Abstract
Arabidopsis thaliana is one of the most intensively studied plant species. More recently, information is accumulating about its closest relatives, the former genus Cardaminopsis. A. thaliana diverged from these relatives, actually treated within three major lineages (Arabidopsis lyrata, Arabidopsis halleri, and Arabidopsis arenosa), ≈5 mya. Significant karyotype evolution in A. thaliana with base chromosome number reduction from x=8 to x=5 might indicate and favor effective genetic isolation from these other species, although hybrids are occurring naturally and have been also constituted under controlled conditions. We tested the evolutionary significance to separate the x=5 from the x=8 lineage using DNA sequence data from the plastome and the nuclear ribosomal DNA based on an extensive, representative worldwide sampling of nearly all taxonomic entities. We conclude that (i) A. thaliana is clearly separated phylogenetically from the x=8 lineage, (ii) five major lineages outside A. thaliana can be identified (A. lyrata, A. arenosa, A. halleri, Arabidopsis croatica, and Arabidopsis pedemontana) together with Arabidopsis cebennensis, and (iii) centers of genetic and morphological diversity are mostly in congruence and are located close to the Balkans in Austria and Slovakia outside glaciated and permafrost regions with few notable exceptions.
Keywords: genetic diversity, phylogenetic relationships, phylogeography, reticulation
The phylogenetic sister relationship of Arabidopsis thaliana with all representatives formerly treated within the genus Cardaminopsis is fully accepted (1, 2) and demonstrated by various phylogenetic studies (3–5). The taxon Cardaminopsis was replaced by Arabidopsis because of their close evolutionary relatedness and morphological similarities. However, genetic and phylogenetic relatedness, which enables even crosses between x=5 and x=8 taxa, provides a huge potential to move from the study of A. thaliana into its wild relatives to answer fundamental evolutionary questions that cannot be addressed in A. thaliana for various reasons (e.g., because of inbreeding system, narrow ecology, unspectacular distribution range, and short life cycle). The transfer from the model system A. thaliana to its wild relatives is attractive and possible because of the currently accepted techniques, resources, and database of information (2, 6).
The evolutionary split between x=5 A. thaliana and x=8 Arabidopsis taxa occurred ≈5 mya (3, 4, 7) and initiated the evolution of A. thaliana with its unique characters compared with the x=8 lineage, and also changes on the chromosome level resulting in its derived genome structure (8–10). On the contrary, there is much more variation in the x=8 lineage (1) resulting in the recognition of several species and subspecies. Because eight wild relatives of A. thaliana on the species level were recognized <8 years ago (11), the number of new taxonomic combinations is increasing (12, 13). An overview on the current status of Arabidopsis taxonomy and synonymy has been given recently (1). In general, three major lineages can be recognized, namely Arabidopsis lyrata, Arabidopsis halleri, and Arabidopsis arenosa (1), and most species or subspecies can be treated within these three lineages. In addition, three species have been described that are not closely related to one of these three species groups: Arabidopsis croatica (Croatia), Arabidopsis cebennensis (France), and Arabidopsis pedemontana (Italy). It can be expected that below the species level the number of taxa will increase further as is the case for A. halleri, which segregates with five subspecies (1). The same will happen to A. arenosa segregates because actual taxonomic treatments are unconvincingly based on comparative cytological, morphological, or genetic analysis (11, 14), and we are still lacking any comparative morphometric analysis.
At the present time, evolutionary studies are restricted to single species or groups of populations (15–18), and no evolutionary framework has been provided yet that comprises the entire genus. High levels of genetic diversity in periglacial regions (17, 19, 20–24), heavy metal tolerance (19, 25), and self-incompatibility and breeding systems (20, 24, 26–35) encompass the completed research on Arabidopsis wild relatives.
In this study we present the first comprehensive phylogenetic framework for the genus. We studied genetic variation of all evolutionary lineages of A. thaliana relatives based on a representative geographic sampling by studying maternally inherited chloroplast DNA (cpDNA) haplotype variation and sequence diversity of the internal transcribed spacer region (ITS) of ribosomal RNA. The cpDNA haplotype data were analyzed phylogeographically, and gene diversity parameters were calculated. The plastid data were compared with the nuclear data, and significant differences among the various evolutionary lineages are highlighted. Finally, we will contribute to the systematic status of some taxonomical–nomenclatoral combinations such as Arabidopsis kamchatica and Arabidopsis arenicola (12, 13).
Results
Nuclear ITS Sequence Data Confirmed Major Lineages in Arabidopsis.
