Abstract
The formation of viable genetic chimeras in mammals through the transfer of cells between siblings in utero is rare. Using microsatellite DNA markers, we show here that chimerism in marmoset (Callithrix kuhlii) twins is not limited to blood-derived hematopoietic tissues as was previously described. All somatic tissue types sampled were found to be chimeric. Notably, chimerism was demonstrated to be present in germ-line tissues, an event never before documented as naturally occurring in a primate. In fact, we found that chimeric marmosets often transmit sibling alleles acquired in utero to their own offspring. Thus, an individual that contributes gametes to an offspring is not necessarily the genetic parent of that offspring. The presence of somatic and germ-line chimerism may have influenced the evolution of the extensive paternal and alloparental care system of this taxon. Although the exact mechanisms of sociobiological change associated with chimerism have not been fully explored, we show here that chimerism alters relatedness between twins and may alter the perceived relatedness between family members, thus influencing the allocation of parental care. Consistent with this prediction, we found a significant correlation between paternal care effort and the presence of epithelial chimerism, with males carrying chimeric infants more often than nonchimeric infants. Therefore, we propose that the presence of placental chorionic fusion and the exchange of cell lines between embryos may represent a unique adaptation affecting the evolution of cooperative care in this group of primates.
Keywords: callitrichid, genetic chimerism, genomic conflict, social behavior
Genetic chimerism, the mingling of two or more genomic lineages within an individual (1), is rare in mammals, but chimerism is prevalent in the hematopoietic tissues of marmosets and other callitrichid primates (2, 3). In these species, fraternal twins exchange cell lines through chorionic fusion during early development (2, 4, 5). On the basis of karyotypic evidence from Callithrix jacchus (2, 3), estimates are that 95% of pregnancies result in the birth of hematopoietic chimeric twins. Chorionic fusion of the twins' placentas begins on day 19 and is complete by day 29, forming a single chorion with anastomoses connecting the embryos, which are still at a presomite stage in development (4–7). The fusion of the chorions and a delay in embryonic development at this stage allows the exchange of embryonic stem cells via blood flow between the twins (2, 8). As a result, the infants are genetic chimeras with tissues derived from self and sibling embryonic cell lineages (2, 3, 8).
Although there is little doubt that tissues derived from hematopoietic origin are universally chimeric (9), the existence of chimeric cells in nonhematopoietic tissues, including germ-line cells, has not been established. Karyotypic analysis of C. jacchus revealed that testes cells express unusual orientation during meiosis, and this evidence suggested that the germ-line cells might include female cells present because of chimerism (8, 10). However, further karyotypic analysis refuted these findings, and an analysis of sex ratios in captive colonies of C. jacchus suggested that germ-line chimerism was not present (11). To investigate whether chimerism occurs in tissues other than those derived from the hematopoietic system, species-specific microsatellite markers were used to examine the extent and distribution of chimerism.
The existence of chimerism throughout somatic and germ-line tissues may have important implications for the evolution of paternal and alloparental care characteristics of this taxon, through genomic conflict or altered perceptions of relatedness between members of a family group (12, 13). Genomic conflict in individuals with genetic heterogeneity has been identified as a possible evolutionary mechanism, influencing behavioral and developmental traits (14–17). Conflict within an individual may influence the development of kin recognition mechanisms. Specifically, how do chimeric organisms identify an individual and determine relatedness to another chimeric individual (12)? Somatic chimerism may provide individuals with self-matching kin recognition cues, causing an overestimate of their relatedness to chimeric offspring. Although the exact mechanisms of kin recognition are unknown in primates, baboons appear to be capable of recognizing paternal offspring, which may involve phenotype matching (18). Phenotype matching has been conclusively demonstrated to occur in at least one mammal species (19). If chimerism in marmosets involves more than hematopoietic tissues, then we predict differential parental behavior toward chimeric and nonchimeric infants and altered estimates of relatedness from those expected for nonchimeric mammals.
