Abstract
Modeling and analysis of genetic regulatory networks is essential both for better understanding their dynamic behavior and for elucidating and refining open issues. We hereby present a discrete computational model that effectively describes the transient and sequential expression of a network of genes in a representative developmental pathway. Our model system is a transcriptional cascade that includes positive and negative feedback loops directing the initiation and progression through meiosis in budding yeast. The computational model allows qualitative analysis of the transcription of early meiosis-specific genes, specifically, Ime2 and their master activator, Ime1. The simulations demonstrate a robust transcriptional behavior with respect to the initial levels of Ime1 and Ime2. The computational results were verified experimentally by deleting various genes and by changing initial conditions. The model has a strong predictive aspect, and it provides insights into how to distinguish among and reason about alternative hypotheses concerning the mode by which negative regulation through Ime1 and Ime2 is accomplished. Some predictions were validated experimentally, for instance, showing that the decline in the transcription of IME1 depends on Rpd3, which is recruited by Ime1 to its promoter. Finally, this general model promotes the analysis of systems that are devoid of consistent quantitative data, as is often the case, and it can be easily adapted to other developmental pathways.
Keywords: budding yeast, computational modeling, meiosis, systems biology, transcriptional networks
Developmental pathways are characterized by a transcriptional cascade that ensures the coordinated expression and activity of a network of genes that is governed by a master activator (1). An important feature is that the expression of regulatory genes occurs during short specific windows in the differentiation pathway. Such short-lived signals are usually accomplished through positive and negative feedback loops (2–6). The importance of a short-lived signal for efficient entry into a developmental pathway is documented in many systems, including lymphocyte differentiation in mice (7) and yeast meiosis (8).
Our objectives were to construct an effective computational model that captures the transient and cascade characteristics of developmental pathways in general, and thereby provides reliable predictions for both its qualitative behavior and the specific interactions between its components. For this purpose, we used the developmental pathway of meiosis in the budding yeast Saccharomyces cerevisiae.
A transcriptional cascade that includes positive and negative feedback loops directs the initiation and progression through meiosis (9, 10). Ime1 is the master transcriptional activator whose transcription and activity is regulated by the meiotic signals (10). Ime1 is recruited to the promoters of the early meiosis-specific genes (EMG) by the DNA-binding protein Ume6 (11). Under conditions leading to vegetative growth, Ume6 recruits two complexes (Isw2, chromatin remodeling; and Rpd3, histone deacetylase) that repress transcription (12, 13). Ime1 possesses two activities: It is required to relieve Rpd3 repression, an activity that is regulated by the meiotic signals, and to activate transcription (14, 15). Ime1 also exhibits positive autoregulation (16). Ime2 is a representative of EMG that functions as a positive regulator of meiosis essential for the transcription of middle and late meiosis-specific genes (10). Ime2 is required for the efficient transcription of all EMG (17, 18). In addition, both Ime1 and Ime2 have negative feedback roles required for the transient transcription of IME1 (8, 17, 18). The mode by which this happens is not known. Additionally, Ime2 phosphorylates Ime1 protein, thereby tagging it for degradation (19).
Numerous analysis paradigms and modeling techniques have been suggested for the elucidation of metabolic, signaling, and regulatory pathways (20–24). Continuous methods that faithfully describe the dynamics of the system require exact numeric values of, e.g., concentrations, expression levels, and timings. However, more often than not, such details are not available (certainly not in their entirety) so as to allow accurate deployment of continuous methods. Still, it turns out that frequently the detailed nature of such methods is not necessary when reasoning about the qualitative behavior of a pathway. Moreover, a discrete method might be more appropriate when focusing on behavioral properties, such as transience, robustness, and stability, which are pertinent to developmental pathways (25, 26). Here we examine the feasibility of a discrete method to faithfully describe and analyze the transient and timely expression of a network of genes.
The simplest kind of a discrete method is a Boolean network. Li et al. (27) have used such a model to show that several properties, such as stability and robustness, can be observed on, e.g., the cell cycle in yeast. However, we found that this method was not effective to our developmental pathway, because it led to an increase without a decline in the transcription of IME1, and the levels of Ime1, IME2 mRNA, and Ime2 oscillated [supporting information (SI) Fig. 5, N = 1]. Hence, we devised an extension to this model that is more expressive in terms of expression levels and transition rules but at the same time preserves the efficiency and ability to effectively refine the way in which specific pathways are treated computationally.
Using our simple yet powerful model, we were able to obtain a clear separation of behaviors, predicting the response of the network to various initial conditions (e.g., a noticeable distinction between conditions leading to transient vs. nontransient behavior was observed). Our simulations show that the meiotic pathway is robust, i.e., it is insensitive to the initial levels of IME1 and IME2 RNA and proteins. The computational results were verified experimentally, showing that our modeling faithfully describes the qualitative behavior of the pathway. Furthermore, we explored variations of the basic network to elucidate the negative feedback mechanism by which Ime1 and Ime2 shut down the transcription of IME1.
Results
Modeling of the Ime1/Ime2 Network.
