Abstract
Advances in tandem mass spectrometry (MS/MS) steadily increase the rate of generation of MS/MS spectra. As a result, the existing approaches that compare spectra against databases are already facing a bottleneck, particularly when interpreting spectra of modified peptides. Here we explore a concept that allows one to perform an MS/MS database search without ever comparing a spectrum against a database. We propose to take advantage of spectral pairs, which are pairs of spectra obtained from overlapping (often nontryptic) peptides or from unmodified and modified versions of the same peptide. Having a spectrum of a modified peptide paired with a spectrum of an unmodified peptide allows one to separate the prefix and suffix ladders, to greatly reduce the number of noise peaks, and to generate a small number of peptide reconstructions that are likely to contain the correct one. The MS/MS database search is thus reduced to extremely fast pattern-matching (rather than time-consuming matching of spectra against databases). In addition to speed, our approach provides a unique paradigm for identifying posttranslational modifications by means of spectral networks analysis.
Keywords: alignment, database searching, posttranslational modifications, tandem mass spectrometry, de novo
Most protein identifications today are performed by matching spectra against databases, using programs like SEQUEST (1) or Mascot (2). Although these tools are invaluable, they are already too slow for matching large tandem mass spectrometry (MS/MS) data sets against large protein databases, particularly when one performs a time-consuming search for posttranslational modifications (PTMs). We argue that new solutions are needed to deal with the stream of data produced by shotgun proteomics projects. Craig and Beavis (3) and Tanner et al. (4) recently developed the X!Tandem and InsPecT algorithms to prune (X!Tandem) and filter (InsPecT) the sequence databases and thus speed up the search. However, these tools still have to compare every spectrum against a (smaller) database.
Here we explore a concept that allows one to perform an MS/MS database search without ever comparing a spectrum against a database. We propose to take advantage of spectral pairs, which are pairs of spectra obtained from overlapping (often nontryptic) peptides or from unmodified and modified versions of the same peptide. Most current protocols try to minimize the number of spectral pairs, because nontryptic and chemically modified peptides further complicate the spectral interpretations and lead to higher running times. MacCoss et al. (5) were the first to realize the potential of overlapping peptides for the identification of modified proteins and have recently demonstrated the increased throughput of modified digestion schemes (6). Also, even samples digested with trypsin typically have many peptides that differ from each other by a deletion of terminal amino acids (semitryptic peptides). In addition, the existing experimental protocols already unintentionally generate many chemical modifications, and it has been shown that existing MS/MS data sets often contain modified versions for many peptides (7, 8).
Although seemingly redundant, spectral pairs open up previously unexplored computational avenues. Having a pair of spectra (one of a modified and another of an unmodified peptide) allows one to (i) separate the b (prefix) and y (suffix) ion mass ladders, (ii) greatly reduce the number of noise peaks, and (iii) propagate the identification of modifications from spectrum to spectrum, thereby detecting unanticipated and multiple modifications. Thus, spectral pairs allow one to generate a small number of peptide reconstructions that are very likely to contain the correct one. Instead of generating covering sets of short 3–4-aa tags (4, 9), this approach generates a covering set of peptides 7–9 aa long. This set typically has a single perfect hit in the database that can be instantly found by hashing and thus eliminates the need to ever compare a spectrum against the database.‡ Other approaches (10–13) that compare de novo peptide sequences against a database of protein sequences obtain their query sequences from individual MS/MS spectra (instead of from spectral pairs) and thus suffer from relatively low accuracy of de novo peptide sequencing (14–16). In addition to improvements in de novo peptide sequencing, spectra denoising and propagation of modifications also improve the standard MS/MS database search.
Let S(P) and S(P*) be spectra of an unmodified peptide P and of its modified version P* (spectral pair). The crux of our computational idea is a simple observation that a “database” consisting of a single peptide P is everything one needs to interpret the spectrum S(P*).§ Thus, if one knows P there is no need to scan S(P*) over the database of all proteins. Of course, in reality, one does not know P, and only S(P) is readily available. Below we show that a spectrum, S(P), is almost as useful as the peptide P for interpreting S(P*) and can thus eliminate the need for database search. This observation opens the possibility of substituting an MS/MS database search with finding spectral pairs and further interpreting the peptides that produced them. We show that these problems can be solved by using a unique combination of de novo and spectral alignment techniques (7, 17) to transform any given spectral pair (S1, S2) into virtual spectra S1,2 and S2,1 of extremely high quality, with nearly perfect b- and y-ion separation and the number of noisy peaks reduced 12-fold.
In addition to fast peptide identification, our approach also provides a unique paradigm for the identification of chemical and posttranslational modifications without any use of a database. Recently, it was argued (7, 18, 19) that the phenomenon of modifications is much more widespread than previously thought, and blind database search was advocated for identification of these modifications. In particular, blind database search recently resulted in the most comprehensive set of PTMs identified in aged human lenses (8). The surprising conclusion of our approach is that we can discover almost all modifications in cataractous lenses (previously identified by blind database search) and even detect some PTMs missed in ref. 8.