Results
We examined the prevalence of chimerism in tissues derived from different embryonic origins by analyzing genotypes of microsatellite loci with a probability of detecting chimerism of 98% based on parental genotypes for these loci. A total of 92 intergenerational individuals that included 36 twin sets of Callithrix kuhlii (Wied's black tufted-ear marmosets) and their parents were assessed. The samples were genotyped in an appropriate blind fashion such that the identity of the individual and the tissue type were unknown. All alleles were noted for each locus, and samples were identified as potentially chimeric if they contained three or four allelic variants at a single locus. The samples were then matched to identity, and twins were noted to be chimeric at a tissue only if the alleles were found to match both the parents as well as their twin. Further, a majority rule approach was used to assign alleles as “self” (i.e., diploid and inherited vertically from the parents) and “sibling” (inherited horizontally from the twin in utero) [see example in supporting information (SI) Fig. 3]. Of the 36 twin sets surveyed, 26 (72.2%) were determined to carry chimeric tissues. Exchange of alleles between twins was not always bidirectional. In 14 twin sets, only one twin was found to have chimeric tissue types, whereas the other 12 chimeric twin sets revealed chimerism in both twin's tissues.
Chimerism was found to be present in every tissue that was analyzed, and the occurrence of chimerism in tissues harvested from marmoset cadavers differed significantly across liver, spleen, kidney, heart, lung, brain, muscle, skin, and hair (“Samples from deceased animals” in Table 1; n = 25, Cochran's Q = 51.6, df = 8, P < 0.001). To determine which tissue type accounted for the variance among the tissues, the tissues were ranked according to percentage of occurrence of chimerism and then grouped. A comparison of liver and spleen revealed a nonsignificant difference between the tissue types (n = 25, Q = 4, df = 1, P = 0.15). The addition of hair samples revealed a significant difference between the tissue types (n = 25, Q = 14.3, df = 2, P < 0.001). The grouping of all other tissues (heart, hair, lung, kidney, skin, brain, and muscle) resulted in no significant difference between the tissue types for the presence of chimerism (n = 25, Q = 10.3, df = 6, P > 0.1). Hematopoietic tissues were significantly more likely to be chimeric than all other tissue types (χ2 = 4.88, df = 1, P < 0.05). The assessment of chimerism in tissues collected from living marmosets revealed nonsignificant differences between the tissue types (“Samples from living animals” in Table 1).
Table 1.
The number of Callithrix kuhlii individuals chimeric for each tissue type
| Tissue | Tissue type | Genotyped, no. | Chimeric, no. | Chimeric, % |
|---|---|---|---|---|
| Samples from deceased animals | ||||
| Placenta | H | 7 | 7 | 100.0 |
| Blood | H | 2 | 2 | 100.0 |
| Spleen | H | 28 | 14 | 50.0 |
| Liver | H | 39 | 15 | 38.5 |
| Heart | S | 30 | 7 | 23.3 |
| Hair | S | 35 | 6 | 17.1 |
| Lung | S | 30 | 4 | 13.3 |
| Kidney | S | 33 | 4 | 12.1 |
| Gonad | G | 21 | 2 | 9.5 |
| Skin | S | 36 | 2 | 5.6 |
| Brain | S | 31 | 1 | 3.2 |
| Muscle | S | 34 | 1 | 2.9 |
| Samples from living animals | ||||
| Sperm | G | 7 | 4 | 57.1 |
| Saliva | S | 31 | 16 | 51.6 |
| Blood | H | 45 | 22 | 48.9 |
| Hair | S | 50 | 13 | 26.0 |
| Fecal | S | 22 | 2 | 9.09 |
H, hematopoietic; S, other somatic; G, germ line.
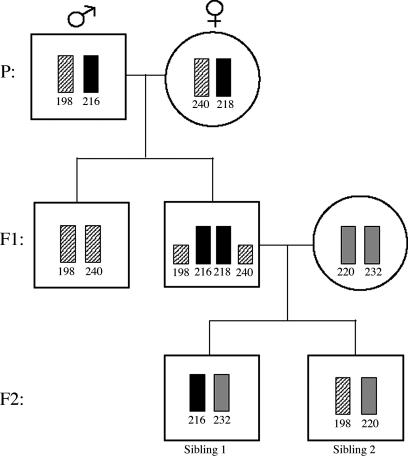
The presence of sibling-derived alleles in multiple tissues suggested that all embryonic cell lineages in C. kuhlii might be affected by chimerism, including gametic tissue. In fact, gonadal tissue was found to be chimeric (2/21), and sperm samples were also chimeric (4/7). Additionally, the 36 twin sets analyzed for chimerism comprised multiple generations within 15 family lines. We determined that individuals in 5 of the 15 families passed on alleles to their offspring that represented gene lineages inherited horizontally from the sibling (see examples in Fig. 1 and SI Fig. 4). One breeding female, whose uterine twin was a male, produced offspring that inherited her sibling's alleles. This documents the possibility that an XY primordial germ cell is capable of maturing and producing viable eggs in a female, a phenomenon that has not been documented for primates. Although we are not currently able to document the fate of the Y chromosome during development of the female's oocytes, our data suggest the intriguing possibility that a female may pass on a Y chromosome to her offspring.