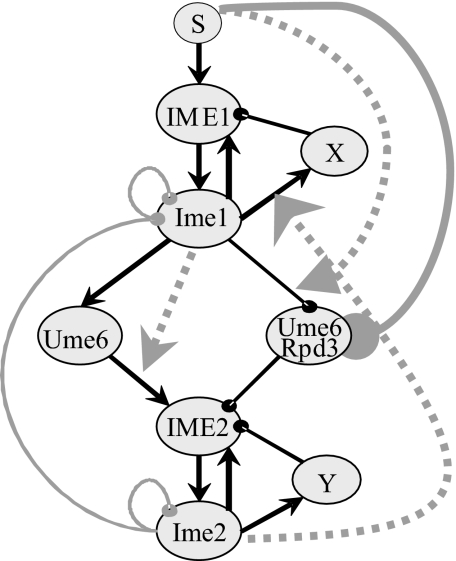
Fig. 1 shows our working hypothesis network that captures the key activities of Ime1 and Ime2. For simplicity, some of the details are represented at a higher level of abstraction. The network includes two putative nodes, X and Y, which are used to shut down the transcription of IME1 and IME2, respectively (as detailed below). We introduced the logical AND operation to describe conjunctive conditional dependence between functions, namely, the activity of a regulator is enabled (beyond direct regulation) only if some other regulator in the system is active (e.g., Ume6 activates IME2 only if Ime1 exists).
Fig. 1.
Working hypothesis network describing the relationship between the expression of IME1 and IME2. Names of RNA molecules are in all capitals. In protein names, only the first letter is capitalized. Solid black arrows represent activation, and solid black lines with a knob represent repression. Solid gray lines with a knob reflect degradation. The dotted gray arrows (going from a node to an edge) represent gating by conjunction (AND). The node S represents the signal that induces entry into meiosis, in our case, nitrogen depletion in the presence of acetate as the sole carbon source.
Our computational model is a discrete transition system. A pathway is represented by a graph whose nodes are RNAs or proteins, and its weighted edges denote regulation (positive weights for activation and negative weights for repression). A current state of the system is represented by a vector of RNA and protein expression levels associated with the nodes, in which each entry can assume one of a small number of integer values ranging from 0 to N. The system's next state is determined by a set of simple transition rules that reflect both the interactions that govern regulation and the conjunctive conditions that enable them. The synchronous application of the transition rules, first to an initial vector and then repeatedly onwards, constitutes a simulation of the system's behavior. The simulation ends when no more changes are observed (in which case the system is said to have reached a “steady state”) or when the network enters an infinite loop of repeated states (in which case it is oscillatory). The series of states that the network assumes during a simulation is called a trajectory.
In the simulations described below we observed that the minimum number of values for an individual state must be 4 (N = 3): When N = 1 (the Boolean model), the system did not reach a steady state, and an infinite loop occurred (SI Fig. 5). When N = 2, no transient behavior for the expression of IME1 and IME2 RNA and proteins was observed (SI Fig. 5). We used N = 9 because this choice was convenient for technical reasons and for the sake of presentation; similar results were observed for several other values of N (e.g., 3 and 15; see SI Fig. 5). The positive and negative edges were assigned weights of +1 and −1, respectively; because the kinetics of meiosis in different strain backgrounds varies, reliable quantitative data for the actual levels of RNA and proteins throughout the meiotic pathway is not comprehensive. Still, for the self-degradation loops, the one on Ime1 had a weight of −1, and the weight of the one on Ime2 was −2 (for reasons to be explained below). The conditional dependence in all cases is positive [in symbols, e.g., cIme1(Ume6, IME2) = +1].
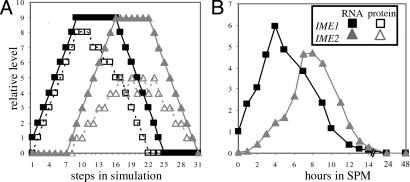
We applied the model to the single initial vector representing the normal conditions under which meiosis is initiated. Under these conditions, IME1 mRNA was in its basal level (i.e., state 1), whereas Ime1 protein and IME2 (mRNA and protein) were absent (i.e., in state 0). Consequently, Ume6, X and Y were not expressed/activated (in state 0), whereas the Ume6/Rpd3 complex was fully active (in state 9). The resulting trajectory showed transient and sequential expression of IME1 and IME2 (mRNA and proteins) (Fig. 2A). This pattern of behavior fits the normal behavior of these genes as reported (8, 17, 28, 29) and as was evident by our quantitative PCR analysis (Fig. 2B).
Fig. 2.
The model faithfully describes the meiotic pattern of expression of IME1 and IME2. (A) Simulation of the network presented in Fig. 1 when N = 9. The initial state of IME1 RNA was 1, that of Ume6/Rpd3 was 9, and the rest were 0. (B) Wild-type diploid cells (Y422) were shifted to meiotic conditions (SPM), and RNA was isolated at the indicated hours. The level of IME1 and IME2 RNA was measured by quantitative PCR. Filled squares, IME1 RNA; open squares, Ime1; filled triangles, IME2 RNA; open triangles, Ime2.
