Abstract
Numerous mutations in the cystic fibrosis (CF) transmembrane conductance regulator (CFTR, a Cl− channel) disrupt salt and fluid transport and lead to the formation of thick mucus in patients' airways. Obstruction by mucus predisposes CF patients to chronic infections and inflammation, which become gradually harder to control and eventually fatal. Aggressive antibiotic therapy and supportive measures have dramatically lengthened CF patients' lives. Here, we report that sphingomyelinases (SMase) from human respiratory pathogens strongly inhibit CFTR function. The hydrolysis of sphingomyelin by SMase makes it more difficult to activate CFTR by phosphorylation of its regulatory domain. By inhibiting CFTR currents, SMase-producing respiratory tract bacteria may not only aggravate pulmonary infection in some CF patients but may also elicit a condition, analogous to CFTR deficiency, in non-CF patients suffering from bacterial lung infection.
Keywords: β-hemolysin, cystic fibrosis, lung infection, phospholipid, sphingomyelinase
Genetic defects in the cystic fibrosis (CF) transmembrane conductance regulator (CFTR) Cl− channel (1–5) can lead to a reduction in Cl− flux and fluid movement into the airways, thickening mucus and causing airway obstruction. CF patients typically have a strong predisposition to bacterial infection and, perhaps as a consequence, have inflamed and fibrosed lungs.
The CFTR Cl− channel is activated when its R (regulatory) domain is phosphorylated by cAMP-dependent protein kinase A (PKA) (6). Activation is generally assayed from the Cl− current that flows through activated channels in voltage–clamp experiments. The R domain, in the middle of the CFTR peptide chain, contains many phosphorylation sites. Activation increases with phosphorylation up to a point, but phosphorylation of all of the sites is not necessary for full activation of current (7). If the N- and C-terminal peptides formed from genetic excision of the R domain are covalently linked together, the resulting single peptide forms a Cl− channel that is constitutively active but with very low open probability (Po) (8). If, on the other hand, the N- and C-terminal peptides are expressed as separate transcripts in Xenopus oocytes, the two unlinked peptides form a channel that is constitutively active with much higher Po but not as high as the fully activated wild-type channel (Po = 0.13 vs. 0.36) (9). The channel formed by the unlinked N- and C-terminal peptides will be referred to as CFTR-ΔR (see ref. 10 for CFTR-ΔR heterologously expressed in the Chinese hamster ovary cell line).
As described below, a screen of venoms from small invertebrates led us to discover that, besides activating voltage-gated K+ channels (11), sphingomyelinase (SMase) D from the Brown spider venom suppresses the Cl− current that flows through the activated CFTR. This discovery, in turn, led to the finding that SMases from respiratory tract bacteria also profoundly suppress CFTR current.
Results
Discovery and Identification of CFTR-Inhibiting Activity in Brown Spider Venom.
We screened venoms from >100 invertebrates and found those from Loxosceles reclusa (Brown spider) and Loxosceles arizonica to have CFTR-inhibiting activity. An example with Brown spider venom is shown in Fig. 1A. Current (assayed during voltage steps) was activated by elevating intracellular cAMP with 0.1 mM isobutylmethylxanthine (IBMX) (a phosphodiesterase inhibitor) and was dramatically reduced on addition of the venom. The venom also inhibited current through the constitutively active mutant CFTR-ΔR channel, as shown in Fig. 1B.
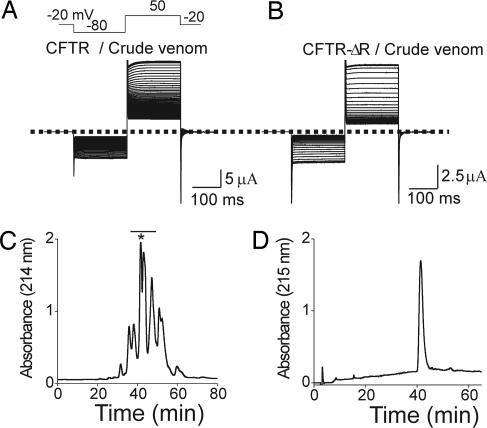
Fig. 1.