Fig. 1.
Vertical transmission of sibling alleles in C. kuhlii, shown for microsatellite locus CK2. The grandfather and grandmother (P) are individuals that had self alleles (198/216) and (218/240), respectively. They gave birth to male twins (F1) with self genotypes of (198/240) and (216/218). One male (216/218) was found to have sibling alleles (198/240) present in his heart, spleen, and lung samples, which represented ≈50% of cells in those tissues and were not present in the hair, skin, and brain samples. This male was paired with a female (220/232). The pair's twin infants (F2) were both heterozygous and nonchimeric; sibling 1 inherited one allele from the father (216) and one from the mother (232). Sibling 2 inherited 220 from the mother and the sibling allele (198) that the father had acquired from his twin through horizontal exchange.
The presence of cells derived from different lineages within an individual may impact behavioral decisions. Genetic chimerism may give rise to genomic conflict such that an individual's decision to cooperate within a group and care for members of the group may depend on the true, or perceived, genetic relatedness between the individuals (12, 16). To illustrate this with a simple example, we consider the increased proportion of shared alleles, because of genetic chimerism, between male twins produced by nonchimeric parents. A chimeric individual's coefficient of relatedness to his twin could increase from the expected fraternal twin value of r = 0.5 to as much as r = 1 in certain tissues. Based on the prevalence of chimerism, the proportion of cells within a tissue that carry sibling alleles, and the probability of the direction of exchange obtained from our data, we estimate that male twins are on average related by r = 0.574 (see SI Text for calculations). More specifically, in a case of unidirectional exchange in which the soma of the donating twin is nonchimeric, he is related to the sperm of the recipient twin by an average r of 0.625 (see SI Text). The relatedness calculations suggest that chimeric callitrichid siblings are more closely related to each other than typical nonchimeric mammalian siblings. Calculations of relatedness under more complex scenarios and involving parental chimerism are beyond the scope of this report; thus, at this stage, it is not known how parental–offspring relatedness may be affected by chimerism.
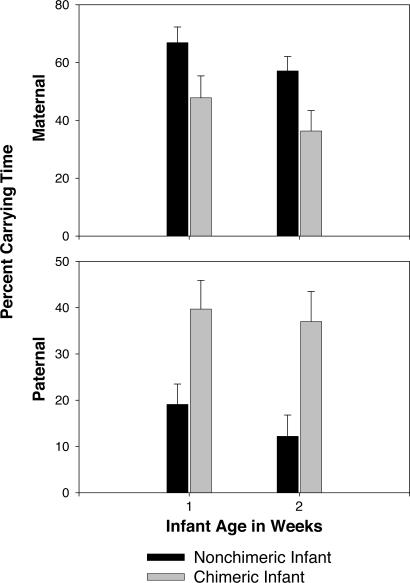
A different issue, but also with potential behavioral implications, is perceived relatedness through kin recognition. To investigate whether the presence of chimeric epithelial tissues, those most likely to mediate kin recognition, are associated with changes in parental care, maternal and paternal infant carrying effort for the first 2 weeks of life was compared between infants determined to be epithelial chimeras (n = 10) and those determined to be nonchimeric (n = 20). Alloparents do not typically provide care during this early period (20) and thus were not included in the analysis. Females carried chimeric infants significantly less than nonchimeric infants (F1,28 = 7.61; P = 0.01), but fathers carried chimeric infants significantly more than nonchimeric infants (F1,28 = 12.08; P = 0.002) (Fig. 2). No significant differences in carrying time by mothers or fathers were found due to parity (F1,28 = 2.124, not significant) or family (F8,28 = 1.865, not significant). An intriguing possibility is that chimeric offspring might match the father at more kin recognition alleles, elevating the perceived relatedness and thus, the confidence of paternity in fathers. An alternative, but not mutually exclusive explanation for this behavior is that mothers actively avoid chimeric offspring. Although the behavioral data do not allow us to discriminate between these two hypotheses, they demonstrate a significant correlation between the chimeric status of offspring and altered patterns of maternal and paternal care in marmosets.
Fig. 2.