We further applied the model to determine the sensitivity of the meiotic pathway to a growing number of combinations of initial states of IME1 and IME2 (mRNA and proteins) (Table 1). The initial vectors were classified according to the shape of the trajectories to which they give rise (see SI Fig. 6), namely (i) normal transient expression; (ii) restrained transient behavior in which the levels of expression were low but expression was sequential and transient; and (iii) abnormal behavior in which there was either no expression or unordered expression, i.e., IME1 was expressed after IME2. Simulations of most initial vectors led to a normal pattern of expression, even when Ime1 and Ime2 were allowed to assume “nonphysiological conditions,” i.e., approaching state 9 (Table 1). We conclude, therefore, that the transcriptional cascade that governs the initiation of meiosis is robust, i.e., it is insensitive to the initial expression levels of IME1 and IME2.
Table 1.
The meiotic system is insensitive to initial conditions, thus it is robust
| Initial levels of Ime1 and Ime2 | Normal behavior, % | Restrained transient, % | Abnormal behavior, % |
|---|---|---|---|
| 0 | 100.0 | — | — |
| 0, 1 | 100.0 | — | — |
| 0, 1, 2 | 95.1 | 4.9 | — |
| 0, 1, 2, 3 | 93.8 | 3.9 | 2.3 |
| 0, 1, 2, 3, 4 | 90.2 | 1.6 | 8.2 |
| 0, 1, 2, 3, 4, 5 | 85.8 | 2.2 | 12.0 |
| 0, 1, 2, 3, 4, 5, 6 | 84.4 | 1.0 | 14.6 |
| 0, 1, 2, 3, 4, 5, 6, 7 | 82.7 | 2.0 | 15.3 |
| 0, 1, 2, 3, 4, 5, 6, 7, 8 | 81.5 | 5.2 | 13.3 |
| 0, 1, 2, 3, 4, 5, 6, 7, 8, 9 | 81.0 | 6.9 | 12.1 |
The initial vectors were classified according to the shape of their resulting trajectories (SI Fig. 6).
A practical consequence of the findings above was that from here onward, in all of the simulations, it was sufficient to use initial vectors containing only a small number of levels for the four nodes of IME1 and IME2 mRNA and protein, e.g., initial vectors with all combinations of 0, 1, and 2, yielding 81 such vectors.
The Sensitivity of the Network to the Presence of Nutrients.
Ectopic expression of IME1 in exponentially growing cells does not promote the transcription of EMG and entry into meiosis (11, 14, 31). This was reflected in our simulations when the node S, which represents the signal initiating meiosis (i.e., the absence of nitrogen in the presence of a nonfermentable carbon source), was initialized to be in state 0. Under these conditions, IME2 was not expressed, even when Ime1 was present (Table 2, column 2). We further observed that the system is well tuned to prevent meiosis in the presence of nutrients. Deletion of the conditional AND edge going from the node S to the edge from Ime1 to Ume6/Rpd3 resulted, under conditions that favor growth, in a mix of normal and abnormal meiosis (Table 2, column 3). However, deleting the direct edge from S to Ume6/Rpd3 had no effect (Table 2, column 4). Experimentally, the effect of the AND edge is validated by the observation that, under meiotic conditions, phosphorylation on Ime1-Y359 by Rim11 is essential to relieve Ume6/Rpd3 repression and to promote meiosis (14). The effect of the “direct edge” is experimentally revealed by the function of Rim15. Under meiotic conditions, depending on Rim15, Rpd3 and Ume6 dissociate (11). This event is not crucial, because deletion of RIM15 results merely in a reduction in spore formation (11).
Table 2.
Inhibition of meiosis in the presence of nutrients is attributed to the AND edge going from node S to the edge from Ime1 to Ume6/Rpd3
| Pattern of expression | Complete control, % | Deletion of AND edge from S, % | Deletion of edge from S to Ume6/Rpd3, % |
|---|---|---|---|
| No expression | 43.2 | 16.1 | 43.2 |
| Nontransient expression of IME1 (IME2 was not expressed) | 56.8 | 0 | 56.8 |
| Transient | 0 | 56.8 | 0 |
| Restrained transient | 0 | 14.8 | 0 |
| Abnormal | 0 | 12.3 | 0 |
Data is shown for the simulation of the 34 (= 81) initial vectors in the presence of nutrients (i.e., the signal S is in state 0). See SI Fig. 6 for types of behavior.
Stability of Proteins (Self-Loops and Their Weights).
Transient transcription of genes does not necessarily result in transient expression of their proteins (32). Therefore, negative self-loops that reflect the intrinsic stability of the proteins in the network are required. Accordingly, the elimination of the self-loop on Ime1 resulted in a nontransient increase in IME1 and IME2 RNA as well as Ime1 protein, whereas the level of Ime2 protein oscillated (SI Fig. 7A). When the Ime2 self-loop was omitted, two types of trajectories were observed: In 50.6% of the initial vectors, IME1 mRNA and proteins showed transient behavior, whereas IME2 RNA and protein remained high rather than declining to state 0, and in the other 49.4%, an abnormal pattern of expression was observed (SI Fig. 7B). Thus, the simulations suggest that the decline in both IME1 and IME2 (RNA and proteins) requires the negative self-loops on both proteins. Moreover, it suggests that the decline in Ime1 alone is not sufficient for the shutting down of IME1 and IME2 RNA as well as Ime2 protein.
Because the half life of Ime2 is shorter than that of Ime1 (29), the weights on the corresponding self-loops were assigned −2 for Ime2 and −1 for Ime1. When the former weight was changed to −1, for 93.8% of the initial vectors the expression of IME2 was nontransient. The rest showed the same behavior as above, but expression was restrained, thereby validating the network.