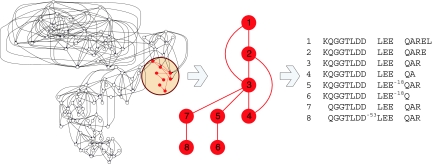
We further combine spectral pairs into a spectral network in which each vertex corresponds to a spectrum and each edge to a spectral pair. Fig. 1 shows a spectral network of 945 MS/MS spectra [corresponding to different peptides from a nuclear factor κB kinase β subunit (IKKβ) protein sample], illustrating the key advantage of spectral networks over the traditional MS/MS database search. Traditional approaches to peptide identification consider each of these spectra separately without attempting to correlate different spectra from related peptides. As a result, the important insights that can be derived from the structure of the spectral network are lost. Our approach consolidates all of these spectra into 117 clusters (vertices of the network) and reveals many spectral pairs (edges of the network). This results in the analysis of all spectra at once and thus increases the confidence of peptide identifications, reinforces predictions of modifications by using correlated spectra, and eliminates the need to “guess” modifications in advance. Moreover, the spectral network even allows one to assemble these spectra into an intact 34-aa segment of the IKKβ protein, thus opening the door for shotgun protein sequencing (20).
Fig. 1.
Spectral network constructed by aligning spectra from overlapping peptides. (Left) Spectral network for 945 spectra representing different peptides from the fragment IVDLQRSPMGRKQGGTLDDLEEQARELYRRLREK of the human IKKβ protein. The spectral network is constructed without any knowledge of the peptide annotations. Each of 117 vertices in the spectral network corresponds either to a single MS/MS spectrum or to a consensus spectrum of multiple MS/MS spectra from the same peptide (derived by clustering). Two vertices are connected by an edge whenever the corresponding spectra form a spectral pair. (Center) A subnetwork of the entire spectral network spanning the fragment KQGGTLDDLEEQAREL (shown by red vertices Left). (Right) Paired peptides found by analyzing the Center spectral subnetwork with our paired spectra detection procedure.
Results
Interpretation of Spectral Pairs/Stars.
The set of all spectra pairing with a spectrum S in the spectral network is called a spectral star. For example, the spectral star for the spectrum derived from peptide 3 in Fig. 1 consists of multiple spectra from five different peptides. The high quality of the virtual star spectra derived from spectral pairs and spectral stars makes de novo interpretation of these spectra straightforward [see supporting information (SI) Fig. 4 and SI Table 2]. Because star spectra feature excellent separation of b- and y-ion ladders and only a small number of noise peaks, de novo reconstructions of these spectra produce reliable (gapped) sequences that usually contain long correct tags.¶ On average, de novo reconstructions of our star spectra correctly identify 72% of all possible “cuts” in a peptide [i.e., on average, 0.72 × (n − 1) b ions (or y ions) in a peptide of length n are identified]. This is a very high number, inasmuch as the first (e.g., b1) and last (e.g., bn−1) b ions are rarely present in the MS/MS spectra, which makes it nearly impossible to explain >80% of all cuts in the IKKβ sample. Moreover, on average, unexplained peaks account for only 5% of the total score of the de novo reconstruction.
Benchmarking in MS is inherently difficult because of a shortage of manually validated large MS/MS samples that represent “gold standards.” Although the ISB data set (21) represents such a gold standard for unmodified peptides, large validated samples of spectra from modified peptides are not currently available. As a compromise, we benchmarked our algorithm by using a set of 11,760 spectra from the IKKβ data set that were annotated using InsPecT and extensively studied in recent publications (4, 7), including comparisons with SEQUEST, Mascot, and X!Tandem. Our entire spectral networks analysis (starting from clustering and ending with interpretations) of this IKKβ data set took 9 min on a regular desktop computer (Intel Pentium 4; 2.8-GHz clock speed). We compared our performance to that obtained with InsPecT, which was previously shown to be 2 orders of magnitude faster than SEQUEST for restricted database search (4). Even when searching against a moderately sized database, such as Swiss-Prot's set of 13,749 human proteins, InsPecT's running time was 55 min (complete running-time results are given in SI Appendix A). Thus, our spectral networks approach (which finds both unmodified and modified peptides) is six times faster than InsPecT (in the mode that searches for unmodified peptides only). Below we give identification results for both spectral pairs and spectral stars.
InsPecT identified 515 unmodified peptides in the IKKβ sample, 413 of which have some other prefix/suffix or modified variant in the sample and are thus amenable to pairing. We were able to find spectral pairs for 386 of these 413 peptides. Moreover, 339 of these 386 peptides had spectral pairs coming from two (or more) different peptides, i.e., pairs (S1, S2) and (S1, S3) such that spectra S2 and S3 come from different peptides.
