absence or presence of 4-thiouridine-5′-triphosphate (TriLink BioTechnologies, San Diego, CA) using pGEM Express Positive Control Templates (Promega, Madison, WI) and RNA Century Marker Templates (Ambion, Austin, TX). RNA was generated by T7-mediated in vitro transcription by using the Ampliscribe T7 high-yield transcription kit (Epicentre; Biozym, Hess Oldendorf, Germany), supplemented with [5-3H] CTP (20 Ci/mmol, Amersham, Buckinghamshire, U.K.). This yielded RNAs of the following lengths: 0.2, 0.3, 0.4, 0.5, 0.75, 1.065, and 2.346 kb. RNA was purified by using the GeneChip Sample Cleanup Module (Affymetrix), precipitated with PelletPaint and 7.5 M ammonium acetate and quality analyzed by the RNA6000 Nano assay (Bioanalyzer 2100, Agilent Technologies).
Primary mouse embryonic fibroblast cells (embryonic day 14.5, ATF1+/−) were grown using high-glucose DMEM (Invitrogen, Karlsruhe, Germany) with 1% penicillin/streptomycin (Invitrogen) and 10% dialyzed FBS (cutoff, 10,000 Da; Sigma, St. Louis, MO) to near confluency. Cells were serum-starved for 48 h with 1% dialyzed FBS and then incubated for 2 h with media containing: (i) 1% FBS (serum-starvation control), (ii) 1% FBS, 200 μM s4U (Sigma) and 1 μCi 3H-cytidine (20 Ci/mmol; BIOTREND, Köln, Germany); (iii) 10% FBS (stimulation control), and (iv) 10% FBS/200 μM s4U/1 μCi 3H-cytidine.
RNA was isolated according to the RNeasy Midi protocol (Qiagen, Valencia, CA), yield and quality were assessed as described above. Incorporation efficiencies were measured by UV/VIS wavescanning and approximate incorporation rates calculated according to ref. 9.
NT-RNA Isolation by Organomercurial Affinity Matrix.
Organomercurial matrix manufacture was made on request by Squarix Biotechnology (Marl, Germany). For a detailed protocol, see SI Methods and Protocols. Briefly, 100 μg of RNA was diluted in binding buffer and loaded onto ≈150 μl of packaged volume of the equilibrated organomercurial matrix loaded to a Small Spin Column (Qiagen). After 4-h incubation under constant gentle rotation and light protection at 4°C, nonbound RNA was eliminated by sequential washing steps. Bound (thiolated) RNA was eluted using binding buffer supplemented with 20 mM 2-mercaptoethanol and ethanol-precipitated. Specific activity of eluted RNA was calculated by 3H-cytidine measurement on a Beckman Coulter (Krefeld, Germany) LS6500.
GeneChip Hybridization.
RNA was reverse-transcribed, linearly amplified, and biotin-labeled with a T7-based strategy as described (16) and hybridized to MU74Av2 or MOE430A GeneChips (Affymetrix), according to manufacturer's instructions.
Statistical Analysis and Data Mining.
Statistical analysis and data mining were performed with the SAS MicroArray Solution 1.0, by using standard settings except the following: consistence within experimental groups (stimulus and labeling combination) was determined by array group correlation. Log-linear mixed models (17) with Bonferroni corrections were fitted for values of perfect matches, only with stimulus (10% FBS or IRI treatment) and labeling (s4U or no s4U) considered to be constant and the chip-ID as random. To identify pathways and functional groups exhibiting increased numbers of significantly expressed genes (based on Gene Set Enrichment Analysis; ref. 18), Kolmogorov–Smirnov or Fisher's exact tests were performed comparing genes from given (Kyoto Encyclopedia of Genes and Genomes, Gene Ontology, and Biocarta) or self-curated pathways. For Fisher's exact test, the overall number of significantly/nonsignificantly expressed genes per chip was cross-tabulated against the number of significantly/nonsignificantly expressed genes of a pathway. Analysis was performed with SAS procedure PROC FREQ. Canonical pathway and gene network analyses were performed by the Ingenuity pathways analysis software package (Ingenuity Systems, Redwood City, CA).
Analysis of qPCR-Based Time-Resolved mRNA Transcription, Degradation, and Stability.
