Abstract
Small molecule inhibitors provide powerful tools to characterize highly dynamic and complex eukaryotic cell pathways such as those mediating membrane traffic. However, a lack of easy and generalizable assays has constrained identification of novel inhibitors despite availability of diverse chemical libraries. Here, we report a facile growth-based strategy in yeast to screen for pathway-specific inhibitors. The approach uses well characterized synthetic genetic growth defects to guide design of cells genetically sensitized for inhibition of chosen pathways. With this strategy, we identified a family of piperazinyl phenylethanone compounds as inhibitors of traffic between the trans-Golgi network (TGN) and endosomes that depends on the clathrin adaptor complex AP-1. The compounds did not significantly alter other trafficking pathways involving the TGN or endosomes, indicating specificity. Compound treatment also altered localization of AP-1 in mammalian cells. These previously uncharacterized inhibitors will be useful for future studies of clathrin-mediated transport in yeast, and potentially in other organisms. Furthermore, the easily automated technology should be adaptable for identification of inhibitors of other cellular processes.
Keywords: clathrin, small molecule, AP-1, GGA, yeast
The study of membrane traffic has long relied on the use of a limited set of small molecules that inhibit transport of lipids and proteins between organelles. A number of trafficking pathways connect the trans-Golgi network (TGN) and endosomes, major sorting stations within the secretory and endocytic pathways, yet no small molecule inhibitors with established specificity for a single pathway between these organelles have been identified. Development of such inhibitors will be instrumental in dissecting contributions of individual pathways and understanding how this traffic is integrated with other cellular processes.
Clathrin, a polymeric vesicle coat protein, plays a largely structural role in endocytosis and traffic between the TGN and endosomes. Clathrin function in these pathways requires cytoplasmic adaptor proteins that recruit clathrin to specific membranes and also concentrate transmembrane cargo into nascent clathrin coated vesicles (1). Adaptor protein-1 complex (AP-1), a heterotetrameric complex, is one of several highly conserved clathrin adaptors involved in traffic between the TGN and endosomes. AP-1-mediated transport is required for proper subcellular distribution of proteins that traffic between the TGN and endosomes, including medically relevant molecules such as the tumor suppressor insulin-like growth factor II receptor (IGF-IIR) and, in polarized cells, the low-density lipoprotein receptor (2, 3). Furthermore, AP-1-dependent traffic may play a role in evasion of HIV-infected cells from the immune response (4). Yet, although AP-1 plays a fundamental role in trafficking between the TGN and endosomes in species as divergent as yeast and mammals, there are still conflicting models for where and how AP-1 functions (5). Thus, to develop additional tools to analyze AP-1-dependent transport pathways and possibly establish leads for therapeutic molecules, we sought to identify small molecule inhibitors that mimic genetic defects in AP-1 dependent traffic in yeast.
Growth based chemical screening offers advantages including identification of compounds that are cell permeant and functional in metabolically active cells (6). Furthermore, such screens require simple readouts, common reagents, and easy optimization. However, to observe growth effects of a compound the target must be involved in cell proliferation, and even in this case, it is difficult to ascribe reduced or enhanced growth to a particular pathway. We reasoned that a common genetic phenomenon, synthetic lethality, would provide a way to focus a growth based assay on a particular cellular process. Synthetic lethality describes a genetic relationship in which combination of two mutant genes elicits a much more severe reduction in growth than either mutation alone (Fig. 1a). Synthetic lethality can result from several situations (7); for instance, gene products may act in two parallel pathways that together are essential so that eliminating a single gene product still leaves one pathway intact whereas elimination of both is lethal. Alternatively, two gene products may act in a single essential pathway. Eliminating either gene product alone leaves sufficient activity for viability but eliminating both completely blocks the pathway.
Fig. 1.