The average number of (gapped) de novo reconstructions (explaining at least 85% of the optimal score) for star spectra was 10.4. Although star spectra generate a small number of gapped reconstructions, these gapped sequences are not well suited for fast membership queries in the database. We therefore transform every gapped de novo reconstruction into an ungapped reconstruction by substituting every gap with all possible combinations of amino acids. On average, this approach results in 165 sequences of length 9.5 aa per spectrum; for 86% of all peptides, one of these tags is correct.
Although checking the membership queries for 165 sequences can be done very quickly with database indexing (at most, one of these sequences is expected to be present in the database), there is no particular advantage in using such superlong tags (9.5 aa on average) for standard database search: a tag of length 6–7 aa will also typically have a unique hit in the database. However, the long 9–10-aa tags have distinct advantages in difficult nonstandard database searches, e.g., discovery of new alternatively spliced variants via MS/MS analysis. Moreover, for standard search, one can generate a smaller set of shorter (6–7-aa) tags based on the original gapped reconstruction and use them for membership queries. We used the obtained gapped reconstruction to generate such short 6-mer tags. On average, each consensus spectrum generates ≈50 6-mer tags. It turned out that 82% of spectra derived from spectral stars contain at least one correct 6-mer tag.
Using Spectral Networks for PTM Identification.
Our approach allows one to detect modifications without any reference to a database. The difference in parent masses within a spectral pair corresponds to either a modification offset or a sum of amino acid masses. Although not every difference in parent mass corresponds to a modification offset (some spectral pairs may be artifacts), a histogram of parent mass differences (Fig. 2a) reveals the modifications present in the IKKβ sample. Indeed, seven of the eight most frequent parent mass differences in Fig. 2a are listed among the eight most common modifications in the IKKβ data set (7). We emphasize that Fig. 2a was obtained without any reference to a database, whereas Tsur et al. (7) found these modifications by database search. The only frequent modification identified by Tsur et al. (7) and not represented in Fig. 2a is deamidation with a small mass offset of 1 Da that is difficult to distinguish from parent mass errors and isotopic peaks artifacts. Interestingly, our approach reveals an offset of +34 (present in thousands of spectral pairs) that was not reported in ref. 7.
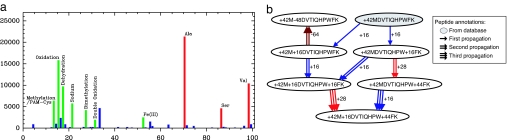
Fig. 2.
Discovery of modifications by using spectral networks. (a) Histogram of absolute parent mass differences for all detected spectral pairs on the IKKβ data set; the y axis represents the number of spectral pairs with a given difference in parent mass. For clarity, we only show the mass range 1–100 Da. The peaks at masses 71, 87, and 99 Da correspond to amino acid masses, and the peaks at masses 14, 16, 18, 22, 28, 32, and 53 Da correspond to known modifications that were also found by Tsur et al. (7) using blind database search. The peak at mass 34 Da corresponds to a putative modification that remains unexplained to date. (b) Modification network for peptide MDVTIQHPWFK from the Lens data set. The shaded node was annotated as peptide + 42MDVTIQHPWFK by database search of the tag VTIQHP; the remaining nodes were annotated by iterative propagation. On each propagation, the source peptide annotation is combined with the modification determined by the spectral product to yield a new peptide annotation (different modifications are shown as edges with different colors).
Additionally, spectral networks can make a contribution for the detection of rare modifications. These modifications usually occur on only a very small number of peptides and are thus unlikely to be detected by the PTM frequency matrix approach from ref. 7. Furthermore, these modifications can co-occur with other more frequent modifications and thus completely escape identification. We addressed these cases by focusing on modification networks, which are subnetworks of the spectral network connecting multiple modification states of the same peptide.
We illustrate our modification networks approach to PTM identification by using the Lens data set. Lens proteins, because of a very low turnover, tend to accumulate many PTMs over time and often result in increased opaqueness and cataracts (7, 13). Of all 11,932 spectra, 2,001 were found to be paired, resulting in the identification of 280 unmodified peptides (88% of all unmodified peptides that have some pair in the data set).