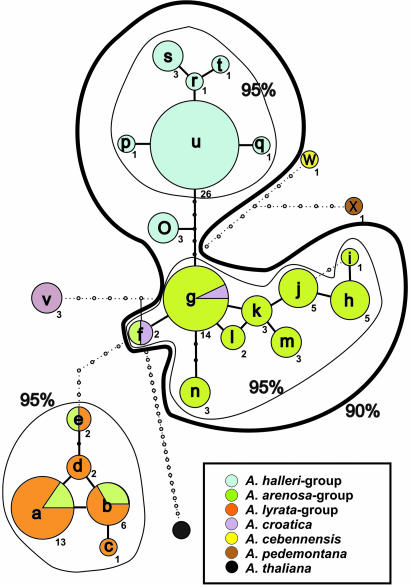
In total we obtained 103 different ITS sequences with varying levels of sequence ambiguities [supporting information (SI) Table 1]. The initial analysis with TCS recognized 24 different groups of sequences, which are coded alphabetically (a–x). The network analysis (Fig. 1) resolves five major lineages of general ITS sequence types (further named as ITS types) that correlate with morphological species delimitation: (i) A. halleri and all its subspecies are combined to a single well defined group represented by seven ITS types and 36 different sequences in total. (ii) All segregates within A. arenosa (inclusive Arabidopsis nitida, Arabidopsis carpatica, Arabidopsis petrogena, Arabidopsis borbasii, Arabidopsis neglecta, and A. arenosa sensu stricto) are combined to one larger cluster with nine ITS types and 35 single sequences in total. Two additional sequences grouping into ITS types g and f are found in A. croatica. (iii) A. croatica is separated with a single ITS type v represented by three ITS sequences. (iv) A. cebennensis and A. pedemontana are combined within one genetically distinct lineage represented by two ITS types (w and x). (v) A. lyrata and all of its segregates (A. kamchatica, A. arenicola, and A. lyrata ssp.) are represented by a group of five ITS types with 19 individual sequences. Five additional unique sequences (found only in one accession, respectively) grouping to three of these five ITS types (a, b, and e) are found in A. arenosa samples. This phylogenetic information content is confirmed by maximum-likelihood analysis (SI Fig. 4). The different groups are recognized with similar significance levels as with TCS. A. halleri and A. lyrata are characterized with high bootstrap support. A. pedemontana and A. cebennensis are also combined to one clade with highly significant statistical support. All other ITS types from A. arenosa representatives are not defined to any of these groups and form a much less resolved cluster. The ITS type v from A. croatica is as shown in the network analysis more closely related to A. arenosa types than to A. lyrata or A. halleri, respectively.
Fig. 1.
General ITS type network generated with TCS. The ITS type codes (a–x) are provided with the respective total number of ITS sequence types summarized within a general ITS type. In the case of types a, b, and e (A. arenosa) and g and f (A. croatica), corresponding individuals show intermediate morphological characters indicating introgression and hybridization.
We only briefly comment on the position of A. thaliana as the internal root, because the ITS sequence data do not sufficiently resolve relationships at the base of the major lineages best explained by an early radiation event ≈2 mya. It is important to mention that there are no other taxa from the mustard family that are positioned phylogenetically between A. thaliana and its relatives analyzed herein. It should also be noted that the position of A. thaliana in the TCS network indicates that ITS types of A. arenosa are more ancestral than those of the other segregates. This finding is important when comparing the data with results from the cpDNA analysis.
Arabidopsis suecica has been excluded from our analysis. This taxon has been fully confirmed as a hybrid between A. thaliana and A. arenosa, and it evolved with a unique origin <400,000 years ago in Fennoscandinavia (36). Another taxon, A. lyrata ssp. kamchatica, with a proven hybrid status in Japan (A. halleri ssp. gemmifera × A. lyrata), carried exclusively A. lyrata-specific ITS types a and b. However, it should be noted that the hybrid origin of A. lyrata ssp. kamchatica accessions from Russia (type locality is Kamchatka) or Alaska and Canada is not proven yet and remains questionable. It is most likely that A. lyrata ssp. kamchatica (or A. kamchatica) from Japan, Korea, and Taiwan is a distinct taxon of hybridogenous origin and not the same species or subspecies as distributed in Russia, Alaska, and Canada.
Chloroplast Haplotypes Indicate Ancient Shared Polymorphisms.
In total 34 cpDNA suprahaplotypes have been characterized. Considering length variation and additional single-nucleotide polymorphisms in the highly dynamic 3′ region of the trnL-F intergenic spacer carrying the trnF pseudogenes polymorphisms these 34 suprahaplotypes consist of 153 haplotypes (numbered from 1 to 153 in SI Table 1). When compared with the ITS data the resulting network analysis provided a different, not species-specific distribution pattern of genetic variation (SI Fig. 5). The most ancestral haplotypes (interior in the network) are shared in various combinations by almost all species lineages as characterized by the ITS analysis. Only haplotypes at the various tips of the cpDNA haplotype network are species-specific. This significant incongruence between nuclear and plastid data sets can be explained in two ways. First, reticulation and hybridization among lineages have transferred cpDNA types from one lineage into the other. However, if this scenario is true we would not expect such a clear evolutionary signal as is provided with the ITS data. The second explanation would expect that ancestral cpDNA type diversity predates separation of the main evolutionary lineages. This scenario is more likely and correlates well with the fact that suprahaplotypes from internal position of the network consists of more haplotypes than those at the tips of the network (SI Fig. 5). However, this hypothesis requires that an old center of genetic diversity is congruent to a center of origin of the various lineages (see below). Chloroplast types from A. cebennensis and A. pedemontana, but also A. croatica, are congruent with such a hypothesis, because these haplotypes (N, T, and Z) are directly connected to the ancestral type A.
As for A. suecica, previous extensive analysis of cpDNA variation (37) has demonstrated that this hybrid taxon (A. thaliana × A. arenosa) is carrying exclusively A. thaliana cpDNA types.
Phylogeographic Data and Gene Diversity Statistics Demonstrate High Levels of Genetic Variation.