Compound synthetic lethal identification of small molecules that inhibit membrane traffic. (a) Schematic of synthetic lethality. Pathways are symbolized by squares and circles on the left; cells are shown on the right. Inactive pathways and reduced cell growth are indicated by dashed outline and gray object. Cells with both pathways intact grow robustly (Top) as do cells lacking one or the other pathway (Middle Two), however cells lacking both pathways grow poorly or are inviable (Bottom). Pathway inhibition can be induced by mutations in components of pathways or, in the case of CSL, by chemical inhibition of these components. (b) Chemicals identified by CSL screening, which restored chitin rings in chs6Δ cells. Cells lacking CHS6 (MDY335) were grown overnight in the presence of DMSO or indicated compounds (25 μM), harvested, and stained for chitin rings with ccfw (arrowheads). Wild-type (MDY326) and chs6Δ apl2Δ cells (MDY573) are shown for comparison. (c) Chemical structures and Chembridge identification numbers of active compounds shown in b.
A small molecule that inhibits a particular pathway should display the same spectrum of synthetic lethal interactions as mutations that inactivate the pathway. We refer to synthetic lethal interactions between a chemical compound and a genetic mutation as “composite synthetic lethality” (CSL). CSL has been used as a method to identify potential targets of bioactive compounds in yeast (8–12). Likewise, CSL should also be effective in identifying novel bioactive compounds that inhibit a particular pathway. In this case, the desired target pathway must have a known genetic synthetic lethal partner. Cells lacking the synthetic lethal genetic partner will serve as a specificity flag by growing poorly when the target pathway is inhibited. In contrast, wild-type cells should be relatively impervious to the same treatment.
Results
Identification of Inhibitors That Mimic Synthetic Growth Interactions of AP-1 Mutations.
In yeast, mutations in the AP-1 β subunit gene display severe synthetic growth effects when combined with mutations of genes encoding a second class of clathrin adaptor; the GGA (for Golgi-localized, γ ear-containing, ARF-binding) proteins (13). The GGA proteins, Gga1p and Gga2p, are closely related proteins which act in traffic between the TGN and endosomes (14). GGA proteins carry out many of the same activities as AP-1 such as clathrin and cargo binding and therefore are thought to be partially redundant with AP-1. Thus, to identify AP-1 pathway inhibitors we screened for compounds that inhibited the growth of gga1/ 2Δ, but not wild-type cells. To maximize sensitivity to inhibitors, we performed screens in strains lacking PDR5 and SNQ2, encoding two ABC transporters known to contribute to drug resistance in yeast. From a 30,000 compound library, 17 chemicals were identified by these criteria. Of these 17, 3 displayed additional synthetic interactions resembling mutations affecting AP-1-mediated pathways: the compounds inhibited the growth of cells carrying a temperature sensitive allele of the clathrin heavy chain (chc1-ts) but were innocuous to cells lacking functional AP-1 (Table 1). The other compounds may target unknown pathways that are synthetic lethal with the GGA pathway.
Table 1.
Effects of chemicals identified in CSL screen
| Chembridge ID | WT | gga1/2 | apl2/4 | chc1-ts | Bud scars |
|---|---|---|---|---|---|
| 5405958 | +++ | + | +++ | +++ | No |
| 6632436 | +++ | ++ | +++ | +++ | No |
| 5587518 | +++ | ++ | +++ | +++ | No |
| 5108486 | +++ | + | +++ | +++ | No |
| 5459862 | +++ | + | +++ | +++ | No |
| 5573168 | +++ | + | +++ | +++ | No |
| 5144321 | +++ | + | +++ | +++ | No |
| 5550761 | +++ | + | +++ | +++ | No |
| 5784306 | +++ | + | +++ | +++ | No |
| 5650622 | +++ | + | ++ | ++ | No |
| 5112303 | +++ | + | ++ | ++ | No |
| 5656277 | +++ | + | ++ | + | No |
| 5669602 | +++ | + | ++ | + | No |
| 5374773 | +++ | ++ | ++ | +++ | No |
| 5431942 | +++ | + | +++ | ++ | Yes |
| 5353860 | +++ | ++ | +++ | ++ | Yes |
| 5416437 | +++ | + | +++ | ++ | Yes |
Listed are chemicals that reproducibly reduced growth of cells lacking GGA1 and GGA2 (gga1/2; MDY327) and relative growth of chemical-treated cells lacking APL2 and APL4 genes encoding the two large subunits of AP-1 (apl2/4Δ: MDY334), or carrying a chc1-ts allele (MDY330). The last column indicates the ability of compounds at 25 μM to induce chitin rings in cells lacking CHS6(MDY335). +++, 70–100% of wild-type growth; ++, 50–70% of wild-type growth; +, <50% of wild-type growth.