Although at a first glance the number of annotations (280) may seem small when compared with the number of paired spectra (2,001), it should be noted that many of these paired spectra come from modified peptides and thus may not generate sufficiently long tags to match the correct peptide in the database. However, most spectra from modified peptides were correctly paired with their unmodified counterparts and were thus already linked to the correct peptide. Additionally, as illustrated in Fig. 3e, the spectral alignment between any two spectra promptly provides both the location and mass of the modification. Thus, suppose that an identified spectrum S was annotated with a peptide p1 … pn and paired with a nonannotated spectrum S′. Using our spectral alignment approach, we can determine on which amino acid pi the modification occurred and readily annotate S′ with p1 … pi−1pi* pi+1 … pn, where pi* stands for a modification of pi. This operation is defined as the propagation of a peptide annotation by means of spectral pairs. To use propagation on any given spectral network, we need to consider two additional conditions: (i) some nonannotated spectra may not be directly connected to an annotated spectrum (e.g., spectra with two modifications), and (ii) some nonannotated spectra may be connected to multiple annotated spectra (e.g., different prefix/suffix variants). We therefore use an iterative procedure that, at each step, propagates peptide annotations from every annotated spectrum onto all its nonannotated neighbors. If a nonannotated spectrum happens to gain more than one putative annotation, then we simply choose that which best explains the spectrum. The neighbors are then marked as annotated and are allowed to propagate their annotations on the next iteration. For example, the propagation procedure starts from 58 (of 117) annotations of unmodified peptides in the spectral network shown in Fig. 1, adds 53 annotations with a single modification on the next iteration, and finally adds 6 annotations with two modifications on the final iteration. Fig. 2 illustrates this iterative propagation on the Lens data set with the modification network for peptide MDVTIQHPWFK. We remark that the existing peptide identification tools have difficulties in identifying and validating peptides with multiple modifications. Modification networks open up the possibility of reliably identifying such heavily modified peptides (which may be common in heavily modified proteins involved in cell signaling like the IKK complex) via cross-validation with other modified peptides as exemplified in Fig. 2.
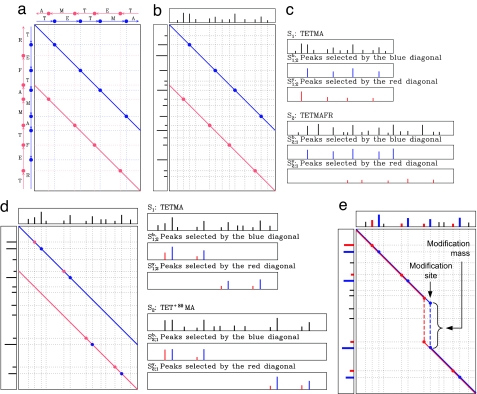
Fig. 3.
Spectral products for terminal and internal modifications. (a) Spectral product for the theoretical spectra of the peptides TETMA and TETMAFR (all points at the intersections between the vertical and horizontal lines). The blue circles correspond to matching b ions in the two spectra; the red circles correspond to matching y ions. The blue and red circles are located on the blue and red diagonals. (b) Spectral product for uninterpreted spectra of the peptides TETMA and TETMAFR. The two diagonals in the spectral product matrix still reveal the points at which peaks from the spectrum at the top match peaks from the spectrum on the left. (c) Spectra S1,2b and S1,2y defined by the blue and red diagonals. (d) Spectral product for uninterpreted spectra with one internal modification. The top spectrum corresponds to an unmodified peptide, and the left-side spectrum corresponds to a modified peptide. In these cases it is not appropriate to construct Si,jb/Si,jy by simply selecting peaks on the diagonals. (e) The algorithm described in the text allows for modifications to occur in the middle of the peptide and separates the overlapping series of b and y ions (blue and red diagonals, respectively). The peaks selected from each spectrum by the blue/red diagonals are shown in the corresponding color.
Overall, the spectral networks analysis of the Lens data set found all but one of the modification types previously identified by blind database search and provided evidence for six previously undetected modifications types (see Table 1). The only modification listed by Tsur et al. (7) and not rediscovered here was again deamidation on N,Q, for the same reasons described above for the IKKβ data set.
Table 1.
Putative modifications identified by spectral networks on the Lens data set
| Location | Modification mass, Da | Type | Putative annotation | Comment | Reference |
|---|---|---|---|---|---|
| M | −48 | Neutral loss | Loss of methane sulfenic acid | Reported on the same site | 24 |
| W | 4 | PTM | Kynurenine | Reported in cataractous lenses | 22 |
| S | 30/73 | Unknown | Unknown | ||
| W | 32 | PTM | Formylkynurenine | Reported in cataractous lenses | 23 |
| N terminus | 57 | Unknown | Carboxyamidomethylation | Possible chemical artifact | 26 |
| N terminus | 229/271 | Unknown | Unknown |
All of these modifications occur on peptides that were previously identified in this sample. However, most of these modifications are rare in that they occur only on specific sites and thus tend to have low spectral counts. Two of these modifications can be explained as artifacts, two are known to occur in the context of lens, and two remain unexplained to date (see Discussion). Spectral networks and annotated MS/MS spectra figures supporting these modifications can be found in SI Appendix B. Our approach identified all previously found modifications except deamidation on N,Q (7).