Carrying effort during the first 2 weeks of infant life for epithelial (hair and saliva) chimeric infants (n = 10) and nonchimeric infants (n = 20), as a percentage of the total time + SEM during which infant was carried. Mothers with chimeric infants carry significantly less than those with nonchimeric infants (F1,28 = 7.61; P = 0.01), and fathers with chimeric infants carry significantly more than those with nonchimeric infants (F1,28 = 12.08; P = 0.002).
Discussion
This study thoroughly characterizes the extent and distribution of genetic chimerism throughout the tissues of callitrichid primates, which has not been done at this level previously. Hematopoietic chimerism in callitrichids was unambiguously documented in the 1960s (2), but it was unknown whether callitrichids displayed chimerism in tissues other than blood-derived tissues. Using highly variable genetic markers, we found that all tissue types sampled contain sibling alleles inherited via horizontal cell exchange. Perhaps most importantly, the germ line is also chimeric. Molecular genotyping analyses revealed that sperm can be genetically chimeric, and genealogical analyses demonstrated that marmosets can pass on sibling alleles, acquired in utero from their twin, to their offspring.
Several lines of evidence were used to determine that the chimerism noted in nonhematopoietic tissues was not simply due to contamination of tissues with blood products. First and foremost, if chimerism was limited to blood products, then the sperm samples that were genotyped after they were separated from the ejaculate material should not have been chimeric. Additionally, the presence of hematopoietic chimerism alone cannot account for the finding of transmission of chimeric cell lineages across generations. Further, tissues known to be rich in blood supply such as the heart and lung tissue, as well as those likely to have high white blood cell counts because of immune function such as lungs, saliva, and skin, should have had equal intensity and prevalence of chimerism as the blood samples, which they did not. Finally, 12 animals were chimeric for nonhematopoietic tissues, yet they were not chimeric for hematopoietic tissues.
Although chimerism in other mammals, such as cows, cats, and humans, usually leads to sterility and appears to be selected against (1, 21), marmosets exhibit high rates of placental fusion and genetic chimerism. The delay in embryonic development at the time of chorionic fusion (6, 7) increases the chance that stem cell exchange between twin embryos occurs before advanced differentiation of embryonic tissues, thereby facilitating genetic exchange between the twins. All species in the subfamily Callitrichinae share, as a derived character, a high investment of males in infant care, alloparental care, and obligate fraternal twinning (22, 23). Potential effects of chimerism for a marmoset include an increase in self-matching phenotypes between offspring and family members, which may lead to greater investment by other group members in offspring.
Chimerism may help to explain the unusual attraction of males to infants in callitrichids, although chimerism itself may not explain cooperative breeding and paternal care in other organisms. Genetic chimerism may serve as a genetic determinant influencing behavioral decisions involving cooperation and conflict in callitrichids, either through genomic conflict or direct and indirect fitness. The quantification of chimerism in callitrichids provides the basic knowledge to develop future field and captive studies to examine reproductive success, kin recognition systems, genomic conflict, and the impacts of relatedness on social behavior.
Materials and Methods
Study System.
The only North American breeding colony of C. kuhlii (Wied's black tufted-ear marmosets) was established in 1991 at the University of Nebraska at Omaha (UNO) by Jeffrey French. The colony at UNO provided a complete known breeding history, and multiple tissue samples were available for the majority of individuals because all carcasses of deceased animals were archived. We identified twin sets with known parentage, using colony breeding histories. We selected families in which a single female and male were housed together, with no subordinate males of breeding age, to ensure known paternity of the offspring. Thirty-six twin sets were available that fit these criteria, and all 36 twin sets and their parents (15 breeding pairs) were available for DNA sampling for this study. Tissues chosen to be sampled represented differing developmental cell lines, including hematopoietic tissue (blood, spleen, and liver), germ-line tissue (sperm and gonad), and other somatic tissues (lung, muscle, skin, and kidney). Although all tissues were not available from all animals, this sampling strategy allowed a thorough assessment of the extent and distribution of chimerism in individuals.
The UNO colony maintains a noninvasive research ethic (24); therefore, tissues collected from living animals were limited to those that could be collected in a noninvasive manner, or that were available after veterinary checkups. Samples that were collected from living animals included blood, hair, sperm, and epithelial cells as found in saliva and feces. Sperm samples were collected by penile stimulation, using FertiCare (Multicept ApS, Rungsted, Denmark) instrumentation (25). Sperm was collected in a 0.2-μl centrifuge tube and then placed in Hepes buffer to allow the sperm to swim up; the top layer was removed to a fresh tube with fresh buffer and frozen at −20°C until it could be extracted. Samples that were collected from deceased animals included skin, muscle, liver, spleen, lung, hair, heart, kidney, brain, gonadal tissue, and fecal samples.