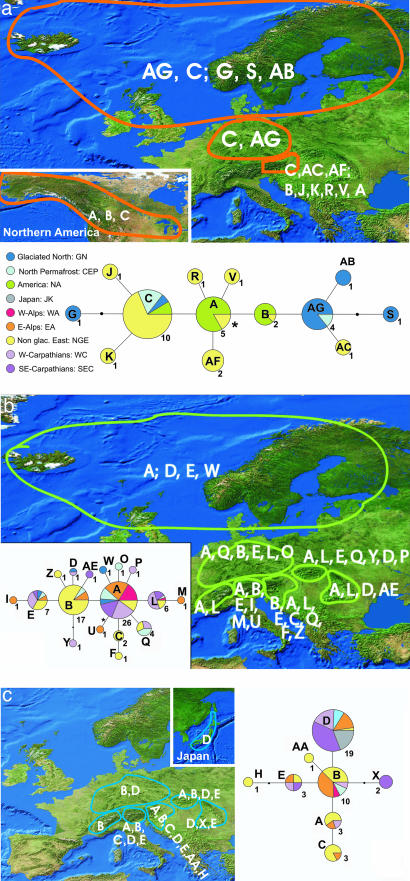
To demonstrate species-specific distribution of cpDNA variation, the suprahaplotype network (SI Fig. 5) has been redrawn for the three major lineages (A. arenosa, A. halleri, and A. lyrata) separately, highlighting only those haplotypes occurring in each lineage, respectively (Fig. 2). The northern hemisphere has been divided into nine major regions: (i) glaciated north (GN), comprising the areas of Europe that were glaciated at the maximum extend of Pleistocene glaciation cycles (Iceland, Norway, Sweden, Fennia, Denmark, coastal areas around the Baltic Sea, northern Ireland, and northern Great Britain); (ii) North America with Alaska, Canada, and the northern U.S.; (iii) Japan, Taiwan, and Korea; (iv) central European permafrost regions comprising the region in central Europe between the GN and the Alps; (v) the western Alps (WA); (vi) the glaciated part of the eastern Alps (GEA); (vii) the nonglaciated east (NGE), comprising the nonglaciated part of the eastern Alps and the nonglaciated area between the eastern Alps, the northern permafrost region, and the western Carpathians; (viii) the western Carpathians (WC); and (ix) the southeastern Carpathians (SEC).
Fig. 2.
Geographic distribution of cpDNA suprahaplotypes. The networks are redrawn from Fig. 3. The regional color code is given in a. (a) A. lyrata. (b) A. arenosa. (c) A. hallen. The geographic regions are defined as described in Results.
In A. halleri eight suprahaplotypes comprising 42 haplotypes were detected (Fig. 2a). The most ancestral suprahaplotype was type A from Austria (GEA and NGE) and Slovakia (WC). Two suprahaplotypes, B and D, were commonly found in more than three regions. The most widespread suprahaplotype D occurred also in Japan and in all European regions except the WA. D is the only suprahaplotype found in Japan. Regional suprahaplotype sharing (SI Table 2) was highest between the NGE and the GEA (five types). Four suprahaplotypes were shared between the NGE and the WC and between the WC and the GEA, respectively. The CE permafrost region had two suprahaplotypes in common with the GEA, the NGE and the WC. Two shared suprahaplotypes were also found between the SEC and the WC, the NGE, or the GEA. Besides the high amount of haplotype sharing between the NGE and the GEA, most haplotypes occurred in one or two neighboring regions (Fig. 2a). Pairwise ΦST estimates showed that the NGE was clearly differentiated from Japan, the SEC, and the WC (SI Table 3). The WA and Japan were totally different because they had no suprahaplotypes in common. Japan was significantly differentiated from all European regions except the SEC. Pairwise FST estimates showed a similar picture. Additionally, the WA were significantly differentiated from the SEC. Nucleotide diversity was highest in the NGE (0.0021), followed by the SEC (0.0014) and the WC (0.0012) (SI Table 4). In the WA and in Japan nucleotide diversity was zero because we detected only one suprahaplotype. Nearly all suprahaplotypes were found in the NGE (seven suprahaplotypes, R = 3.69). The contacting regions, the GEA and the WC, also showed high numbers of different suprahaplotypes (GEA, five suprahaplotypes, R = 2.86; WC, four suprahaplotypes, R = 2.77). Effective genetic diversity was also highest in the NGE (3.69) followed by the GEA (2.42) and the WC (2.38). Private suprahaplotypes were found in the NGE and the SEC. The corresponding diversity estimates for all haplotypes (SI Table 5) were congruent. Here the GEA shows the highest number of haplotypes (12 haplotypes, R = 4.82) and the highest effective genetic diversity (8.6). Similar high values are found for the SEC (11 haplotypes, R = 4.75, va = 6.23) and the NGE (12 haplotypes, R = 4.62, va = 6.26). The WA and Japan again have the lowest diversity estimates. Private haplotypes are found in all regions. Interestingly, 93% of the SEC haplotypes are private; in the WC and in Japan even 100% of the haplotypes are private.