Two of the six putative modifications were recently identified in cataractous lenses by other groups (22, 23), thus reinforcing our predictions. One more modification was previously reported as a loss of methane sulfenic acid on the same site (24). The discovered N-terminal modification with an offset of 57 Da is potentially interesting: it occurs only on two semitryptic peptides whose nontryptic ends were previously reported as degraded N terminii of βB1-crystallins (25), thus also reinforcing our predictions. Moreover, given that all protein N terminii are expected to be acetylated (as has generally been observed), this could hypothetically correspond to a previously undetected in vivo modification of the degraded N terminii. Note that this 57-Da offset would normally be attributed to a common experimental artifact caused by the cysteine alkylation (26). However, the fact that this 57 Da is not observed on any other peptides, and the lack of corroborating peptide fragmentation evidence (i.e., characteristic loss of 57 Da from precursor mass), suggest that this modification is a localized event that could warrant further investigation. As an additional confirmation step, we modified the traditional database search parameters to consider all our discovered putative modifications and observed a complete agreement with our proposed annotations with large Xcorr and ΔCn scores.
It should be noted that all of these putative modification types occur on peptides that had been identified previously in this data set (7, 8). However, most of these modifications are rare in that they occur only at specific sites and thus tend to have low spectral counts, which is the major reason why they are hard to detect through blind database search. By independently comparing each MS/MS spectrum against a database, blind database search generates many false-positives that are usually filtered by requiring a minimum number of occurrences of each modification. Although successful in detecting multiple-site modifications, this approach leads to difficulties in the detection of single-site and less-common modifications.
The spectral networks approach remedies this limitation of blind database search by being more selective in the assignment of modified peptide annotations. Spectral pairs provide additional evidence that two spectra were derived from the same peptide (in the form of correlated ion peaks and intensities) and thus add significance to otherwise difficult spectrum identifications. As illustrated in Fig. 2, this increased sensitivity is particularly evidenced on modification networks by the grouping of multiple spectra from different modification states of the same peptide. SI Appendix B contains supporting evidence for all the modifications listed in Table 1, in the form of modification networks and annotated MS/MS spectra.
Discussion
We have demonstrated the utility of spectral networks for the identification of proteins and modifications. The key idea of our approach is that correlations between MS/MS spectra of modified and unmodified peptides allow one to greatly reduce noise in individual MS/MS spectra, thus making de novo interpretations so reliable that they can substitute for the time-consuming matching of spectra against databases. We have also shown how the correlated spectral content on modification networks can provide consistent evidence to support the identification of rare modifications and highly modified peptides. Our spectral networks software is freely available from www-cse.ucsd.edu/groups/bioinformatics/software.html. A current limitation of our approach is its restricted applicability to spectra with parent charges 1 and 2; two further algorithmic developments are necessary to allow for the integration of spectra with higher parent charges into spectral networks. First, although spectral alignment works for two spectra of precursor charge 3 (or higher), it generally does not work for comparison of a spectrum of precursor charge 1 or 2 with a spectrum of precursor charge 3. The main reason is that spectra of higher precursor charge tend to generate b and y ions of higher charge that do not align to the singly charged variants predominant in spectra of precursor charge 1 or 2. Second, even if two spectra with parent mass 3 (or higher) are aligned, reliable de novo algorithms for interpreting multicharged spectra are still unknown.
Tandem mass spectra are inherently noisy, and mass spectrometrists have long been trying to reduce the noise and achieve reliable de novo interpretations by advancing both instrumentation and experimental protocols. In particular, Zubarev and colleagues (27) recently demonstrated the power of using both collision-induced dissociation and electron capture dissociation spectra. We emphasize that, in contrast to our approach, this technique, as well as the recent approach described in Frank et al. (28), require special instrumentation or highly accurate Fourier transform MS. Another approach for reducing the complexity of spectra involves stable isotope labeling (29). However, the impact of this approach (for peptide identification) has been restricted, in part by the cost of the isotope and the high mass resolution required. Alternative end-labeling chemical modification approaches have disadvantages such as low yield, complicated reaction conditions, and unpredictable changes in ionization and fragmentation. As a result, the impact of these important techniques is mainly in protein quantification rather than identification (29). The key difference between our approach and labeling techniques is that, instead of trying to introduce a specific modification in a controlled fashion, we take advantage of multiple modifications naturally present in the sample. Our spectral networks approach allows one to decode these modifications (without knowing in advance what they are) and thus provides a computational (rather than instrumentation-based) solution to the problem of MS/MS spectra identification.
Materials and Methods
Data Sets.
We describe our algorithm by using MS/MS spectra from human IKKβ and lens proteins, two particularly challenging samples for PTM analysis. The IKKβ data set consists of 45,500 spectra acquired from a digestion of the inhibitor of IKKβ protein by multiple proteases, thereby producing overlapping peptides. [Spectra were acquired on a ThermoFinnigan (San Jose, CA) LTQ mass spectrometer.] The IKKβ complex represents an ideal test case for algorithms that search for PTM peptides. Until recently, phosphorylations were the only known PTMs in IKK, which is insufficient to explain all mechanisms of signaling and activation/inactivation of IKK by >200 different stimuli. Revealing the combinatorial code responsible for PTM-controlled signaling in IKK remains an open problem. Previous analyses of this IKKβ data set resulted in 11,760 identified spectra and 1,154 annotated peptides (4, 7). This IKKβ sample presents an excellent test case for our protocol because 77% of all peptides in this sample have spectral pairs.