Identification of CFTR-inhibitory activity. (A and B) IBMX (0.1 mM)-activated CFTR (A) and constitutively active CFTR-ΔR (B) currents elicited at 3-s intervals (only every fifth trace is shown) with the voltage protocol shown and gradually disappearing after venom addition (1:3,000 dilution). The dotted line indicates zero current. (C) Size-exclusion chromatography of 10 μl of venom; the bar identifies active fractions. (D) The asterisk-marked peak in C was further purified on a reverse-phase column, which yielded a major peak containing the activity.
To isolate CFTR-inhibiting activity, we subjected 10 μl of Brown spider venom to size-exclusion FPLC, which yielded several active fractions (Fig. 1C). The most abundant fraction was subjected to reverse-phase HPLC (Fig. 1D), and the large peak obtained was run on SDS/PAGE. The single band from SDS/PAGE was digested into fragments with trypsin. Of these fragments, 9 were identified by liquid chromatography tandem MS (HPLC coupled with tandem MS) as emanating from the Lr1 isoform of SMase D and 12 from the Lr2 isoform [supporting information (SI) Fig. 8] (12). The observed masses of the active materials matched the theoretical masses of the two isoforms (for Lr1, 31.2 kDa theoretical and 31.2 kDa observed; and for Lr2, 31.4 kDa theoretical and 31.3 kDa observed).
To confirm that the CFTR-inhibiting activity of the venom was from SMase D, we produced a recombinant version of Lr2SMase D using the Escherichia coli expression system (see Materials and Methods). The recombinant Lr2SMase D inhibited currents through both wild-type CFTR and CFTR-ΔR (Fig. 2 A–D). To be certain that Lr2SMase D rather than extraneous material in the bacterial product was responsible, two histidine residues known to be essential to the enzymatic activity of SMase D were replaced by alanine (13). This mutant of Lr2SMase D had no effect on CFTR current (Fig. 3A), confirming that the observed CFTR inhibition by the venom was caused by Lr2 and/or other isoforms of SMase D. A further test, consistent with this conclusion, is that exposure of Lr2SMase D to low pH, a procedure known to inhibit or inactivate the enzyme (13, 14), eliminated its ability to inhibit CFTR current (Fig. 3B).
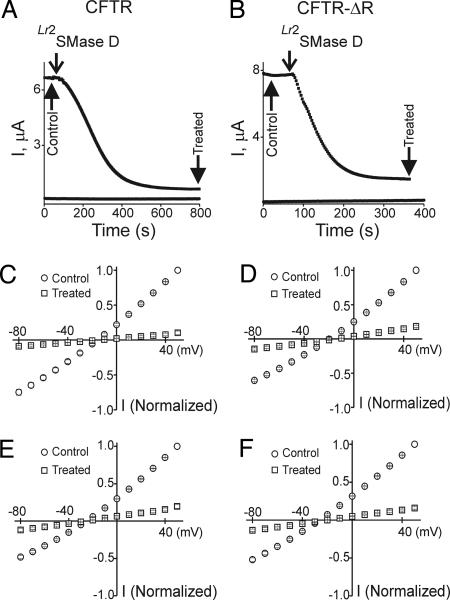
Fig. 2.
Inhibition of CFTR and CFTR-ΔR currents by recombinant spider and bacterial SMase D. (A and B) Plots of currents elicited by repeated (0.33-Hz) pulses to 50 mV (see Fig. 1A) against time. Top traces are from oocytes injected with cRNA of CFTR (A) or CFTR-ΔR (B); lower traces (A and B) were obtained from uninjected oocytes. In all four cases, recombinant Lr2SMase D was added at the time indicated (8 ng/μl was used throughout the study). Current–voltage (I–V) relations were collected at the beginning (control) and end (treated) of six similar recordings for each case. (C–F) Normalized I–V relations of CFTR (C and E) and CFTR-ΔR (D and F) before and after treatment with recombinant Lr2SMase D (C and D) or CpSMase D (8 ng/μl; E and F). Data shown are presented as mean ± SEM (n = 6); error bars are generally smaller than the symbols.