In A. lyrata 13 different suprahaplotypes comprising 31 haplotypes were detected in Europe and North America (Fig. 2b). The most ancestral suprahaplotype A was found in Alaska and Canada, but also in the NGE. Two major lineages evolved from suprahaplotype A. Derived suprahaplotypes from the tips of the network were distributed only in Europe. Three rare suprahaplotypes from Austria (R, V, and AF) were directly connected to the most ancestral suprahaplotype A. In North America the most frequent and ancestral suprahaplotypes A, B, and C were found.
Regional suprahaplotype and haplotype sharing is quite low in A. lyrata (SI Table 2). A maximum of two suprahaplotypes and haplotypes, respectively, is shared among the four different regions. Pairwise ΦST and FST estimates showed a clear differentiation for all regions except the NGE and CE permafrost region (SI Table 3), which provide some additional evidence for periglacial survival in permafrost dominated areas during the Pleistocene (1). SI Table 4 summarizes suprahaplotype frequencies and genetic diversity indices of A. lyrata. The formerly glaciated north of Europe (GN) appears to be the region with highest genetic diversity estimates. Nucleotide diversity (0.0028), effective diversity (2.81), and the number of different haplotypes corrected for sample size (3.8) are higher than in any other region. These estimates are different when calculating diversity parameters taking all haplotypes into account (SI Table 5). Both regions, the formerly GN and the NGE, have the same number of different haplotypes (5.3). However, the NGE shows the highest effective genetic diversity (7.09). Among the remaining regions, the lowest effective diversity and haplotype frequency for suprahaplotype estimates and haplotype estimates was observed for the northern permafrost region. In summary, we observed that in general genetic diversity in A. lyrata is lower than in A. halleri, and in both species groups NGE outside the glaciers and permafrost areas played an important role as refuge area and center of genetic diversity. However, in contrast to A. halleri, periglacial survival in A. lyrata might have also played a substantial role in maintaining genetic diversity throughout its northern distribution range.
In A. arenosa 17 suprahaplotypes comprising 72 haplotypes were detected (Fig. 2c). Ancestral suprahaplotype A is widely distributed in Europe, whereas type B is restricted to Austria and Slovenia. Suprahaplotype and haplotype sharing among adjacent regions is extensive (SI Table 2). Pairwise ΦST and FST estimates showed that among the regional groups for A. arenosa the NGE is differentiated strongest from the WA (SI Table 3). Additionally, the NGE is clearly differentiated from the other regional groups (except GN for ΦST). Among the remaining regions there is significant differentiation of the WA compared with the GN for both ΦST and FST estimates. Considering FST estimates, only the WC are differentiated from the WA and the GEA. Nucleotide diversity based on suprahaplotypes was highest in the WC (0.0016) followed by the GN (0.0015) and the NGE (0.0014) (SI Table 4). However, the differences are mostly minor and depend largely on sample size. In any case, again the NGE and adjacent areas showed the highest levels of genetic diversity. This result is much more obvious when all haplotypes are considered (SI Table 5). Here we observe extremely high levels of genetic diversity in the NGE and, although lowered, still high levels of genetic variation in the Carpathians, in the GEA, and in central European permafrost regions. In summary, it has to be concluded that genetic diversity is much higher than in A. lyrata and A. halleri, and, as concluded for A lyrata and A. halleri, the NGE plays a dominant role as the center of genetic diversity.
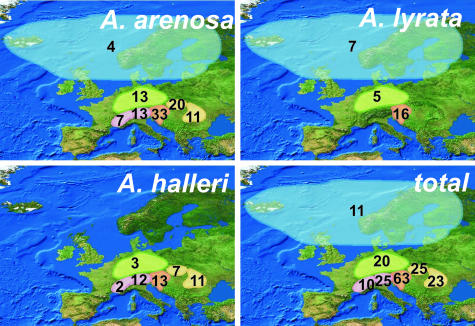
Taking into consideration that cpDNA haplotype variation to some extent predates evolution of the several evolutionary lineages the overall distribution of haplotypes among the nine defined regions is remarkable, with strong gradients of decreasing number of haplotypes in any geographical direction starting from a center in Slovakia and eastern Austria (Fig. 3). It is also obvious from ΦST/FST comparisons (SI Table 3) that for all three species groups ΦST does not generally exceed FST, which means that haplotype phylogeny does not fit haplotype distribution significantly. This again favors the assumption that an old stock of cpDNA haplotypes had existed before species diversification and Pleistocene migration.
Fig. 3.
Distribution of total numbers of cpDNA haplotypes in A. halleri, A. lyrata, and A. arenosa.
Gene diversity statistics for the nuclear ribosomal RNA (ITS) (SI Table 6) do not correspond perfectly with cpDNA data. However, one general conclusion is also obvious and can be drawn: Total genetic diversity is highest in A. arenosa, followed by A. lyrata and finally A. halleri. Other incongruencies are best explained by the marker system itself and the type of data: (i) The ITS loci are subjected to concerted evolution. However, the direction and final result of the homogenization process are not predictable. (ii) The data set is biased by the various sequence ambiguities. The ITS data also demonstrated for all three species that ΦST does not exceed FST, which means that ITS phylogeny does not fit ITS type distribution significantly (SI Table 7).
Comments on A. kamchatica, A. arenicola, A. cebennensis, A. pedemontana, and A. croatica.