The Lens data set (13) consists of 27,154 MS/MS spectra from a trypsin digestion of lenses from a 93-yr-old male (spectra were obtained on a ThermoFinnigan LCQ Classic ion trap mass spectrometer). This data set was studied extensively (7, 8, 13), resulting in the identification of 416 unmodified peptides and 450 modified peptides. Furthermore, 318 unmodified peptides had spectral pairs and 343 modified peptides had an unmodified version in the sample.
Spectral Pairs.
Peptides P1 and P2 form a peptide pair if either (i) P1 differs from P2 by a single modification/mutation or (ii) P1 is either a prefix or suffix of P2.‖ Two spectra form a spectral pair if their corresponding peptides are paired. Although the peptides that give rise to a spectral pair are not known in advance, we show below that spectral pairs can be detected with high confidence by using uninterpreted spectra.
Our approach for detecting spectral pairs is similar in spirit to the blind search for modified peptides first described by Pevzner et al. (17) and further developed by Tsur et al. (7). Hansen et al. (30) and Tang et al. (31) have alternatively proposed enumeration- and preindexing-based approaches to blind database search, and Savitski et al. (19) recently complemented blind database search by taking into account the retention time. It should be noted that the retention time analysis imposes the constraint that both spectra must come from the same sample, whereas our approach seamlessly enables detection of spectral pairs from multiple MS/MS sample runs (e.g., different cell states or diseased/healthy tissue samples).
For two spectra S1 and S2, the spectral product of S1 and S2 is the set of points (x, y) in 2D for every x ∈ S1 and y ∈ S2 (where S1 and S2 are represented as sets of masses). Fig. 3a shows the spectral product for the theoretical spectra of two peptides. The similarity between the two spectra is revealed by two diagonals in the spectral product: one is formed by matching b ions (blue) and the other by matching y ions (red).
Fig. 3 b and d shows pairs of uninterpreted spectra, denoted S1 and S2, and their spectral product. Although the “colors” of the peaks are not known, in this case we still take the liberty of naming one diagonal “blue” and the other “red.” One can use circles (matching peak masses) on the blue diagonal to transform the original spectrum S1 into spectrum S1,2b (Fig. 3c) with a much smaller number of peaks (a peak in S1 is retained in S1,2b only if it generates a circle on the blue diagonal). Similarly, one can transform S1 into a spectrum S1,2y by using circles on the red diagonal. The peak scores in both spectra S1,2b and S1,2y are inherited from spectrum S1. Similarly, the spectrum S2 is transformed into spectra S2,1b and S2,1y.††
Intuitively, if two spectra are unrelated, the blue and red diagonals represent random matches, and the number of circles appearing on these diagonals is small. Paired spectra, to the contrary, are expected to have many circles on these diagonals. Although this simple criterion (number of circles on the diagonals) would already allow one to roughly distinguish paired spectra from unrelated spectra, we describe below a more accurate spectral alignment test for finding spectral pairs. See SI Appendix C and ref. 17 for the advantages of spectral alignment over simpler spectral convolution approaches similar to FFT cross-correlation analysis.
Fig. 3b illustrates case (ii) in the definition of spectral pairs. The situation becomes less transparent in case (i), namely when modification/mutation occurs in the middle of the peptide (Fig. 3d). In this case, both detecting spectral pairs (Si, Sj) and further processing them into spectra Si, jb and Si, jy is rather complicated. In SI Appendix D we describe the antisymmetric spectral alignment algorithm for deriving virtual spectra Si, j from spectral pairs that also covers this case of internal modifications/mutations.
For the sake of simplicity, the above description hides many details that turn interpretation of spectral pairs into a rather difficult algorithmic problem. The original algorithm from Pevzner et al. (17) considered only b-b (or y-y) pairs of matching peaks and was not able to consider all three types of matching peaks (b-b, y-y, and b-y) when computing the spectral alignment. This complication was addressed by Tsur et al. (7) for the case “spectrum vs. peptide” comparison. In spectral networks we face a more difficult case of “spectrum vs. spectrum” comparison‡‡ and take into account the antisymmetric path condition (15, 33) that further complicates the spectral alignment algorithm (even in the case of a single internal modification).
Spectral Networks.