Genetic Analysis.
All samples were assigned a random letter/number combination to perform all genotyping analyses under an appropriate experimental blind. DNA extractions were done by using proteinase K digestion, phenol/chloroform purification, and ethanol precipitation. All samples were resuspended in water. Hair, saliva, sperm, and blood samples were extracted by using a QIAmp DNEASY extraction kit (Qiagen, Valencia, CA) to ensure high quality and quantity DNA. Fecal samples were extracted by using the QIAGEN DNEasy stool kit. A GeneQuant II spectrophotometer was used to quantify DNA from the extracted sample. All DNA was then diluted to 20 ng/μl. DNA was PCR-amplified by using markers CJ1, CJ6, CJ13, and CJ14 (26), as well as species-specific marker CK2 developed for this project. Amplification products were analyzed with an ABI310, and genotypes were scored by using GeneScan software (Applied Biosystems, Foster City, CA). To determine the individual's self genotype for each locus a majority rule analysis was applied. In cases where tissue genotypes varied within an individual, it was assumed that the diploid genotype found most prevalently across tissues most likely represented the alleles present in the individual because of vertical inheritance from the parents (self), rather than horizontal transfer from the twin (sibling alleles). In fact, there were no cases in which two different diploid genotypes were found among tissues in a single individual; all cases of chimerism involved three or four alleles in the tissue samples. Chimerism was verified for an individual when the blind was lifted and the putative sibling alleles were shown to be the majority rule genotype of the individual known to be the twin, and all alleles noted were also found in the parents of the twins. Heterozygous and chimeric genotypes were amplified at least three times and were confirmed as chimeric if all genotypes matched. Additionally, duplicate samples were collected for tissues such as the liver; all duplicate sample genotypes matched in 100% of the replicates. The allele frequencies for the microsatellite loci CJ1, CJ6, CJ13, CJ14, and CK2 are shown in SI Table 2.
Behavioral Data.
Carrying data were collected daily via scan samples recorded throughout the day during the first 2 weeks of life for each infant. Each infant was identified by using unique markings, such as white stripes on the tail, and was assigned a unique name. The identity of the individual and the family member carrying the individual were noted at the time of observation. These scores were then tallied and the average carrying effort of each family member was calculated.
Statistical Analyses.
Cochran's Q test was used to evaluate whether there were significant differences between tissues that were identified as chimeric. Cochran's Q provides a method for testing whether three or more matched frequencies or nominal data differ significantly among themselves (27). χ2 tests were used to further examine the differences between the prevalence of chimerism in tissue types. Carrying effort of the caregivers of thirty infants that had been genetically assessed for chimerism were analyzed by using an analysis of variance, specifically 2 (time: week1/week2) × 2 (chimeric/nonchimeric). Parity effects on care giving behaviors was assessed by a 2 (parity: multiparous/primiparous) × 2 (time: week1/week2) × 2 (carrier: dam/sire) ANOVA. Carrying differences between family groups was assessed with a 2 (time: week1/week2) × 2 (carrier: dam/sire) × 9 (family group) ANOVA.
Supplementary Material
Acknowledgments
We thank D. Hightower, K. Patera, and H. Jensen for animal care and maintenance of breeding records; J. Sommer and M. Bessert for help with microsatellite development; J. Meeker, A. Startzer, D. Beyer, and J. Brozek for help with sample and data collection; J. Fite for help with behavioral data collection; R. Gibson for help with statistical analyses and calculations of relatedness, as well as comments on earlier drafts; and G. Veomett and A. Kamil and two anonymous reviewers for their comments on drafts. This research was supported by the National Science Foundation Grants IBN 0417202 (to G.O., C.N.R., and J.A.F.) and IBN 00-91030 (to J.A.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607426104/DC1.