The Role of Ime1 in the Negative Autoregulation of IME1 Transcription.
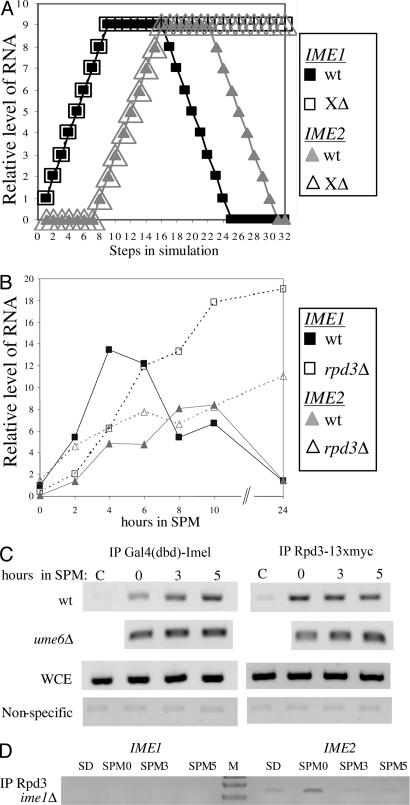
The network reflects the assumption that the decline in the transcription of IME1 is through autoregulation. This assumption is based on the observation that in cells expressing the ime1–3 allele [a nonsense mutation at amino acid 349, resulting in a temperature-sensitive (ts) phenotype] at 25°C meiosis is completed, but the transcription of IME1 is nontransient (8). Because IME1 encodes a transcriptional activator, we assumed that its negative role depended on an additional yet unidentified protein or complex of proteins, namely X. Indeed, deleting node X (and its incident edges) from the network led to a nontransient behavior for the transcription of IME1 and IME2 and to an infinite loop in the expression of their proteins (Fig. 3A and data not shown). We conclude, therefore, that Ime1 is a key player in the negative feedback loop. When the state of X was 1 in the initial vectors (rather than 0), meiosis was not initiated in 28.4% of the initial vectors, mainly when IME1 mRNA started in state 0 (i.e., absent). A restrained transient expression was observed in 14.8% of the initial vectors, and in the rest (56.8%), a normal transient behavior was observed (SI Fig. 8). These results are expected, because premature activation of X should indeed lead to premature shutting down of the system.
Fig. 3.
Rpd3 is recruited by Ime1 to its promoter, serving as a negative regulator. (A) Simulation of the network upon deletion of the outgoing edge from X. The initial state of IME1 RNA was 1, that of Ume6/Rpd3 was 9, and the other nodes were in state 0. (B) Wild-type diploid cells (Y1635-1) and their isogenic rpd3Δ/rpd3Δ cells were shifted to meiotic conditions (SPM), and RNA was isolated at the indicated hours. The level of IME1 and IME2 RNA was measured by quantitative PCR. Filled squares, IME1 wild type; filled triangles, IME2 wild type; open squares, IME1 rpd3Δ (XΔ); open triangles, IME2 rpd3Δ (XΔ). (C and D) Samples for ChIP analysis were taken from cells incubated in SPM for the indicated times or from synthetic dextrose (SD)-grown cells. The PCRs amplified either the IME1 gene (C and D) or the IME2 gene (D). The diploid strains used were Y422 (wild type), Y449 (ime1Δ), and Y1326–1 (ume6Δ), carrying on 2 μ vectors the following chimeric genes: IME2 (YEp1791), RPD3–13xmyc (YEp2546), and pADH1-gal4(dbd)-ime1(id) (YEp2780). Antibodies directed against myc and Gal4(dbd) were used for IP. WCE, whole-cell extract.
Further validation of the network required the identification of X. The network structure and the above analysis suggested two predictions: (i) deletion of X, similar to the deletion of IME1, will lead to nontransient transcription of IME1 and IME2 (Fig. 3A and SI Fig 9), and (ii) X's function will depend on Ime1. We assumed that one component in X might be the histone deacetylase, Rpd3. In accord with our first prediction, in cells deleted for RPD3, the transcription of IME1 was nontransient (Fig. 3B). Moreover, apparently at early meiotic stages, Rpd3 functioned as a positive rather than a negative regulator (Fig. 3B). We suggest that this effect is indirect and mediated by Ime2. This suggestion is based on the observations that Rpd3 represses the transcription of IME2 (12) and that Ime2 represses the transcription of IME1 (8, 17, 18). The prediction that X is activated by Ime1 is validated by the observation that Ime1 recruits Rpd3 to its own promoter, as evident from ChIP assays (Fig. 3 C and D). Moreover, this recruitment is independent of Ume6 (Fig. 3C).
The Role of Ime2 in the Negative Autoregulation of IME2 Transcription.
The network assumes that the decline in the transcription of IME2 is through autoregulation. Because IME2 encodes a kinase, we suggest that its negative role depends on an additional yet unidentified protein or protein complex, designated Y in the network. Deleting Y from the network led to a transient expression of IME1 and a nontransient expression of IME2 (pattern as in SI Fig. 7B). We conclude, therefore, that transient expression of Ime1 is not sufficient for the transient expression of IME2. This result validates the structure of the network and suggests that Ime2 is also a negative regulator of its own transcription. Note that the effect of Ime2 may also be indirect through its effect on the transcription of another gene, for instance one of the middle meiosis-specific genes. Starting the simulation with Y = 1 had no significant effect on the normal behavior of the system. We suggest that Rpd3 is also a component of Y, because its deletion led to a nontransient transcription of IME2 (Fig. 3B).