The herein analyzed accessions of A. kamchatica from Japan are characterized by cpDNA suprahaplotype AD, which derived from A. halleri haplotypes. The same accessions are defined by ITS type b, which is characteristic for A. lyrata. This confirms the hypothesis that A. kamchatica from Japan, Korea, and Taiwan indeed represents a hybrid between A. halleri ssp. gemmifera and A. lyrata. However, all A. kamchatica (A. lyrata ssp. kamchatica) accessions from outside these countries analyzed herein are characterized by cpDNA suprahaplotype B and ITS type b. Thus, they are very similar to any other A. lyrata accessions from North America analyzed herein not favoring any hybridization scenario as proven for Japanese accessions. However, the taxonomy of the two subspecies A. lyrata ssp. lyrata and A. lyrata ssp. kamchatica in Russia and North America is even more complicated by descriptions that all plants of ssp. lyrata might be diploid, while all plants of ssp. kamchatica have been described to appear tetraploid (38, 39). Considering these data our results favor autopolyploidization of A. lyrata ssp. lyrata resulting in A. lyrata ssp. kamchatica distributed actually in Russia, Alaska, and Canada. Additional research with material of known ploidy level is badly needed.
A. arenicola as analyzed by Warwick et al. (12) is characterized by cpDNA suprahaplotype A and ITS type e (12), which supports closest relatedness to A. lyrata. We think that both taxa, A. arenicola and A. kamchatica from outside Japan, Taiwan, and Korea, are best summarized within a broadly defined A. lyrata, maybe best on the subspecies level. If future research will demonstrate that Japanese hybrids, also treated as A. kamchatica, are not related to North American and Russian A. lyrata ssp. kamchatica, taxonomic rules will require a new name for these Japanese hybrids, e.g., Arabidopsis kawasakiana (13). A. cebennensis and A. pedemontana are old diploids (confirmed by chromosome counts and microsatellite analysis; M.A.K. and R. Schmickl, unpublished data) and genetically well defined species with a relictual distribution in southeast France and northwest Italy, respectively. A. croatica is distantly related only to A. arenosa, but in this case secondary contact with A. arenosa in Croatia resulted in genetic admixture.
Discussion
Comparative DNA sequence analysis of a plastid and a nuclear locus across the whole genus Arabidopsis has allowed us to introduce a phylogenetic framework for all known closest relatives of A. thaliana. If we consider that ancestors of these closest relatives diverged from the A. thaliana ancestor ≈5 mya (3, 4) and count the mean number of mutational steps in the ITS network (Fig. 1) from A. thaliana to any other tip of the network (30 steps), we obtain a rough estimate for the age of the inner part of the network of ≈2 million years, which is close to the beginning of the Pleistocene and its various glaciation and deglaciation cycles. A similar value is also obtained for ITS phylogenetic reconstructions enforcing a molecular clock (data not shown). In addition, cpDNA data favored a primary center of genetic diversity in the eastern part of its European distribution. During all of the Pleistocene, this area might have served also as an important refuge area for the various segregates of A. halleri, A. lyrata, and A. arenosa. Consequently, these refuge areas have served as a genetic reservoir for the generation of new taxa mostly within the A. halleri and A. arenosa lineages, which is reflected by the numerous taxa described from this region. Few taxa have been forced very early during their evolution into relictual areas in southeastern France and northwestern Italy (such as A. cebennensis and A. pedemontana) or Croatia (A. croatica), but theses taxa did not expand back into their (unknown) original distribution areas. Interestingly, all of these species are highly endemic with narrow distribution ranges and consequently exhibit less genetic variation than any other species. However, throughout the Pleistocene A. arenosa, A. lyrata, and A. halleri evolved differently in terms of ecological adaptation (1) and range expansion. A. lyrata was the most successful colonizer of northern regions, and our data demonstrate that A. lyrata survived glaciation periods north of the central European ice sheets in permafrost regions (21). High levels of genetic variation have been maintained because of large effective population sizes and the self-incompatible breeding system. However, more recent colonization of formerly glaciated areas such as in North America resulted in much lower levels of genetic variation. In case of A. halleri we have a significant preference for higher altitudes rather than simply harsh environments. This is also reflected by its mainly central to east European distribution in mountainous to subalpine habitats, with only one successful colonizer in eastern Asia in Japan and adjacent regions (A. halleri ssp. gemmifera). The situation in A. arenosa and its various segregates is more complex and not resolved in detail by our data. We can conclude that in A. arenosa neither our ITS nor the cpDNA data reflect any intraspecific differentiation as demonstrated by morphological or cytological variation (1). Furthermore, the ITS data demonstrate extensive genetic contact of A. arenosa with A. croatica but also A. lyrata (M.A.K., unpublished data), but not with A. halleri.
Our data provide a first comparative overview on genetic diversity on all Arabidopsis segregates on a representative geographic scale. The most important finding here is that A. arenosa carries much higher levels of genetic diversity than any other species. This is best explained by a breeding system that is dominated by self-incompatibility (M.A.K., unpublished data). An additional alternative explanation for these high levels of genetic variation is past and ongoing hybridization and reticulation with A. lyrata. Such a complex suture zone has been circumscribed in Austria outside the range of the last maximum glaciation and outside the permafrost areas (M.A.K., R. Schmickl, and M.M., unpublished data).