The correlation score of spectra S1 and S2 is defined as the total score of all peaks in spectra S1,2b and S1,2y: score(S1, S2) = score(S1,2b) + score(S1,2y). Similarly, score(S2, S1) = score(S2,1b) + score(S2,1y). We accept S1 and S2 as a putative spectral pair if both the ratio [score(S1, S2)]/[score (S1)] and the ratio [score(S2, S1)]/[score (S2)] exceed a predefined threshold (0.4 in the examples below), where score(Si) is the summed score of all peaks in Si. In addition, we assume that the correlation score between a given spectrum S and any unrelated spectrum S′ approximately follows a Gaussian distribution. Thus, a correlation score is only considered significant if the probability of this score appearing by chance is <0.05. The combined filtering efficiency of these criteria allowed us to retain 78.4% of all correct spectral pairs at a precision level of 95% and to find several different variants for most unmodified peptides. The main reason why the remaining spectral pairs were not detected by our alignment procedure was the change in fragmentation patterns between these closely related peptides. The spectral pairs that satisfy the tests above form the spectral network on the set of all spectra (see Fig. 1 for an example). The spectral network for the whole IKKβ data set has 43 connected components with 1,021 vertices and 1,569 edges. The small number of connected components is not surprising because overlapping peptides in this data set can be assembled into a small number of contigs [an effect previously explored in the context of shotgun protein sequencing (20)].
By reducing the number of noise peaks by a factor of 8, spectra pairs provide a dramatic increase in signal-to-noise ratio (see SI Table 2), even when compared with consensus spectra obtained from clusters (20) (let alone individual spectra). Moreover, spectral pairs provide a nearly perfect separation between b- and y-ion ladders, the key condition for successful de novo reconstruction.
Spectral Stars.
Even though for a single spectral pair (S1, S2) the spectra S1,2b and S1,2y already have high signal-to-noise ratio, we show that spectral stars allow one to further enrich the b- and y-ion ladders. A spectral star consisting of spectral pairs (S1, S2), (S1, S3), …, (S1, Sn) allows one to increase the signal-to-noise ratio by considering 2(n − 1) spectra S1, ib and S1, iy for 2 ≤ i ≤ n. We combine all these spectra into a star spectrum S1*, as in our clustering approach (details provided in SI Appendix D).
Supplementary Material
Acknowledgments
We thank Ebrahim Zandi (University of Southern California, Los Angeles, CA) and Brian Searle (Oregon Health and Science University, Portland, OR) for providing the MS/MS data sets and Vineet Bafna, Stephen Tanner, and Phillip Wilmarth for insightful discussions. This work was supported by National Institutes of Health Grant NIGMS 1-R01-RR16522.
Abbreviations
- MS/MS
tandem mass spectrometry
- PTM
posttranslational modification.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701130104/DC1.
‡The peptide sequence tag approach reduces the number of considered peptides but does not eliminate the need to match spectra against the filtered database.
§In a blind database search, the list of possible modifications is not known in advance, and P suffices to interpret S(P*) (7). In a restrictive database search, one also needs the list of possible modifications in order to interpret S(P*).
In contrast to the standard de novo algorithms, we do not insist on reconstructing the entire peptide and often shorten the found path by removing its prefix/suffix if the path does not explain any peaks. As a result, the found path does not necessarily start/end at the beginning/end of the peptide.
Condition (ii) can be viewed as a variation of condition (i) if one considers extending a peptide by a few residues as a single “mutation” (such variations are common in MS/MS samples). More generally, peptides P1 and P2 form a peptide pair if either (i) P1 is a modified/mutated version of P2 or (ii) P1 and P2 overlap. Although our techniques also work for this generalization, we decided to limit our analysis to simple peptide pairs, as described above. We found that such simple pairs alone allow one to interpret most spectra. Adding pairs of spectra that have more subtle similarities further increases the number of spectral pairs but slows down the algorithm.
We remark that the assignments of upper indexes to spectra S1,ib and S1,iy are arbitrary, and it is not known in advance which of these spectra represents b ions and which represents y ions.
In the case of spectrum vs. peptide, one knows the sets of b and y ions in the theoretical spectrum of the peptide, whereas in the spectrum vs. spectrum case this partition is unknown. A similar problem was considered by Zhang and McElvain (32) in case of MS2 and MS3 spectra comparison.