Modeling the Negative Feedback Loop.
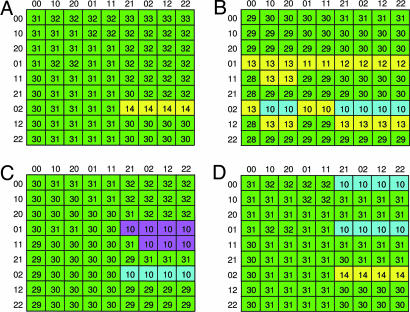
Ime2 plays a pivotal role in shutting down the transcription of IME1: In cells deleted for IME2, the transcription of IME1 is nontransient (SI Fig. 9 and refs. 17 and 18). This finding was verified by simulations of a network in which the edges coming out of Ime2 were deleted (SI Fig. 9). However, a major unsolved issue is the manner in which this is accomplished. Fig. 1 shows a schematic illustration of the null hypothesis network regarding the mode by which Ime2 inhibits the transcription of IME1. Because phosphorylation of Ime1 by Ime2 targets Ime1 to degradation and because Ime1 is a positive autoregulator of its own transcription (30), it is possible that the effect of Ime2 on the transcription of IME1 is only through its effect on the level of Ime1, as suggested in ref. 30. To evaluate the feasibility of this hypothesis, we compared a simulation of the original network with one in which the AND edge from Ime2 to the edge from Ime1 to X was deleted. The number of steps (the length of the trajectory) required to reach a steady-state was used as a primary discriminator, followed by checking the transient behavior displayed by the trajectories themselves. We used Karnaugh-like maps (33) as a powerful visualization tool to demonstrate these modes of behavior (Fig. 4). Our simulations suggest that this hypothesis is less favorable, because it resulted in sensitivity to initial conditions. In some cases, when Ime1 protein was prematurely expressed but IME1 mRNA was not, abnormal, or restrained transient, expression of IME1 and IME2 was observed (compare Fig. 4B to 4A). Furthermore, deletion of the edge from Ime2 to Ime1 led to a nontransient expression of both Ime1 and Ime2. Thus, the effects of Ime2 on the stability and activity of Ime1 are essential for the transient expression of these genes.
Fig. 4.
Karnaugh-like maps. Shown is the functional dependence between initial states and the type of behavior of the trajectory, for four hypothetical networks. (A) Null hypothesis network as illustrated in Fig. 1. (B) Deletion of edge from Ime2 to the edge from Ime1 to X. (C) Addition of negative edge from Ime2 to IME1; Ime2 affects IME1 directly. (D) Addition of a positive edge from Ime2 to X; Ime2 affects X directly. Each cell in the tables represents a specific initial vector. The rows of the tables are indexed by the initial states of IME1 RNA and proteins, respectively, and the columns are indexed by the similar states of IME2. The numbers in the cells state the length of the trajectory of the corresponding initial vector. The cells are marked by different colors representing the following four types of trajectories' behavior (see SI Fig. 6): normal transient expression (green); restrained expression with weak transience (yellow); short trajectories, expression not induced (purple); and IME1 expression is weak but transient, whereas IME2 is not expressed or expressed before Ime1 (turquoise).
It is also possible that Ime2 has an Ime1-independent effect on the transcription of IME1, either directly or through X. In both cases, simulation resulted in normal expression but also abnormal or no expression (Fig. 4 C and D). This analysis supports our initial hypothesis, namely that Ime1's negative autoregulation indeed depends on Ime2 and that Ime2, in turn, does not affect IME1 or X directly (Figs. 1 and 4A).
Discussion
We presented a simple but powerful computational model that can be effectively used to describe the wave-like pattern of expression in networks of genes within a developmental pathway. Our model is able to predict/elucidate missing regulatory elements in a network. This is a general model that can be easily adapted to any developmental or signaling pathway regardless of the availability of detailed quantitative data. We used the developmental pathway of meiosis in budding yeast as a tool to study the ability of our computational model to faithfully describe the system and to promote new insights on its structure. We focused on two main regulators: IME1, which encodes the master transcriptional activator, and IME2, which encodes a regulatory kinase whose transcription depends on Ime1 (10). The network structure, as illustrated in Fig. 1, was based on reported data and on several assumptions regarding unknown interactions between its components.