Our aim is to contribute substantially to the poor knowledge on Arabidopsis wild relatives and, therefore, to stimulate further research in these nonmodel plants. Previous studies have already successfully focused on A. halleri and A. lyrata (1, 2), but other taxa such as A. arenosa offer some additional resources of genetic diversity and character variation. Despite differences in ecological niche differentiation and evolutionary history, all three species groups are represented by diploids (but in the case of A. arenosa and A. lyrata tetraploids have been also observed frequently) and a predominantly effectively working self-incompatibility system.
Materials and Methods
Plant Material.
We used various sources of plant material. In total we studied 620 accessions of relatives of A. thaliana. Plant material was obtained from the following herbaria (acronym is indicated): Biologiezentrum Linz (LI), Zürich University (ZT), Natural History Museum and Herbarium London (BM), Natural History Museum and Herbarium Vienna (W), Herbarium of the Botanical Institute at Vienna University (WU), the California Academy of Sciences in San Francisco (CAS), Agriculture and Agri-Food Canada in Ottawa (DAO), New York Botanical Gardens (DH), Gray Herbarium at Harvard University (GH), and Bratislava, Academy of Sciences (SAV). Additional material was collected in the wild, and vouchers were deposited at Heidelberg Herbarium (HEID). See SI Table 1 for detailed sampling information. Sequence data used herein were obtained from 452 accessions, with a complete data set for both selected marker systems, nuclear ITS and plastidic trnL-F, from 365 accessions. For the remaining accessions we added sequence data for the trnL intron or the trnLF intergenic spacer only. For taxon designation we followed a conservative approach because we are still lacking careful morphometric analysis of A. arenosa and A. lyrata and its corresponding segregates. We largely followed the species concept of Al-Shehbaz et al. (11), but we treated A. neglecta within the A. arenosa aggregate (1). This is also true for any other segregate of A. arenosa [compare A. petrogena, A. nitida, and other (1)]. We also did not follow Shimizu et al. (13) and Warwick et al. (12) and kept A. kamchatica and A. arenicola as representatives of a more broadly defined A. lyrata. However, if possible we provided these various provisory names according to morphological descriptions, voucher labels, or geographic distribution along with SI Table 1. We also provided a geographic map visualizing the worldwide sampling in SI Fig. 6.
DNA Preparation and Sequencing.
DNA extraction from dry leaf samples (either herbarium material or silica gel-dried material collected directly in the wild) followed a simple cetyltrimethylammonium bromide protocol (38). Amplification and sequencing of the ITS and the trnL intron–trnLF intergenic spacer followed the protocols and information provided earlier (trnL-F, refs. 40 and 41; ITS, ref. 42). GenBank accession numbers are provided in SI Table 1.
DNA-Based Phylogenetic Reconstructions and Networks.
Alignments were created manually because of nearly identical sequence length, and indels were coded as binary characters. As for the plastid trnL-F region we did not align the 3′ region of the trnL-F intergenic spacer because of extensive trnF pseudogene copy number variation and resulting ambiguities in the alignment. This reduced amount of DNA variation has been used to define “suprahaplotypes,” which are mostly based on single-nucleotide polymorphisms. The DNA sequence information from the pseudogene-rich region has been used to subdivide these suprahaplotypes into a significant higher number of haplotypes (41, 43) without any further phylogenetic calculations. The cpDNA data (suprahaplotypes) have been subjected to network analysis. Suprahaplotype networks were constructed for the trnL intron–trnLF intergenic spacer. For this purpose all indels (except polyT stretches) were coded as additional single binary characters. Haplotype networks were constructed by using TCS version 1.21 (44) according to acceptance criteria outlined earlier (45). DNA sequence data from the ITS were obtained from a direct sequencing approach. In principle this DNA region is subjected to a process called concerted evolution (42), and multiple copies might indicate species-specific naturally occurring variation among loci or might demonstrate the result of more recent hybridization and reticulation. We obtained numerous sequences with ambiguous sites and not totally homogenized ITS copies. Therefore, we used the TCS program to group the 103 different sequences in total. TCS recognized 24 groups of ITS types (further named as “general types”), and we selected manually one representative sequence for each type with the lowest number (or even zero) of ambiguous sites. These 24 sequences (ITS types a–x) were used for phylogenetic reconstructions running a maximum-likelihood analysis with PAUP*4.0 (46) [options: exhaustive search, Multrees (save multiple trees), and TBR (tree bisection and reconnection) branch swapping]. Substitution models were selected by MODELTEST 3.5 (47) under the Akaike information criterion, with parameters estimated during ML exhaustive searches. The same alignment has been subjected to a network analysis by using TCS as outlined for the plastid data starting initially with a 95% confidence interval (three internal steps allowed) and adding remaining groups of sequences with the 90% confidence interval option (four steps) and finally running standard settings (allowing the maximum number of steps). Sequences from A. thaliana served as outgroup in various calculations (ITS, GenBank accession no. AJ232900; trnL intron, GenBank accession no. DQ313522; trnLF intergenic spacer, GenBank accession no. DQ528960).
Gene Diversity Statistics and Phylogeographic Inference.