For a detailed protocol, see SI Methods and Protocols. Briefly, RNA of serum-starved cells treated with 10% FBS at t = 0 was harvested at different time points. To follow the half-life of mRNAs, serum-induced cells were treated with 2 μg/ml Actinomycin D (ActD) at t = 15 min and RNA harvested at t = 30, 45, 75, 105, and 120 min. To determine changes in mRNA stability, serum-starved cells were treated with 2 μg/ml ActD, and total RNA was harvested at t = 90 min. To follow mRNA stability after serum induction, cells were induced with 10% FBS (t = 0) for 15 min before addition of 2 μg/ml ActD (t = 15min) and RNA harvested at t = 105 min. Two micrograms of RNA each was used for qRT-PCR analyses.
In Vivo NIAC-NTR.
Surgical methods have been described (23). Briefly, C57/Bl6 mice (males, ≈20 g body weight) were anesthetized with diethylether. After flank incision, the renal artery and vein of the left kidney were occluded by a vascular clamp for 45 min. The clamp was removed, and the organ was allowed to reperfuse (time 0). Animals were allowed to recover with free access to nutrition. At time 0 + 1 h, 200 μM s4U (bidistilled water as control) together with 34 μCi 3H-cytidine (20 Ci/mmol; 1 Ci = 37 GBq; American Radiolabeled Chemicals, St. Louis, MO) were injected i.p. in a total volume of 1 ml of 0.9% NaCl solution. At time 0 + 2 h, injection was repeated. At time 0 + 3 h, mice were killed and kidneys removed for histopathologic examination and RNA isolation. Total RNA (150 μg) was processed by organomercurial affinity matrix capturing. All experiments were done with permission of the local university and government commission for animal experiments.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of M.H., a passionate and excellent scientist, who unexpectedly died during the final preparation phase of this manuscript. This work was supported by the Deutsche Forschungsgemeinschaft Graduiertenkolleg (GRK) 886, Sonderforschungsbereich 405 and 636, Collaborative Research Centre 488 (Molecular and Cellular Bases of Neural Development, Heidelberg, Germany), Forschergruppe 302, GRK 791/1.02, GRK 484 and Sachbeihilfe Schu-51/7-2, by the Fonds der Chemischen Industrie, the European Community through Grant QLG1-CT-2001-01574, the Bundesministerium für Bildung und Forschung through Nationales Genomforschungsnetz grants FZK-01GS01117 and KGCV-1/01GS0416 and project number 0313074C (systems biology), and by the Alexander von Humboldt Stiftung through the Max-Planck-Forschungspreis für Internationale Kooperation 1998.
Abbreviations
- s4U
4-thiouridine
- NT-RNA
newly transcribed RNA
- NIAC-NTR
noninvasive application and capture of NT-RNA
- IRI
ischemia reperfusion injury
- qPCR
quantitative PCR.
Footnotes
Conflict of interest statement: The authors declare competing financial interests. A patent application relating to this work has been filed.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession nos. GSE6697 and GSE6698).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610439104/DC1.