CSL Inhibitors Affect AP-1-Dependent Transport.
As a more direct test for inhibition of AP-1-dependent traffic, we examined the effects of compounds on localization of the chitin synthase Chs3p. Chs3p deposits a ring of chitin around emerging buds that remains as a scar after bud release (15). Cell surface localization and function of Chs3p depends on Chs6p. In chs6Δ cells, Chs3p is retained intracellularly, reducing chitin incorporation at the bud site and conferring resistance to the cell wall toxin calcofluor white (ccfw) (16). Intracellular retention in chs6Δ cells requires AP-1; inhibition of AP-1-mediated traffic restores cell surface localization of Chs3p, the appearance of chitin rings, and ccfw sensitivity (17). Of the 17 compounds, the 3 that displayed AP-1-like synthetic lethal interactions were the only ones that induced chitin rings in chs6Δ cells and also were the only ones that shared a piperazinyl phenylethanone backbone (Fig. 1 b and c). We refer to these compounds as A5, G3 and O3. By growth inhibition of gga1/2Δ cells and ccfw-treated chs6Δ cells, A5 was the most potent (Fig. 2a), with an estimated EC50 of 4 μM for gga1/2Δ growth inhibition. Neither A5 nor G3 inhibited growth of wild-type or AP-1-deficient cells at concentrations up to 300 μM. Treatment with A5 was as effective in inhibiting growth of gga1/2Δ cells carrying wild-type SNQ2 and PDR5 as it was with the original gga1/2Δ snq2Δ pdr5Δ cells, indicating that these mutations are not necessary for sensitivity to this set of compounds (data not shown).
Fig. 2.
Synthetic growth and traffic inhibition by CSL active compounds. (a) Synthetic growth effects. Compounds A5 (Top), G3 (Middle), and O3 (Bottom) were added at different concentrations to wild-type (MDY326), AP-1Δ (apl2Δ apl4Δ; MDY334), chc1-ts (MDY330), and gga1/2Δ (MDY327) cells, or chs6Δ (MDY335) cells with 60 μM ccfw. Values represent the fold growth inhibition observed after overnight incubation with compound compared with growth of the same strains with DMSO. (b) Synthetic effects on α-factor maturation. Wild-type (MDY326) or gga2Δ (MDY380) cells were treated with indicated compounds (25 μM) or DMSO (Φ) overnight, metabolically labeled for 10 min, and subjected to a 30-min chase. Secreted α-factor was immunoprecipitated from the media and imaged by SDS/PAGE followed by autoradiography.
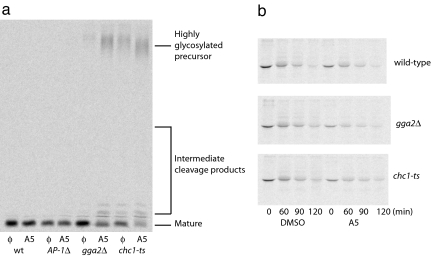
Clathrin dependent TGN-endosome traffic in yeast can also be monitored by measuring the maturation of the mating pheromone α-factor precursor. The efficiency of maturation serves as a sensitive indicator of the integrity of clathrin-dependent TGN-endosome pathways necessary for localization of the TGN protease Kex2p that initiates maturation (18). Alone, genetic inactivation of AP-1 pathway components does not affect α-factor maturation but when AP-1-dependent traffic is perturbed together with chc1-ts or gga2Δ, strong defects in maturation ensue (13, 19). Like genetic inactivation of AP-1 pathway components, the three compounds inhibited α-factor maturation in gga2Δ cells but not wild-type cells (Fig. 2b). In agreement with the growth-based assays, A5 exhibited the most potent effects (Fig. 2b) and exacerbated α-factor maturation defects in gga2Δ and chc1-ts cells but not in cells already lacking functional AP-1 (Fig. 3a).