The feasibility of the model to faithfully describe the network structure was validated by its simulation under normal meiotic conditions, as well as under abnormal conditions or upon deletion of specific edges and nodes. The following results validate our model: (i) Simulation under normal meiotic conditions resulted in a transient and sequential pattern of expression of IME1 and IME2, i.e., the induction in the expression of IME1 occurred before IME2 and its decline depended on Ime2's presence (Fig. 2A), in agreement with experimental results (Fig. 2B and refs. 8, 17, and 28–30). (ii) In the presence of nutrients, i.e., when the state of the signal node was 0, IME2 was not expressed, whereas IME1 was either not expressed or expressed in a nontransient mode (Table 2). These results are in agreement with the reports that ectopic expression of Ime1 in exponentially growing cells does not promote the transcription of early meiosis-specific genes and entry into meiosis (14, 31, 34, 35). Moreover, our simulations suggest that the effect of nutrients (signal node) on shutting down the activity of Ume6/Rpd3 is mainly mediated through Ime1 (Table 2). The nutrient effect responsible to relieve Rpd3 repression is transmitted through two kinases, Rim11 and Rim15. Phosphorylation of Ime1 by Rim11 on Tyr-359 is essential for the relief of Rpd3 repression and initiation of meiosis (14). However, Rim15, which is required to dissociate the Ume6/Rpd3 complex (11), is not essential for meiosis (11, 36). Thus, the structure of the network regarding the effect of the signal node was experimentally validated. (iii) Deleting all edges coming out of Ime1 resulted in a nontransient expression of IME1 and in no expression of IME2 (SI Fig. 9). Deleting all edges coming out of Ime2 resulted in a nontransient expression of both IME1 and IME2 (SI Fig. 9). Furthermore, deleting Y from the network led to a transient expression of IME1 and to a nontransient expression of IME2. Thus, both Ime1 and Ime2 are required to shut down the transcription of IME1, and Ime2 is also required to shut down its own transcription. These results are in agreement with the observations that deletion of either IME1 or IME2 resulted in a nontransient transcription of IME1 and that, in the absence of Ime2, IME2 transcription was also nontransient (SI Fig. 9 and refs. 8, 17 and 18).
Several important and previously unrecognized predictions for the regulation of yeast meiosis emerged from applying this computational model. (i) The meiotic pathway is robust, i.e., it is insensitive to the initial expression levels of IME1 and IME2 (RNA and proteins) (Table 1). This prediction suggests that the system is well tuned to deal with unfavored situations such as premature expression of Ime2. Thus, under meiotic conditions, meiosis will be executed even when the induction in the transcription of IME1 and IME2 is abnormal. This conclusion is partially revealed by the observation that cells expressing either IME1 or IME2 from the GAL1 promoter enter and complete meiosis (8, 37). Moreover, in cells carrying one, two, three, and five copies of IME1 the initial relative level of IME1 mRNA was 1.0, 3.5, 6.6, and 15.0, respectively, whereas the percentage of asci at 48 h was 80, 72, 80, and 76%, respectively. The sensitivity of meiosis to the initial levels of Ime2 can be similarly tested by modulating the copy number of IME2 ectopically expressed from the IME1 promoter. (ii) The simulation predicts that the intrinsic stability of Ime1 is required to accomplish transient expression of both IME1 and IME2. This is a notable prediction, because overexpression of IME1 is deleterious, resulting in an increase in nondisjunction (8). Currently, an IME1 allele that results in a stable Ime1 is not available. (iii) The simulation predicts that the intrinsic stability of Ime2 is required to shut down its own transcription. We conclude, therefore, that the decline in Ime1 alone is not sufficient for shutting down the transcription of IME2, and thus the transient expression of IME2 is due to active repression rather than lack of activation. (iv) Previously it was assumed that the decline in the transcription of both IME1 and IME2 is a direct result of the Ime2-dependent degradation of Ime1 (29), which is required to relieve repression in the transcription of IME1 (38) and EMG (11, 15). Our simulation predicts that this decline alone is not sufficient, because the deletion of X resulted in a nontransient transcription of IME1. Ime1 may function as both a positive and a negative regulator of its own transcription, and accordingly, in the computational model, X could represent a time delay between the positive and negative roles of Ime1. However, it is also possible that X represents a protein or protein complex that shuts down the transcription of IME1, depending on Ime1. The discrimination between these two hypotheses required the identification of X. We assumed that X might be Rpd3, and we examined, therefore, its effect on the transcription of IME1. We show that the decline in the transcription of IME1 depends on Rpd3 (Fig. 3B). We further show that activation of X (Rpd3) is due to the fact that it is recruited by Ime1 to the IME1 promoter (Fig. 3 C and D). Thus, our computational prediction is verified experimentally. (v) The network structure (Fig. 1) assumes that activation of X by Ime1 depends on Ime2. However, the only experimental data supporting this hypothesis are the observations that Ime2 phosphorylates Ime1 (19, 29) and that, at early meiotic times, Ime1 relieves repression by Rpd3 rather than promoting it (11). We used our model to examine this assumption. Simulation of a network deleted for this dependency resulted in a less robust behavior, namely sensitivity of the network to the initial levels of Ime1 and Ime2 (compare Fig. 4A with Fig. 4B). We suggest, therefore, that either the association between Ime1 and Rpd3 depends on Ime2 or that the ability of Ime1 to overcome Rpd3 repression is inhibited by Ime2. We further suggest that phosphorylation of either Ime1 and/or Rpd3 (itself or a different protein in the complex) by Ime2 mediates this effect. (vi) As described above, Ime2 is required to shut down the transcription of IME1 (SI Fig. 9 and refs. 17 and 18). It is not known whether, in addition to the above-described effect of Ime2, it is also directly required to activate X or to shut down IME1's transcription. We used our model to examine the feasibility of these options, employing Karnaugh-like maps as a tool to discriminate between these alternative possibilities. The robust nature of our favored network (Figs. 1 and 4A) in comparison with the putative networks (Figs. 4 C and D) suggests that Ime2 is required only for the degradation of Ime1 and for its ability to recruit the repression activity of X. (vii) Two isomorphic modules, Ime1-X-IME1 and Ime2-Y-IME2 (Fig. 1) were used to describe the decline in the transcription of IME1 and IME2, respectively. It is tempting to speculate that such modules may be part of any transcriptional cascade. (viii) Finally, experimental results suggests that Rpd3 is present in both X and Y complexes. It is tempting to speculate that X and Y might correspond to the Rpd3 S and Rpd3 L complexes, because only the latter includes Ume6 (ref. 39 and Fig. 3C).