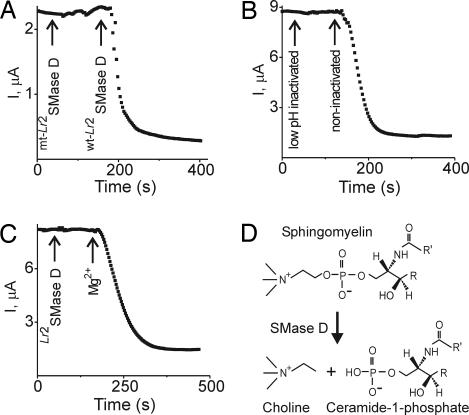
Fig. 3.
Effects of histidine mutations, pH, and Mg2+ on SMase D activity. Shown are time courses of CFTR-ΔR currents. (A) Arrows indicate successive addition of SMase D with or without the H11A and H47A mutations. (B) Successive addition of SMase D that was or was not pretreated with 0.1% trifluoroacetic acid (pH <3) for14 h. (C) No Mg2+ was present in the bath during the first ≈3 min. Arrows indicate successive additions of SMase D and 1 mM Mg2+. For each of the three cases (A–C), similar results were observed in three to four experiments. (D) Reaction scheme of SMase D-catalyzed sphingomyelin hydrolysis. R and R′ represent acyl chains.
Does SMase D inhibit CFTR current by virtue of its lipase activity, as suggested by the above, or by directly binding to CFTR? Irreversible enzymatic hydrolysis is suggested by the fact that no recovery occurred after washout of Lr2SMase D for 30 min (Fig. 4A). Also, the active site of SMase D contains a Mg2+ ion (15) and a bound Mg2+ is essential for lipase activity. The CFTR-inhibiting effect of Lr2SMase D, like its lipase activity, requires Mg2+ (Fig. 3C). Altogether, these observations strongly suggest that the lipase activity of SMase D is important in suppressing CFTR current. Are the products of sphingomyelin hydrolysis responsible for the inhibition? The enzyme catalyzes removal of the choline group of sphingomyelin (which is naturally present mainly in the outer leaf of the membrane bilayer). The hydrolysis products are choline and ceramide-1-phosphate (16, 17) (Fig. 3D). Acute addition of either to the bath solution (with ceramide-1-phosphate predissolved in methanol/dodecane) caused no significant inhibition of CFTR-ΔR current (Fig. 4 B and C). Acute addition of sphingomyelin (predissolved in methanol) failed to restore the current after SMase D treatment (Fig. 4D).
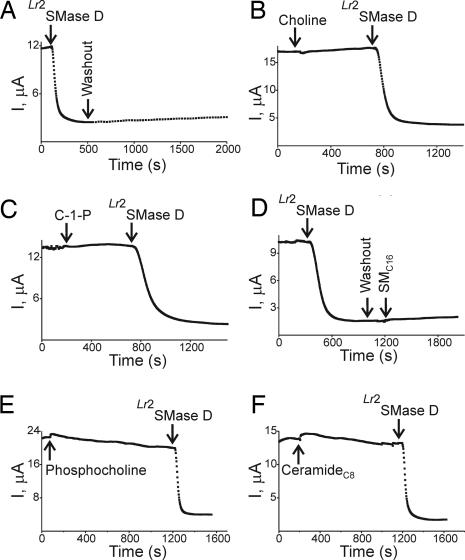
Fig. 4.
Effects of sphingomyelin and its hydrolysis products on CFTR-ΔR currents. (A) Arrows indicate addition and washout of SMase D (after 500 s, the data were collected at 30-s intervals instead of 3 s). (B) Arrows indicate successive additions of choline (10 mM) and Lr2SMase D. (C) Arrows indicate successive additions of ceramide-1-phosphate (C-1-P; 0.25 mg/ml; the stock was in 49:1 methanol/dodecane) and Lr2SMase D [the bath solution contained 2% of methanol/dodecane (49:1)]. (D) Arrows indicate successive addition and washout of Lr2SMase D and addition of sphingomyelin (C16-SM; 0.5 mg/ml; stock was in methanol), where the bath solution contained 2% methanol. (E) Arrows indicate successive additions of phosphocholine (1 mM) and Lr2SMase D. (F) Arrows indicate successive additions of ceramide-C8 (0.25 mg/ml; the stock was in ethanol) and Lr2SMase D, where the bath solution contained 2% ethanol. Similar results were observed in at least three experiments for each case. Lr2SMase D was used as a positive control for CFTR inhibition in B, C, E, and F. Concentrations of all chemical and lipid stocks are 50 times the final.