Fig. 3.
Compound A5 specifically inhibits AP-1-dependent traffic. (a) Synthetic α-factor maturation defects caused by A5. Cells of the indicated genotype (same strains as in Fig. 2a) were treated with compound, and α-factor was analyzed as in Fig. 2b. (b) Degradation of Ste3p is not affected by compound A5 treatment. Cells were treated and labeled as in Fig. 2b, and Ste3p was immunoprecipitated at the indicated chase points from cell lysates and visualized by SDS/PAGE and autoradiography.
AP-1 Pathway Inhibitors Are Specific.
We also examined the effects of A5 on other traffic pathways involving the TGN and endosomes. The pheromone receptor Ste3p is transported through the secretory pathway to the plasma membrane where it undergoes constitutive endocytosis, transport through endosomes, and delivery to the vacuole for degradation (20). By pulse–chase immunoprecipitation, turnover of Ste3p was undisturbed by A5 treatment of wild-type, gga2Δ, and chc1-ts cells (Fig. 3b), indicating that A5 did not impede Ste3p transport through either the secretory or endocytic pathways. We further assessed the secretory pathway by monitoring processing and secretion of α-factor in wild-type cells. Precursor α-factor is translocated into the endoplasmic reticulum and core glycosylated. The core oligosaccharides are elaborated during transport through the Golgi, yielding the heterogeneous highly glycosylated precursor. Proteolytic maturation at the TGN produces the mature peptide. Processing and secretion of α-factor is not dependent on AP-1 (19). Likewise, processing and secretion of α-factor in wild-type cells were unaffected by A5 (Fig. 4a).
Fig. 4.
Compound A5 does not significantly affect other pathways through the TGN. (a) Secretion of α-factor is unaffected in A5-treated cells. Wild-type cells (MDY 326) were grown in the presence of 25 μM A5 or DMSO overnight. Cells were labeled for 10 min, and then α-factor was immunoprecipitated from the media (e, extracellular) or from cell lysates (i, intracellular) at indicated chase points. Immunoprecipitates were imaged by SDS/PAGE followed by autoradiography. (b) Maturation of ALP is unaffected by compound A5 treatment. Wild-type cells (SEY6210) were grown in the presence of 25 μM A5 or DMSO overnight. Cells were labeled for 5 min, and ALP was immunoprecipitated from cell lysates at indicated chase points. For comparison, apl6Δ (GPY1783-25a) cells were analyzed in parallel. ALP precursor (p) and mature (m) forms are indicated. (c) Maturation of CPS is only slightly affected by A5 treatment. Wild-type cells (MDY 326) or gga1Δ gga2Δ cells (MDY327) were treated and analyzed by pulse–chase immunoprecipitation as in a. CPS precursor (p) and mature (m) forms are indicated.
Vacuolar alkaline phosphatase (ALP) is synthesized as a precursor that is proteolytically matured in the vacuole after delivery from the Golgi by a clathrin-independent pathway that requires another adaptor complex, AP-3 (21, 22). Treatment of wild-type cells with A5 resulted in no accumulation of precursor ALP by pulse–chase immunoprecipitation, in contrast to cells lacking a subunit of the AP-3 complex (Fig. 4b). The vacuolar protease carboxyl peptidase S (CPS) depends on the GGA proteins for transport to the vacuole and proteolytic maturation. A5 elicited only a subtle delay in CPS maturation (<2-fold; Fig. 4c). By comparison, deletion of the GGA genes completely prevented maturation over the same time period (Fig. 4 c). The slight effect of A5 on CPS maturation may reflect activity of AP-1 and associated proteins in CPS transport as AP-1 and Gga proteins physically interact (13). Together our results provide evidence that CSL screening effectively identified specific inhibitors of AP-1 dependent membrane traffic.