References
- 1.Eng J, McCormack A, Yates J. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 2.Perkins D, Pappin D, Creasy D, Cottrell J. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Craig R, Beavis R. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 4.Tanner S, Shu H, Frank A, Wang L, Zandi E, Mumby M, Pevzner P, Bafna V. Anal Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 5.MacCoss M, McDonald W, Saraf A, Sadygov R, Clark J, Tasto J, Gould K, Wolters D, Washburn M, Weiss A, et al. Proc Natl Acad Sci USA. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klammer AA, MacCoss MJ. J Proteome Res. 2006;5:695–700. doi: 10.1021/pr050315j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsur D, Tanner S, Zandi E, Bafna V, Pevzner PA. Nat Biotechnol. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- 8.Wilmarth PA, Tanner S, Dasari S, Nagalla SR, Riviere MA, Bafna V, Pevzner PA, David LL. J Proteome Res. 2006;5:2554–2566. doi: 10.1021/pr050473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mann M, Wilm M. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 10.Shevchenko A, Loboda A, Sunyaev S, Shevchenko A, Bork P, Ens W, Standing K. Anal Chem. 2001;73:1917–1926. doi: 10.1021/ac0013709. [DOI] [PubMed] [Google Scholar]
- 11.Liebler D, Hansen B, Davey S, Tiscareno L, Mason D. Anal Chem. 2002;74:203–210. doi: 10.1021/ac0155512. [DOI] [PubMed] [Google Scholar]
- 12.Han Y, Ma B, Zhang K. IEEE Computational Systems Bioinformatics Conference; August 16–19, 2004; Stanford, CA. 2004. pp. 206–215. [DOI] [PubMed] [Google Scholar]
- 13.Searle BC, Dasari S, Wilmarth PA, Turner M, Reddy AP, David LL, Nagalla SR. J Proteome Res. 2005;4:546–554. doi: 10.1021/pr049781j. [DOI] [PubMed] [Google Scholar]
- 14.Taylor J, Johnson R. Rapid Commun Mass Spectrom. 1997;11:1067–1075. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1067::AID-RCM953>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 15.Dancík V, Addona T, Clauser K, Vath J, Pevzner P. J Comput Biol. 1999;6:327–342. doi: 10.1089/106652799318300. [DOI] [PubMed] [Google Scholar]
- 16.Frank A, Pevzner P. Anal Chem. 2005;77:964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 17.Pevzner P, Dancík V, Tang C. J Comput Biol. 2000;7:777–787. doi: 10.1089/10665270050514927. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen ML, Savitski MM, Zubarev RA. Mol Cell Proteomics. 2006;5:2384–2391. doi: 10.1074/mcp.M600248-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Savitski MM, Nielsen ML, Zubarev RA. Mol Cell Proteomics. 2006;5:935–948. doi: 10.1074/mcp.T500034-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Bandeira N, Tang H, Bafna V, Pevzner P. Anal Chem. 2004;76:7221–7233. doi: 10.1021/ac0489162. [DOI] [PubMed] [Google Scholar]
- 21.Keller A, Purvine S, Nesvizhskii A, Stolyar S, Goodlett D, Kolker E. OMICS. 2002;6:207–212. doi: 10.1089/153623102760092805. [DOI] [PubMed] [Google Scholar]
- 22.Takikawa O, Truscott RJ, Fukao M, Miwa S. Adv Exp Med Biol. 2003;527:277–285. doi: 10.1007/978-1-4615-0135-0_31. [DOI] [PubMed] [Google Scholar]
- 23.Vrensen GF, van Marle J, Jonges R, Voorhout W, Breipohl W, Wegener AR. Exp Eye Res. 2004;78:661–672. doi: 10.1016/j.exer.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Lapko VN, Smith DL, Smith JB. J Mass Spectrom. 2000;35:572–575. doi: 10.1002/(SICI)1096-9888(200004)35:4<572::AID-JMS971>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 25.David LL, Lampi KJ, Lund AL, Smith JB. J Biol Chem. 1996;271:4273–4279. doi: 10.1074/jbc.271.8.4273. [DOI] [PubMed] [Google Scholar]
- 26.Creasy DM, Cottrell JS. Proteomics. 2004;4:1534–1536. doi: 10.1002/pmic.200300744. [DOI] [PubMed] [Google Scholar]
- 27.Savitski MM, Nielsen ML, Zubarev RA. Mol Cell Proteomics. 2005;4:1180–1188. doi: 10.1074/mcp.T500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Frank AM, Savitski MM, Nielsen ML, Zubarev RA, Pevzner PA. J Proteome Res. 2007;6:114–123. doi: 10.1021/pr060271u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shevchenko A, Chernushevich I, Ens W, Standing K, Thomson B, Wilm M, Mann M. Rapid Commun Mass Spectrom. 1997;11:1015–1024. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1015::AID-RCM958>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 30.Hansen BT, Davey SW, Ham AJ, Liebler DC. J Proteome Res. 2005;4:358–368. doi: 10.1021/pr0498234. [DOI] [PubMed] [Google Scholar]
- 31.Tang W, Halpern B, Shilov I, Seymour S, Keating S, Loboda A, Patel A, Schaeffer D, Nuwaysir L. Anal Chem. 2005;77:3931–3946. doi: 10.1021/ac0481046. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, McElvain J. Anal Chem. 2000;72:2337–2350. doi: 10.1021/ac000226k. [DOI] [PubMed] [Google Scholar]
- 33.Chen T, Kao M, Tepel M, Rush J, Church G. J Comput Biol. 2001;8:325–337. doi: 10.1089/10665270152530872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.