In summary, the presented model is useful for two main reasons: (i) It is simple and easily adapted to any pathway, and (ii) as detailed above, it has strong predictive capabilities that point research toward directions unseen by looking only at the experimental data. Finally, in this report, the model was successfully used to elucidate unknown interactions between Ime1 and Ime2, the two main regulators of meiosis in budding yeast. The model is embodied in code with an appropriate application program interface (API), which is available on request.
Methods
The Computational Model.
To allow the simulation of networks that display transient behavior, we extended the basic model used by Li et al. (27) as described in the sections that follow.
Nodes' states.
A node's state can be any integer number between 0 and N (>1). For example, if N = 9, we allow the nodes to be in states 0–9 throughout the execution.
The transition function.
If aij is the weight of the regulation edge from node j to node i, and sj(t) is the state of node j in time step t, let us denote the scalar product
where j ranges over all nodes that have edges to i as ki(t). The transition function is as follows:
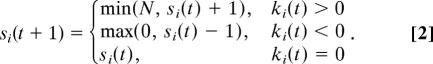 |
Gating transitions by a conjunctive condition (AND).
New types of edges, going from a node to a weighted edge, were used to reflect the observation that sometimes the activity of a regulator is enabled only if some other element in the system is active or is inactive. By ck(i, j), we denote the dependency of an edge from node i to node j on the state of node k. We allow two possibilities: If the effect of i on j requires that node k is in state >0 (active), then ck(i, j) = +1 (positive dependence). If this effect requires that node k is in state 0 (inactive), then ck(i, j) = −1 (negative dependence).
The transition function is applied synchronously during discrete time steps t = 0, 1, 2, …, starting from the initial (t = 0) state to produce a simulation. Our extensions do not increase the complexity of computing a simulation: One transition step for the whole network can be computed in time that is linear in its size, including the test for whether we have reached a steady state.
As noted in Results, the set of interesting and biologically meaningful initial states is often limited to a subspace of the (N + 1)n possible initial states (where n is the number of nodes in the network). However, when trying to observe the trajectories that lead from initial states to final ones, the number of intermediate states does increase considerably [because now all (N + 1)n states are plausible]; the number of final states is also potentially very large. To analyze the large variety of outcomes and discriminate among them, we look at specific properties of the simulations, such as the lengths of the trajectories and the signature of the trajectories (e.g., rising followed by falling) resulting from specific initial states. Moreover, to identify the pattern of the behaviors and establish a functional relation between the initial states and the outcomes, we used Karnaugh-like maps to cluster these behaviors in a systematic fashion.
Strains, Media, and Procedures.
Strains used were as follows: Y1635-1 (MATa/MATα and rpd3Δ::HIS3/RPD3) and its isogenic rpd3Δ::HIS3 rpd3Δ::HIS3 (Y1635-2) and Y1326-1 (ume6Δ/ume6Δ) derivatives; and Y422 (MATa/MATα) and its isogenic Y449 (ime1Δ::hisG/ime1Δ::hisG) derivative. Detailed information on genotypes is available on request. Meiosis was initiated by shifting acetate-grown logarithmic cells to sporulation media as described in ref. 40. RNA was extracted by using hot acidic phenol. cDNA quantitative PCR synthesis was performed according to the manufacturer's instructions (ABGene, Surrey, U.K.). ChIP analysis was done as reported in ref. 11.
Supplementary Material
Acknowledgments
We thank Esti Yeger-Lotem and Yael Mandel-Gutfreund for helpful discussions and for critical reading of the manuscript. This work was supported by a grant from the Center for Complexity Science, Israel (to Y.K. and R.Y.P.) and a Sherman Interdisciplinary Graduate School Fellowship (to A.R.).
Abbreviation
- EMG
early meiosis-specific genes.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611168104/DC1.