Inhibition of CFTR Currents by Bacterial SMase D.
The bacterial pathogen Corynebacterium pseudotuberculosis produces CpSMase D that has ≈30% amino acid identity to SMase D from spider venom (18–20). Recombinant CpSMase D, like Lr2SMase D (Fig. 2 C and D), inhibited currents through both CFTR and CFTR-ΔR (Fig. 2 E and F). The respiratory pathogen Arcanobacterium haemolyticum also produces SMase D, an extracellular product (19–21). Significantly, infection with this bacterium is invariably associated with respiratory symptoms (21).
Inhibition of CFTR Currents by Bacterial SMase C.
The genome of many clinically important pathogenic respiratory tract bacteria, including Bacillus anthracis and Staphylococcus aureus, encodes SMase C instead of SMase D (22–24). SMase C hydrolyzes sphingomyelin to phosphocholine and ceramide (25) rather than to choline and ceramide-1-phosphate (Fig. 5A). The role of SMase C in infection remains largely unknown, in part because studies with “purified” native bacterial SMase C have been notoriously difficult to interpret due to the presence of additional pathogenic factors (26), including other phospholipases. For example, when we analyzed native SaSMase C (from S. aureus) and BcSMase C (from Bacillus cereus) that we obtained from a common commercial source, impurity of the former was indicated by numerous bands on SDS/PAGE, and of the latter by two bands on SDS/PAGE and many peaks in mass spectrum (SI Fig. 9, where the molecular mass of SMase C is 30–35 kDa).
Fig. 5.
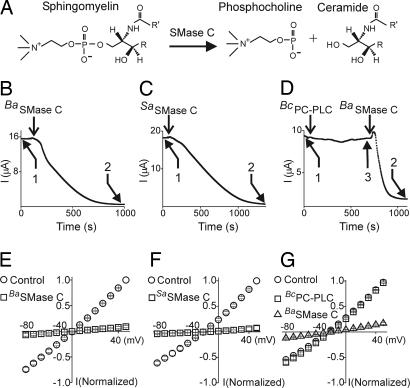
Inhibition of CFTR currents by SMase C and PC-PLC. (A) Reaction scheme of SMase C-catalyzed sphingomyelin hydrolysis. (B–D) CFTR currents at 50 mV plotted against time, before and after addition (arrows) of recombinant BaSMase C (B) or SaSMase C (C) (40 ng/μl was used throughout the study) or “purified” native BcPC-PLC bound with Zn2+ (50 ng/μl) (D). I–V relations for control (1), BaSMase C/SaSMase C (2) or BcPC-PLC (3) were collected from six similar recordings in each case. (E–G) Normalized I–V relations before and after BaSMase C, SaSMase C or BcPC-PLC. SMase C was used as a positive control for CFTR inhibition in D and G. Data shown are mean ± SEM (n = 6); error bars are generally smaller than the symbols.
To circumvent the problem of impurity, we produced recombinant SMase C with cDNAs that we cloned from B. anthracis and S. aureus. Recombinant SMase C from either bacterium inhibited currents through both CFTR (Fig. 5 B, C, E, and F) and CFTR-ΔR (SI Fig. 10 A and B). The inhibitory effect required the presence of Mg2+ (SI Fig. 10 C and D) (27), as did inhibition with Lr2SMase D (Fig. 3C). Acute addition of the SMase C hydrolysis products phosphocholine or ceramide caused no significant current inhibition (Fig. 4 E and F).
The specificity of SMases for sphingomyelin over phosphatidylcholine (PC) is almost certainly not absolute, which raises the question of whether the observed effect of SMases on CFTR might result from enzymatic hydrolysis of PC, which is a major type of phospholipid in the outer leaf of the membrane. We tested this possibility with a phospholipase C (PLC) from B. cereus, which (relatively) specifically removes phosphocholine from PC (BcPC-PLC). We found no significant inhibition of CFTR-ΔR current after addition of the enzyme (Fig. 5 D and G).