For the phylogeographic analysis, individual cpDNA sequences were divided into nine regional groups based on geography and observed haplotypes. Genetic diversity was estimated as haplotype richness (R, the number of different haplotypes corrected for sample size through rarefaction) (48), nucleotide diversity (49), and effective genetic diversity (50). We estimated genetic differentiation between all pairs of regions and among all regions with an analysis of molecular variance using the program ARLEQUIN (51). Both FST, an estimate of differentiation based on allele frequencies, and ΦST, an estimate of differentiation taking into account the molecular distance between haplotypes, were estimated. In an analysis of molecular variance framework these values are estimated as the proportion of variance among groups. These two estimators are analogous to GST and NST, respectively (52). In the case of correspondence between haplotype phylogenies and their geographic distribution, estimates for NST (ΦST) will be greater than the GST (FST) values (52, 53). The program PERMUT (www.pierroton.inra.fr/genetics/labo/Software) tests whether the difference between the two estimates is significant by a permutation test exchanging haplotypes but conserving haplotype frequencies (54). It generates a random distribution for NST, which allows determining a P value for the observed estimate. The average of the random distribution corresponds to GST. GST/NST differs from FST/ΦST in the way they treat differences in sample size (52).
We performed the same analysis for the ITS data. It should be noted that sequence ambiguities due to multiple intraindividual copies might bias the calculated genetic parameters significantly, but these analyses were kept to confirm at least general trends as demonstrated by the cpDNA data.
Supplementary Material
Acknowledgments
We gratefully acknowledge support over the last 4 years from the Austrian Science Fund and more recently from Deutsche Forschungsgemeinschaft and Deutscher Akademischer Austauschdienst (to M.A.K.).
Abbreviations
- cpDNA
chloroplast DNA
- ITS
internal transcribed spacer region
- GN
glaciated north
- WA
the western Alps
- GEA
the glaciated part of the eastern Alps
- NGE
nonglaciated east
- WC
western Carpathians
- SEC
southeastern Carpathians.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database. For a list of accession numbers, see SI Table 1.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701338104/DC1.
References
- 1.Clauss M, Koch MA. Trends Plant Sci. 2006;11:449–459. doi: 10.1016/j.tplants.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell-Olds T. Trends Ecol Evol. 2001;16:693–700. [Google Scholar]
- 3.Koch MA, Haubold B, Mitchell-Olds T. Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 4.Koch MA, Haubold B, Mitchell-Olds T. Am J Bot. 2001;88:534–544. [PubMed] [Google Scholar]
- 5.O'Kane SL, Jr, Al-Shehbaz IA. Ann Missouri Bot Gard. 2003;90:603–612. [Google Scholar]
- 6.Mitchell-Olds T, Al-Shehbaz IA, Koch MA, Sharbel T. In: Diversity and Evolution in Plants: Genotype and Phenotype Variation in Higher Plants. Henry RJ, editor. Cambridge, UK: CABI; 2005. pp. 119–137. [Google Scholar]
- 7.Kuittinen H, Aguadé M. Genetics. 2000;155:863–872. doi: 10.1093/genetics/155.2.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuittinen H, de Haan AA, Vogl C, Oikarinen S, Leppälä J, Koch M, Mitchell-Olds T, Langley CH, Savolainen O. Genetics. 2004;168:1575–1584. doi: 10.1534/genetics.103.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch MA, Kiefer M. Am J Bot. 2005;92:761–767. doi: 10.3732/ajb.92.4.761. [DOI] [PubMed] [Google Scholar]
- 10.Yogeeswaran K, Frary A, York TL, Amenta A, Lesser AH, Nasrallah JB, Tanksley SD, Nasrallah ME. Genome Res. 2005;15:505–515. doi: 10.1101/gr.3436305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Shehbaz IA, O'Kane S, Price RA. Novon. 1999;9:296–307. [Google Scholar]
- 12.Warwick SI, Al-Shehbaz IA, Sauder CA. Can J Bot. 2006;84:269–281. [Google Scholar]
- 13.Shimizu KK, Fujii S, Marhold K, Watanabe K, Kudoh H. Acta Phytotax Geobot. 2005;56:163–172. [Google Scholar]
- 14.Mesicek J. Preslia (Praha) 1970;42:225–248. [Google Scholar]
- 15.Wright SI, Lauga B, Charlesworth D. Mol Ecol. 2003;12:1247–1263. doi: 10.1046/j.1365-294x.2003.01743.x. [DOI] [PubMed] [Google Scholar]
- 16.Ramos-Onsins SE, Stranger BE, Mitchell-Olds T, Aguadé M. Genetics. 2004;166:373–388. doi: 10.1534/genetics.166.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonsell B, Kustås K, Nordal I. Ecography. 1995;18:321–332. [Google Scholar]
- 18.Charlesworth D, Bartolome C, Schierup MH, Mable MK. Mol Biol Evol. 2003;20:1741–1753. doi: 10.1093/molbev/msg170. [DOI] [PubMed] [Google Scholar]