References
- 1.Yu H, Gerstein M. Proc Natl Acad Sci USA. 2006 [Google Scholar]
- 2.Bolouri H, Davidson EH. Dev Biol. 2002;246:2–13. doi: 10.1006/dbio.2002.0617. [DOI] [PubMed] [Google Scholar]
- 3.Freeman M. Nature. 2000;408:313–319. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- 4.Ferrell JE., Jr Curr Opin Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 5.Bowtell DD, Simon MA, Rubin GM. Cell. 1989;56:931–936. doi: 10.1016/0092-8674(89)90626-0. [DOI] [PubMed] [Google Scholar]
- 6.Bouhon IA, Kato H, Chandran S, Allen ND. Brain Res Bull. 2005;68:62–75. doi: 10.1016/j.brainresbull.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Marine JC, Topham DJ, McKay C, Wang D, Parganas E, Stravopodis D, Yoshimura A, Ihle JN. Cell. 1999;98:609–616. doi: 10.1016/s0092-8674(00)80048-3. [DOI] [PubMed] [Google Scholar]
- 8.Shefer-Vaida M, Sherman A, Ashkenazi T, Robzyk K, Kassir Y. Dev Genet. 1995;16:219–228. doi: 10.1002/dvg.1020160302. [DOI] [PubMed] [Google Scholar]
- 9.Chu S, Herskowitz I. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- 10.Kassir Y, Adir N, Boger-Nadja E, Guttmann-Raviv N, Rubin-Bejerano I, Sagee S, Shenhar G. Int J Cytol Surv Cell Biol. 2003;224:111–171. doi: 10.1016/s0074-7696(05)24004-4. [DOI] [PubMed] [Google Scholar]
- 11.Pnueli L, Edry I, Cohen M, Kassir Y. Mol Cell Biol. 2004;24:5197–5208. doi: 10.1128/MCB.24.12.5197-5208.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadosh D, Struhl K. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 13.Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 14.Rubin-Bejerano I, Sagee S, Friedman O, Pnueli L, Kassir Y. Mol Cell Biol. 2004;24:6967–6979. doi: 10.1128/MCB.24.16.6967-6979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Washburn BK, Esposito RE. Mol Cell Biol. 2001;21:2057–2069. doi: 10.1128/MCB.21.6.2057-2069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sagee S, Sherman A, Shenhar G, Robzyk K, Ben-Doy N, Simchen G, Kassir Y. Mol Cell Biol. 1998;18:1985–1995. doi: 10.1128/mcb.18.4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith HE, Mitchell AP. Mol Cell Biol. 1989;9:2142–2152. doi: 10.1128/mcb.9.5.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida M, Kawaguchi H, Sakata Y, Kominami K, Hirano M, Shima H, Akada R, Yamashita I. Mol Gen Genet. 1990;221:176–186. doi: 10.1007/BF00261718. [DOI] [PubMed] [Google Scholar]
- 19.Guttmann-Raviv N. Haifa: Technion–Israel Institute of Tecnology; 2001. PhD thesis. [Google Scholar]
- 20.Barkai N, Leibler S. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 21.Schilling CH, Edwards JS, Palsson BO. Biotechnol Prog. 1999;15:288–295. doi: 10.1021/bp9900357. [DOI] [PubMed] [Google Scholar]
- 22.Smolen P, Baxter DA, Byrne JH. Neuron. 2000;26:567–580. doi: 10.1016/s0896-6273(00)81194-0. [DOI] [PubMed] [Google Scholar]
- 23.Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles ED. Nature. 2002;420:190–193. doi: 10.1038/nature01166. [DOI] [PubMed] [Google Scholar]
- 24.Bolouri H, Davidson EH. BioEssays. 2002;24:1118–1129. doi: 10.1002/bies.10189. [DOI] [PubMed] [Google Scholar]
- 25.Bernot G, Comet JP, Richard A, Guespin J. J Theor Biol. 2004;229:339–347. doi: 10.1016/j.jtbi.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Batt G, Ropers D, de Jong H, Geiselmann J, Mateescu R, Page M, Schneider D. Bioinformatics. 21(Suppl 1):i19–i28. doi: 10.1093/bioinformatics/bti1048. [DOI] [PubMed] [Google Scholar]
- 27.Li F, Long T, Lu Y, Ouyang Q, Tang C. Proc Natl Acad Sci USA. 2004;101:4781–4786. doi: 10.1073/pnas.0305937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kassir Y, Granot D, Simchen G. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- 29.Guttmann-Raviv N, Kassir Y. Mol Cell Biol. 2002;22:2047–2056. doi: 10.1128/MCB.22.7.2047-2056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Genes Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman A, Shefer M, Sagee S, Kassir Y. Mol Gen Genet. 1993;237:375–384. doi: 10.1007/BF00279441. [DOI] [PubMed] [Google Scholar]
- 32.Foiani M, Liberi G, Lucchini G, Plevani P. Mol Cell Biol. 1995;15:883–891. doi: 10.1128/mcb.15.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohavi Z. Switching and Finite Automata Theory. New York: McGraw–Hill; 1978. [Google Scholar]
- 34.Colomina N, Gari E, Gallego C, Herrero E, Aldea M. EMBO J. 1999;18:320–329. doi: 10.1093/emboj/18.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidan S, Mitchell AP. Mol Cell Biol. 1997;17:2688–2697. doi: 10.1128/mcb.17.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell AP, Driscoll SE, Smith HE. Mol Cell Biol. 1990;10:2104–2110. doi: 10.1128/mcb.10.5.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shenhar G, Kassir Y. Mol Cell Biol. 2001;21:1603–1612. doi: 10.1128/MCB.21.5.1603-1612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrozza MJ, Florens L, Swanson SK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Biochim Biophys Acta. 2005;1731:77–87. doi: 10.1016/j.bbaexp.2005.09.005. discussion 75–76. [DOI] [PubMed] [Google Scholar]
- 40.Kassir Y, Simchen G. Methods Enzymol. 1991;194:94–110. doi: 10.1016/0076-6879(91)94009-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.