Inhibition of Natural CFTR Mutants by SMases.
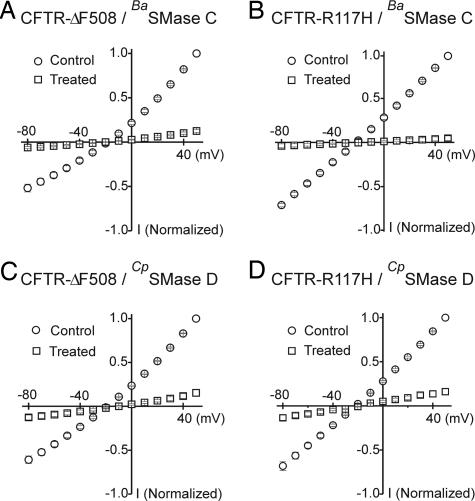
Approximately 70% of CF cases are caused by deletion of residue F508 (CFTR-ΔF508) (4, 5). The mutation impairs folding and trafficking of the protein but, once inserted into the plasmalemma, the ΔF508 mutant channel exhibits robust activity comparable to that of wild type (28). It is unclear whether a small but functionally important amount of CFTR-ΔF508 is present in the airway epithelial cells of these CF patients. In contrast, the product of CFTR-R117H (29), another relatively common mutant, folds properly, is well transported to the cytoplasmic membrane in affected patients but is only partially functional. SMases C and D inhibited currents through both ΔF508 and R117H mutant CFTR channels (Fig. 6).
Fig. 6.
Inhibition of disease-causing CFTR mutants by bacterial SMases C and D. Normalized I–V relations of ΔF508 (A and C) or R117H (B and D) mutants before and after exposure to BaSMase C (A and B) or CpSMase D (C and D). Data shown are mean ± SEM (n = 6); error bars are generally smaller than the symbols. The I–V curves were collected as described Fig. 2.
Dependence of SMase-Induced Inhibition on CFTR Phosphorylation Level.
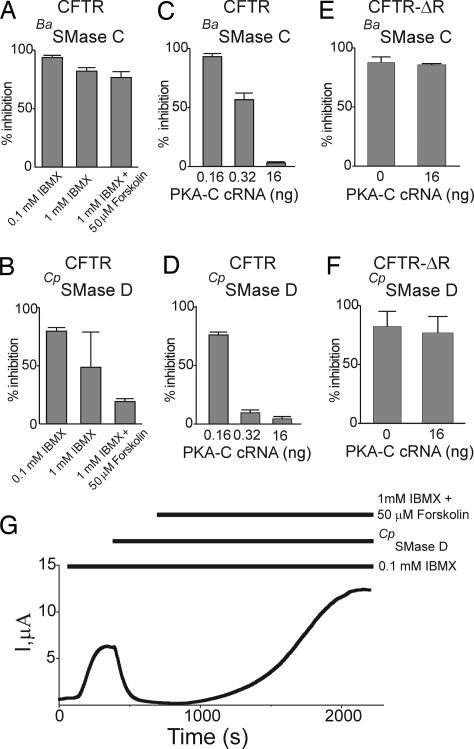
We initially used 0.1 mM IBMX to elevate intracellular cAMP of oocytes. This treatment activated CFTR current to 30–40% of maximum, and SMases typically inhibited 80–90% of the activated current (Fig. 7A and B). A higher IBMX concentration (1 mM) activated more CFTR current (40–90% of maximum), but the extent of inhibition by SMase D became highly variable (Fig. 7B). To test the likely possibility that this variability reflects different cAMP levels in individual oocytes, we further boosted the cAMP level with the combination of 1 mM IBMX and 50 μM forskolin (an adenylate cyclase stimulator) to maximally activate the CFTR channel (9). Under this condition, SMase C (which removes both choline and phosphoryl groups) inhibited 75% of the current (Fig. 7A), whereas SMase D (which removes only the choline group) inhibited only 20% (Fig. 7B), as investigated further below. SMase D inhibition is reversible and can be overcome by high cAMP concentration. In Fig. 7G, CTFR current activated by 0.1 mM IBMX was almost completely inhibited by addition of SMase D; raising the cAMP level with 1 mM IBMX plus 50 μM forskolin not only restored the current but also boosted it to about twice the original level. Such reactivation of SMase D-suppressed current demonstrates that the enzymatic treatment does not render the CFTR channel irreversibly inactive.