References
- 1.Guhaniyogi J, Brewer G. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 2.Gingerich TJ, Feige JJ, LaMarre J. Anim Health Res Rev. 2004;5:49–63. doi: 10.1079/ahr200460. [DOI] [PubMed] [Google Scholar]
- 3.Khodursky AB, Bernstein JA. Trends Genet. 2003;19:113–115. doi: 10.1016/S0168-9525(02)00047-1. [DOI] [PubMed] [Google Scholar]
- 4.Fan J, Yang X, Wang W, Wood WH, III, Becker KG, Gorospe M. Proc Natl Acad Sci USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirayoshi K, Lis JT. Methods Enzymol. 1999;304:351–362. doi: 10.1016/s0076-6879(99)04021-5. [DOI] [PubMed] [Google Scholar]
- 6.García-Martínez J, Aranda A, Pérez-Ortín JE. Mol Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Ono M, Kawakami M. J Biochem. 1977;81:1247–1252. [PubMed] [Google Scholar]
- 8.Woodford TA, Schlegel R, Pardee AB. Anal Biochem. 1988;171:166–172. doi: 10.1016/0003-2697(88)90138-8. [DOI] [PubMed] [Google Scholar]
- 9.Favre A, Moreno G, Salet C, Vinzens F. Photochem Photobiol. 1993;58:689–694. doi: 10.1111/j.1751-1097.1993.tb04953.x. [DOI] [PubMed] [Google Scholar]
- 10.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, Trent JM, Staudt LM, Hudson J, Jr, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 11.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO. PloS Biology. 2004;2:0206–0214. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kidney Int. 2004;66:479–531. [Google Scholar]
- 13.Supavekin S, Zhang W, Kucherlapati R, Kaskel FJ, Moore LC, Devarajan P. Kidney Int. 2003;63:1714–1724. doi: 10.1046/j.1523-1755.2003.00928.x. [DOI] [PubMed] [Google Scholar]
- 14.Lien YH, Lai LW, Silv AL. Life Sci. 2003;74:543–552. doi: 10.1016/j.lfs.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Ciftcioglu N, Kajander EO. Pathophysiology. 1998;4:259–270. [Google Scholar]
- 16.Kenzelmann M, Klären R, Hergenhahn M, Bonrouhi M, Gröne H-J, Schmid W, Schütz G. Genomics. 2004;83:550–558. doi: 10.1016/j.ygeno.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 17.Chu TM, Weir BS, Wolfinger RD. Bioinformatics. 2004;20:500–506. doi: 10.1093/bioinformatics/btg435. [DOI] [PubMed] [Google Scholar]
- 18.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 19.Zaharevitz DW, Anderson LW, Malinowski NM, Hyman R, Strong JM, Cysyk RL. Eur J Biochem. 1992;210:293–296. doi: 10.1111/j.1432-1033.1992.tb17420.x. [DOI] [PubMed] [Google Scholar]
- 20.Moyer JD, Malinowski N, Ayers O. J Biol Chem. 1985;260:2812–2818. [PubMed] [Google Scholar]
- 21.Darnowski JW, Handschumacher RE. Cancer Res. 1986;46:3490–3494. [PubMed] [Google Scholar]
- 22.Canellakis ES. J Biol Chem. 1957;227:701–709. [PubMed] [Google Scholar]
- 23.Furuichi K, Wada T, Iwata Y, Kitagawa K, Kobayashi K-I, Hashimoto H, Ishiwata Y, Asano M, Wang H, Matsushima K, et al. J Am Soc Nephrol. 2003;14:2503–2515. doi: 10.1097/01.asn.0000089563.63641.a8. [DOI] [PubMed] [Google Scholar]
- 24.Zager RA. Circ Res. 1991;68:185–196. doi: 10.1161/01.res.68.1.185. [DOI] [PubMed] [Google Scholar]
- 25.Inoue H, Taba Y, Miwa Y, Yokota C, Miyagi M, Sasaguri T. Arterioscler Thromb Vasc Biol. 2002;22:1415–1420. doi: 10.1161/01.atv.0000028816.13582.13. [DOI] [PubMed] [Google Scholar]
- 26.Benard L. RNA. 2004;10:458–468. doi: 10.1261/rna.5183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lal A, Gorospe M. Cell Cycle. 2006;5:1422–1425. doi: 10.4161/cc.5.13.2902. [DOI] [PubMed] [Google Scholar]
- 28.Kenzelmann M, Rippe K, Mattick JS. Mol Syst Biol. 2006;2:44. doi: 10.1038/msb4100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fog R, Pakkenberg H. J Neurochem. 1976;27:1261–1262. doi: 10.1111/j.1471-4159.1976.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 30.Pakkenberg H, Fog R. Exp Neurol. 1972;36:405–410. doi: 10.1016/0014-4886(72)90001-5. [DOI] [PubMed] [Google Scholar]
- 31.Spector R. J Neurochem. 1985;45:1411–1418. doi: 10.1111/j.1471-4159.1985.tb07207.x. [DOI] [PubMed] [Google Scholar]
- 32.Lee G, Dallas S, Hong M, Bendayan R. Pharmacol Rev. 2001;53:569–596. [PubMed] [Google Scholar]
- 33.Gasser T, Moyer JD, Handschumacher RE. Science. 1981;213:777–778. doi: 10.1126/science.7256279. [DOI] [PubMed] [Google Scholar]
- 34.Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. Nat Biotechnol. 2005;23:232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.