We tested unrelated compounds, sortin-2 and sortin-3, known to affect traffic to the vacuole in yeast and plant cells, for inhibition of AP-1 dependent traffic (23). Neither sortin enhanced α-factor maturation in cells carrying chc1-ts or gga2Δ, nor did they induce ccfw sensitivity in chs6Δ cells (data not shown). Thus, the piperazinyl phenylethanone compounds identified here seem to be uniquely specific for AP-1-dependent traffic.
Several previously uncharacterized synthetic analogues of A5 were prepared [Table 2 and supporting information (SI) Materials and Methods]. Although none of the new compounds were more potent than A5, the different activity levels of highly related compounds such as S8 and S9 (Table 2) suggest that this family of compounds may have a specific cellular target(s).
Table 2.
Compounds tested in structure-activity relationship analysis
| Compound | ID | ccfw growth inhibition, 30 μM | Toxic |
|---|---|---|---|
 |
A5 | 6.7 | No |
 |
S1 | 2.4 | No |
 |
S2 | 3.6 | 150 μM |
 |
S3 | 1.7 | 95 μM |
 |
S4 | 6.6 | 150 μM |
 |
S5 | 1.4 | No |
 |
S6 | 1.0 | No |
 |
S7 | 7.0 | 14 μM |
 |
S8 | 3.6 | No |
 |
S9 | 1.2 | No |
 |
S10 | 4.2 | No |
 |
S11 | 1.0 | 130 μM |
 |
S12 | 1.4 | 150 μM |
 |
S13 | 3.0 | No |
 |
S14 | 1.1 | No |
 |
7701175 | 1.1 | No |
Indicated are chemical structures (Compound), identifying number (ID), relative growth inhibition of cells lacking CHS6 (MDY335) in 10 μM ccfw and 30 μM compound (ccfw growth inhibition), and, for toxic compounds, the concentration at which compound produced a 2-fold growth inhibition of wild-type cells (Toxic).
Compound A5 Alters AP-1 Localization in HeLa Cells.
We determined whether A5 exhibits activity in mammalian cells by investigating localization of AP-1 in HeLa cells. AP-1, visualized by immunofluorescence, is normally distributed throughout the cell with a diffuse perinuclear concentration (Fig. 5a). Incubation with compound A5 resulted in a more distinct perinuclear localization of AP-1 staining (Fig. 5a). Quantitation of staining intensity in A5-treated cells revealed a clear increase in high-intensity regions compared with DMSO-treated cells (Fig. 5b). In particular, substantially fewer weakly staining cells (<0.3% of their area occupied by high-intensity regions) were present in A5-treated cells compared with DMSO-treated cells. Conversely, strongly staining cells (>1.2% of area occupied by high-intensity regions) were more abundant in the A5-treated population. To probe the specificity of this effect, we tested a related compound, 7701175, which displays much reduced activity in yeast (Table 2). AP-1 staining intensity in cells treated with 7701175 resembled that of DMSO-treated cells, with a substantial fraction of weakly stained cells and only a slight increase in highly stained cells (Fig. 5 a and b). These results provide evidence that compound A5 specifically alters the distribution of AP-1 in mammalian cells and raise the possibility that A5 may target an evolutionarily conserved event. Furthermore, the enhanced perinuclear accumulation of AP-1 in the presence of A5 suggests that the drug acts at a stage after AP-1 membrane recruitment. A5 treatment of yeast did not seem to alter the distribution of GFP-tagged AP-1. However in yeast AP-1 is localized as scattered foci, a pattern that may obscure the type of change apparent in HeLa cells where AP-1 is more diffusely distributed.
Fig. 5.
Localization of AP-1 in treated human cells. (a) Compound A5 increases perinuclear staining of AP-1 in HeLa cells. Cells were treated for 8 h with 50 μM A5, 7701175, or DMSO in DMEM. Cells were fixed, and AP-1 was visualized by immunofluorescence microscopy. (b) Quantitation of cell area occupied by high-intensity staining. Samples were imaged, and the percentage of cell area covered by high-intensity staining was determined for each image frame (n = 70). Displayed is the distribution of images within the indicated areas occupied by high-intensity staining.