- 19.Pauwels M, Samitou-Laprade P, Holl AC, Petit D, Bonnin I. Mol Ecol. 2005;14:4403–4414. doi: 10.1111/j.1365-294X.2005.02739.x. [DOI] [PubMed] [Google Scholar]
- 20.Hewitt GM. Philos Trans R Soc London Ser B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clauss MJ, Mitchell-Olds T. Mol Ecol. 2006;15:2753–2766. doi: 10.1111/j.1365-294X.2006.02973.x. [DOI] [PubMed] [Google Scholar]
- 22.Kärkkäinen K, Løe G, Ågren JK. Evolution (Lawrence, Kans) 2004;58:2831–2836. [PubMed] [Google Scholar]
- 23.Van Treuren R, Kuittinen H, Karkkainen K, Baena-Gonzalez E, Savolainen O. Mol Biol Evol. 1997;14:230–238. doi: 10.1093/oxfordjournals.molbev.a025758. [DOI] [PubMed] [Google Scholar]
- 24.Clauss MJ, Cobban H, Mitchell-Olds T. Mol Ecol. 2002;11:591–601. doi: 10.1046/j.0962-1083.2002.01465.x. [DOI] [PubMed] [Google Scholar]
- 25.Charlesworth D, Awadalla P, Mable BK, Schierup MH. Ann Bot. 2000;85:227–239. [Google Scholar]
- 26.Van Rossum F, Bonnin I, Fenart S. Mol Ecol. 2004;13:2959–2967. doi: 10.1111/j.1365-294X.2004.02314.x. [DOI] [PubMed] [Google Scholar]
- 27.Bert V, Macnair MR, de Laguerie P, Saumitou-Laprade P, Petit D. New Phytol. 2000;146:225–233. doi: 10.1046/j.1469-8137.2000.00634.x. [DOI] [PubMed] [Google Scholar]
- 28.Castric V, Vekemans X. Mol Ecol. 2004;13:2873–2889. doi: 10.1111/j.1365-294X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- 29.Kasuba M, Dwyer K, Hendershot J, Vrebalow J, Nasrallah JB, Nasrallah ME. Plant Cell. 2001;13:627–643. [PMC free article] [PubMed] [Google Scholar]
- 30.Schierup MH, Mable BK, Awadalla P, Charlesworth D. Genetics. 2001;158:387–323. doi: 10.1093/genetics/158.1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nasrallah ME, Liu P, Sherman-Broyles S, Boggs NA, Nasrallah JB. Proc Natl Acad Sci USA. 2004;101:16070–16074. doi: 10.1073/pnas.0406970101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mable BK, Robertson A, Dart S, DiBerardo C, Witham L. Evolution (Lawrence, Kans) 2005;59:1437–1448. [PubMed] [Google Scholar]
- 33.Comai L, Tygai A, Winter K, Holmes-Davis S, Reynolds R, Stevens Y, Byers B. Plant Cell. 2000;12:1551–1567. doi: 10.1105/tpc.12.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fiebig A, Kimport A, Preuss D. Proc Natl Acad Sci USA. 2004;101:3286–3291. doi: 10.1073/pnas.0305448101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sall T, Lind-Halldén C, Jakobbson M, Halldén C. Hereditas. 2004;141:313–317. doi: 10.1111/j.1601-5223.2004.01833.x. [DOI] [PubMed] [Google Scholar]
- 36.Jakobsson M, Hagenblad J, Tavaré S, Säll T, Halldén C, Lind-Halldén C, Nordborg M. Mol Biol Evol. 2006;23:1217–1231. doi: 10.1093/molbev/msk006. [DOI] [PubMed] [Google Scholar]
- 37.Säll T, Jakobsson M, Lind-Halldén C, Halldén C. J Evol Biol. 2003;16:1019–1029. doi: 10.1046/j.1420-9101.2003.00554.x. [DOI] [PubMed] [Google Scholar]
- 38.Mulligan GA. Rhodora. 1995;97:109–163. [Google Scholar]
- 39.Mulligan GA. Can Field Nat. 2002;116:611–622. [Google Scholar]
- 40.Dobeš C, Mitchell-Olds T, Koch M. Mol Ecol. 2004;13:349–370. doi: 10.1046/j.1365-294x.2003.02064.x. [DOI] [PubMed] [Google Scholar]
- 41.Koch M, Dobeš C, Matschinger M, Bleeker W, Vogel J, Kiefer M. Mol Biol Evol. 2005;22:1032–1043. doi: 10.1093/molbev/msi092. [DOI] [PubMed] [Google Scholar]
- 42.Koch M, Dobeš C, Mitchell-Olds T. Mol Biol Evol. 2003;20:338–350. doi: 10.1093/molbev/msg046. [DOI] [PubMed] [Google Scholar]
- 43.Koch M, Dobeš C, Kiefer C, Schmickl R, Klimes L, Lysak MA. Mol Biol Evol. 2007;24:63–73. doi: 10.1093/molbev/msl130. [DOI] [PubMed] [Google Scholar]
- 44.Clement M, Posada D, Crandall KA. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 45.Templeton AR, Crandall KA, Sing CF. Genetics. 1992;132:619–663. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swofford DL. Sunderland, MA: Sinauer; 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. [Google Scholar]
- 47.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 48.El Mousadik A, Petit RJ. Theor Appl Genet. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- 49.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ Press; 1987. p. 245. [Google Scholar]
- 50.Gregorius HR. Math Biosci. 1978;41:253–271. [Google Scholar]
- 51.Excoffier L, Laval G, Schneider S. Evol Bioinf Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 52.Pons O, Petit RJ. Genetics. 1996;144:1237–1245. doi: 10.1093/genetics/144.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. Mol Ecol. 2005;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- 54.Burban C, Petit RJ, Carcreff E, Jactel H. Mol Ecol. 1999;8:1593–1602. doi: 10.1046/j.1365-294x.1999.00739.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.