References
- 1.McLaren A. Mammalian Chimeras. Cambridge: Cambridge Univ Press; 1976. [Google Scholar]
- 2.Benirshke K, Anderson JM, Brownhill LE. Science. 1962;138:513–515. doi: 10.1126/science.138.3539.513. [DOI] [PubMed] [Google Scholar]
- 3.Gengozian N, Batson JS, Eide P. Cyto. 1964;3:384–393. doi: 10.1159/000129828. [DOI] [PubMed] [Google Scholar]
- 4.Wislocki GB. Anat Rec. 1932;52:381–392. [Google Scholar]
- 5.Wislocki GB. Amer J Anat. 1939;64:445–483. [Google Scholar]
- 6.Missler M, Wolff JR, Rothe H, Heger W, Merker HJ, Treiber A, Scheid R, Crook GA. J Med Prim. 1992;21:288–298. [PubMed] [Google Scholar]
- 7.Merker H-J., Sames K, Casto W, Heger W, Neubert D. In: Nonhuman Primates, Developmental Biology and Toxicology. Neubert D, Merker H, Hendrickx A, editors. Berlin: Ueberreuta Wissenschaft-Wein; 1988. [Google Scholar]
- 8.Benirshke K, Brownhill LE. Cytogenetics. 1963;2:331–341. doi: 10.1159/000129790. [DOI] [PubMed] [Google Scholar]
- 9.Dantas S, Barros R. Braz J Gen. 1997;20:645–649. [Google Scholar]
- 10.Hampton SH. Am J Phys Anthropol. 1973;38:265–268. doi: 10.1002/ajpa.1330380221. [DOI] [PubMed] [Google Scholar]
- 11.Gengozian N, Brewen JG, Preston RJ, Batson JS. J Med Primatol. 1980;9:9–27. doi: 10.1159/000460118. [DOI] [PubMed] [Google Scholar]
- 12.Haig D. Am J Primatol. 1999;49:285–297. doi: 10.1002/(SICI)1098-2345(199912)49:4<285::AID-AJP1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 13.Goldizen AW. In: Primate Societies. Smuts BB, Cheney DC, Seyfarth RM, Wrangham RW, Struhsaher TT, editors. Chicago: Univ Chicago Press; 1987. [Google Scholar]
- 14.Rinkevich B. J Evol Biol. 2004;17:1178–1179. doi: 10.1111/j.1420-9101.2004.00809.x. [DOI] [PubMed] [Google Scholar]
- 15.Spencer HG. Ann Rev Genet. 2000;34:457–477. doi: 10.1146/annurev.genet.34.1.457. [DOI] [PubMed] [Google Scholar]
- 16.Haig D. Genomic Imprinting and Kinship. New Brunswick, NJ: Rutgers Univ Press; 2002. [Google Scholar]
- 17.Day T, Bonduriansky R. Genetics. 2004;167:1537–1546. doi: 10.1534/genetics.103.026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchan JC, Alberts SC, Silk JB, Altmann J. Nature. 2003;425:179–181. doi: 10.1038/nature01866. [DOI] [PubMed] [Google Scholar]
- 19.Mateo JM, Johnston RE. Proc R Soc Lond B. 2000;267:695–700. doi: 10.1098/rspb.2000.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fite JE, Patera KJ, French JA, Rukstalis M, Hopkins EC, Ross CN. J Human Evol. 2005;49:122–142. doi: 10.1016/j.jhevol.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohno S, Trujillo JM, Stenius L, Christian C, Teplitz RL. Cytogenetics. 1962;1:258–265. doi: 10.1159/000129735. [DOI] [PubMed] [Google Scholar]
- 22.Mittermeier R. Ecology and Behaviour of Neotropical Primates. Washington, DC: World Wildlife Fund; 1988. [Google Scholar]
- 23.Price EC. Am J Primatol. 1992;26:23–33. doi: 10.1002/ajp.1350260106. [DOI] [PubMed] [Google Scholar]
- 24.Schaffner CM, Shepherd RE, Santos CV, French JA. Am J Primatol. 1995;36:185–200. doi: 10.1002/ajp.1350360303. [DOI] [PubMed] [Google Scholar]
- 25.Keuderling J, Schneiders A, Sonksen J, Nayudu PL, Hodges JK. Am J Primatol. 2000;52:149–154. doi: 10.1002/1098-2345(200011)52:3<149::AID-AJP4>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 26.Nievergelt CM, Mundy NL, Woodruff DS. Mol Ecol. 1998;7:1431–1439. [PubMed] [Google Scholar]
- 27.Siegel S, Castellan NJ. Nonparametric Statistics, for the Behavioural Sciences. New York: McGraw–Hill; 1988. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.