Discussion
Our results indicate that piperazinyl phenylethanone-based chemicals identified by CSL inhibit membrane traffic between the TGN and endosomes without apparently altering other related pathways. Thus, we have used the chemical-genetic strategy of CSL to identify previously uncharacterized pathway-specific inhibitors active in living cells. Extensive research has yielded a plethora of synthetic lethal interactions covering the entire spectrum of cellular processes in yeast, including many pathways conserved in multicellular eukaryotes (7, 24). Thus, CSL should be useful in generating probes for a variety of biological functions. The approach should be, in principle, applicable in any cell-type or situation where chemical and genetic inactivation can be combined. In particular, a variety of cancers are hypersensitive to perturbations in pathways that normally may not affect cellular viability (25). Taken together, these considerations suggest that targeted CSL is an effective strategy to identify small molecule inhibitors for investigation of basic cellular processes as well as possible lead compounds for therapeutics development.
Materials and Methods
Strains.
The deletion allele of APL2 was generated as described (26). The chc1-ts allele was generated by homologous recombination of two PCR products. One was an amplified region of plasmid YIpCHC521Δcla containing mutations conferring temperature-sensitive growth; the second encoded a region overlapping the C terminus of CHC1 and containing the URA3Mx cassette and sequences 3′ of CHC1 (27, 28). Primer sequences are listed in SI Materials and Methods. The deletion alleles of PDR5 and GGA1 were generated by using standard PCR-based knockout methods (29). All other alleles were derived from commercially available deletion libraries (Research Genetics, Huntsville, AL). Strains were generated from crosses with MDY326 (MATα his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0, pdr5Δ::URA3, snq2Δ::KanMx) to generate strains MDY327 (gga1Δ::URA3Mx, gga2Δ::KanMx), MDY330 (chc1-ts::URA3Mx), MDY334 (apl 2Δ::URA3, apl4Δ::KanMx), MDY335 (chs6Δ::KanMx), MDY380 (gga2Δ::KanMx), and MDY573 (chs6Δ::KanMx, apl2Δ::URA3, apl4Δ::KanMx, MET15). SEY6210 and GPY1783-25a (apl6Δ) used for ALP analysis have been described (13). Strains used for analysis of compound effect in the absence of drug pumps were SEY6210 and GPY3431 (30).
Media and Reagents.
All media were from GIBCO; chemical reagents were from Sigma unless otherwise noted. YPD medium is 1% Bacto-yeast extract, 2% Bacto-peptone, dextrose added to 2% after sterilization. SCMG is yeast nitrogen base without amino acids or ammonium sulfate, 0.1% monosodium glutamate, 40 mM lithium acetate, 2% dextrose supplemented with amino acids and bases required for growth of auxotrophic strains. HeLa cells were grown in DMEM high glucose with l-glutamine and sodium pyruvate and supplemented with 10% FBS and pen-strep. The Chembridge Diverset E library was used for CSL screens; compound O3, G3, 7701175, and initial samples of A5 were purchased from Chembridge. Initial NMR analysis suggested commercial A5 to be a hydrochloride salt. We generated both the neutral and hydrochloride salt forms and observed that commercial A5 was identical to the synthetic sample of A5-HCl by gas chromatography and NMR analysis (SI Text). Both A5 and A5-HCl demonstrated the activities of commercial A5; however, activity levels of neutral A5 were more stable (data not shown). For experiments in Table 1 and Fig. 2b, A5-HCl was used; for all other experiments, neutral A5 was used.
Screening and CSL Phenotypic Analysis.
For initial compound screening, overnight cultures of MDY326 and MDY327 were diluted to OD600 0.00001 and 0.0003 in YPD, and 30 μl of each were transferred into 384-well plates (Nunc, Rochester, NY). Compounds were transferred from 10-mM stocks by using sterile replica pinners (Genetix, Boston, MA), resulting in approximate screening dosage of 30 μM. Plates were incubated for 18–24 h at 30°C in a humidified chamber. Plates were sealed with Costar sealing tape (Fisher Scientific, Pittsburgh, PA) and vortexed vigorously; seals were removed and absorbance was read at OD490 with a Spectramax 340PC Spectrophotometer. Absorbance was normalized to untreated wells in each plate. For CSL analysis, MDY326, MDY327, MDY330, MDY334, MDY335, SEY6210, or GPY3431 was diluted to 0.0003 in YPD. For ccfw sensitivity determination, MDY335 was diluted into YPD with 60 μM calcofluor white. For dosage determinations, compounds were serially diluted in cultures to obtain the final concentration in 30-μl samples, and cells were grown and analyzed as for compound screening.
Immunoprecipitation and Immunoblotting.
For pulse–chase immunoprecipitation of Ste3p and α-factor maturation assays, cells were grown at 30°C to an OD600 of 0.2 in YPD supplemented with DMSO or compounds. Cells were washed twice with SCMG with 50-μM compound and then 50 μCi (1 Ci = 37 GBq) [35S]methionine (Perkin–Elmer, Waltham, MA) per OD600 was added for labeling. Chase was initiated at 10 min by pelleting the cells and resuspending at 2 OD600 per ml in YPD. At time points indicated for Ste3p and at 30 min of chase for α-factor maturation, 0.5 OD600 in 200 μl were transferred to tubes containing sodium fluoride and sodium azide (10 mM final). Immunoprecipitation of α-factor and cell lysis and immunoprecipitation of Ste3p were performed as described (31, 32). For pulse–chase immunoprecipitations of ALP, CPS, and α-factor, cells were treated as above, except that in the case of ALP cells were labeled for 5 min and in all cases the chase was initiated by addition of 1:10 volume of 2% yeast extract, 0.3% cysteine, and methionine. Immunoprecipitation of CPS, α-factor, and ALP has been described (18, 30).
Microscopy.
For visualization of chitin bud-scars in yeast, MDY336 cells were grown as described for compound screening. Fifteen microliters of cells were mixed with 15 μl of 2.5 mM calcofluor white in YPD and incubated for 10 min. Cells were washed twice with 1 ml of YPD and visualized by fluorescence microscopy. For AP-1 immunofluorescence, cells were fixed in 3% formaldehyde and then incubated with 1:500 dilution of anti-AP-1 antibody 100/3 (Abcam, Cambridge, MA) in 0.44 mg/ml BSA and 0.22% Saponin for 1 h. Cells were washed with PBS followed by a 60-min incubation with 1:500 dilution of Alexa Fluor 594-conjugated goat anti-mouse antibody (Molecular Probes, Invitrogen, Carlsbad, CA). Cells were mounted in ProLong Gold (Invitrogen) and visualized on a Zeiss Axiovert 200M Microscope with a 9100-CCD camera (Hamamatsu, Hamamatsu City, Japan) for HeLa cells and an ORCA II camera (Hamamatsu) for yeast cells. Image analysis was performed on images exported to the .lsm format from Axiovision (Zeiss, Thornwood, NJ). Metamorph software (Zeiss) was used to generate intensity histograms. Cell area was defined as pixels at or above 224, and high intensity was defined as pixels at or above 704. Cell area and pixel intensity were computed for individual image frames usually containing four to eight cells. A total of 70 frames for each treatment were analyzed, and the number of frames in which overall cell area was occupied by given intensity intervals was plotted.
Supplementary Material
Acknowledgments
We thank Dr. Caroline Shamu and the staff at the Institute of Chemistry and Cell Biology, Harvard Medical School, for earlier screens and their assistance in developing screening protocols. Screening was performed at the University of California, Los Angeles (UCLA) Molecular Screening Shared Resource with the help of Joseph Rogers. We thank Todd Lorenz (UCLA) for generation of the chc1-ts allele and Vikram Anand (UCLA) for assistance with ALP assays. HeLa cells were a gift of Alex Van Der Bliek (UCLA). We thank Tom Kirchhausen and members of the J.H. and G.S.P. laboratories for helpful discussions. This work was supported by National Institutes of Health (NIH) Grant GM67911 (to G.S.P.) and NIH National Research Service Award DK062608 (to M.C.D.).
Abbreviations
- TGN
trans-Golgi Network
- ccfw
calcofluor white
- CSL
composite synthetic lethal
- CPS
carboxyl peptidase S
- ALP
alkaline phosphatase
- AP-1
adaptor protein-1 complex
- GGA
Golgi-localized
- γear-containing
ARF-binding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607773104/DC1.
References
- 1.Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- 2.Meyer C, Zizioli D, Lausmann S, Eskelinen EL, Hamann J, Saftig P, von Figura K, Schu P. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folsch H, Pypaert M, Schu P, Mellman I. J Cell Biol. 2001;152:595–606. doi: 10.1083/jcb.152.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roeth JF, Collins KL. Microbiol Mol Biol Rev. 2006;70:548–563. doi: 10.1128/MMBR.00042-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinners I, Tooze SA. J Cell Sci. 2003;116:763–771. doi: 10.1242/jcs.00270. [DOI] [PubMed] [Google Scholar]
- 6.Huang J, Zhu H, Haggarty SJ, Spring DR, Hwang H, Jin F, Snyder M, Schreiber SL. Proc Natl Acad Sci USA. 2004;101:16594–16599. doi: 10.1073/pnas.0407117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guarente L. Trends Genet. 1993;9:362–366. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- 8.Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 9.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 10.Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G. PLoS Genet. 2005;1:e24. doi: 10.1371/journal.pgen.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, et al. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 12.Brown JA, Sherlock G, Myers CL, Burrows NM, Deng C, Wu HI, McCann KE, Troyanskaya OG, Brown JM. Mol Syst Biol 2. 2006;2006:0001. doi: 10.1038/msb4100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costaguta G, Stefan CJ, Bensen ES, Emr SD, Payne GS. Mol Biol Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonifacino JS. Nat Rev Mol Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- 15.Cabib E, Roh DH, Schmidt M, Crotti LB, Varma A. J Biol Chem. 2001;276:19679–19682. doi: 10.1074/jbc.R000031200. [DOI] [PubMed] [Google Scholar]
- 16.Chuang JS, Schekman RW. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valdivia RH, Baggott D, Chuang JS, Schekman RW. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- 18.Payne GS, Schekman R. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- 19.Yeung BG, Phan HL, Payne GS. Mol Biol Cell. 1999;10:3643–3659. doi: 10.1091/mbc.10.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis NG, Horecka JL, Sprague GF., Jr J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowles CR, Odorizzi G, Payne GS, Emr SD. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- 22.Stepp JD, Huang K, Lemmon SK. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zouhar J, Hicks GR, Raikhel NV. Proc Natl Acad Sci USA. 2004;101:9497–9501. doi: 10.1073/pnas.0402121101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ooi SL, Pan X, Peyser BD, Ye P, Meluh PB, Yuan DS, Irizarry RA, Bader JS, Spencer FA, Boeke JD. Trends Genet. 2006;22:56–63. doi: 10.1016/j.tig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Kaelin WG. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 26.Rad MR, Phan HL, Kirchrath L, Tan PK, Kirchhausen T, Hollenberg CP, Payne GS. J Cell Sci. 1995;108:1605–1615. doi: 10.1242/jcs.108.4.1605. [DOI] [PubMed] [Google Scholar]
- 27.Pishvaee B, Munn A, Payne GS. EMBO J. 1997;16:2227–2239. doi: 10.1093/emboj/16.9.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bensen ES, Costaguta G, Payne GS. Genetics. 2000;154:83–97. doi: 10.1093/genetics/154.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Costaguta G, Duncan MC, Fernandez GE, Huang GH, Payne GS. Mol Biol Cell. 2006;17:3907–3920. doi: 10.1091/mbc.E06-05-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan HL, Finlay JA, Chu DS, Tan PK, Kirchhausen T, Payne GS. EMBO J. 1994;13:1706–1717. doi: 10.1002/j.1460-2075.1994.tb06435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan PK, Davis NG, Sprague GF, Payne GS. J Cell Biol. 1993;123:1707–1716. doi: 10.1083/jcb.123.6.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.