Fig. 7.
Dependence of SMase inhibition of CFTR and CFTR-ΔR currents on PKA activity. (A and B) Percent inhibition by BaSMase C and CpSMase D of CFTR current (at 50 mV; mean ± SEM, n = 4–12) activated by 0.1 mM, 1 mM, or 1 mM IBMX plus 50 μM forskolin. (C and D) Percent inhibition of CFTR current by BaSMase C and CpSMase D, plotted against the amount of PKA-C cRNA injected per oocyte (mean ± SEM, n = 3–6). (E and F) Percent inhibition of CFTR-ΔR current by BaSMase C (E) or CpSMase D (F) (at 50 mV; mean ± SEM, n = 4 - 5) with and without coinjection of PKA-C cRNA (16 ng per oocyte). (G) Record of currents elicited by pulses to 50 mV in an oocyte injected with CFTR cRNA, after applications (horizontal bars) of 0.1 mM IBMX, 8 ng/μl CpSMase D, and 1 mM IBMX plus 50 μM forskolin. Similar results were observed in five individual oocytes.
To check whether the antagonistic effect of cAMP on SMase inhibition results from PKA-mediated phosphorylation, we coexpressed CFTR with the catalytic subunit of PKA (PKA-C), which has constitutive kinase activity independent of cAMP. Coinjection of PKA-C and CFTR cRNAs induced robust CFTR current, whereas the background non-CFTR current after PKA-C cRNA injection alone remained minimal (SI Fig. 11). A 0.32-ng dose of PKA-C cRNA produced practically maximal current: the current only increased 5 ± 1% (mean ± SEM; n = 3) on addition of 1 mM IBMX plus 50 μM forskolin. As expected, the ability of SMase to inhibit CFTR current decreased with increasing amounts of coinjected PKA-C cRNA (Fig. 7 C and D). A relatively high dose of PKA-C cRNA was needed to overcome the inhibition caused by BaSMase C, a result, which parallels the finding above that SMase C inhibition was more difficult to overcome with a boost of cAMP than was SMase D inhibition (Fig. 7 A and B).
These findings strongly suggest that overcoming SMase inhibition requires a higher level of R domain phosphorylation. This idea is supported by our finding that SMase inhibition of CFTR-ΔR (which has no phosphorylatable R domain) was not significantly affected by coexpression of PKA-C (Fig. 7 E and F). This finding is consistent with the idea that the open state of CFTR-ΔR expressed in an oocyte is not as stable as that of the wild-type channel “fully” activated by PKA (9).
Discussion
SMase inhibition of CFTR current results from enzymatic hydrolysis of sphingomyelin, which makes it more difficult to activate CFTR by phosphorylation of the R domain. Higher levels of phosphorylation by PKA can overcome the inhibitory effect when the R domain is present but not in its absence (Fig. 7). SMase cleaves sphingomyelin into a small piece (choline or phosphocholine) and a large one (ceramide-1-phosphate or ceramide). The large piece has two long hydrophobic chains and, like sphingomyelin, has a very strong preference to remain within the membrane. In principle, the sphingomyelin hydrolysis products might be the active inhibitors. The highly water-soluble hydrolysis products phosphocholine (of SMase C) or choline (of SMase D) would easily diffuse away after hydrolysis and, were they responsible for inhibition, it would be transient. Furthermore, in Fig. 4, we observed no significant inhibition of CFTR-ΔR current after acute addition of choline or phosphocholine. The poorly water-soluble products ceramide (of SMase C) and ceramide-1-phosphate (of SMase D) would be unlikely to leave the membrane. A persistent attachment of either or both lipid hydrolysis products to CFTR (at least for the duration of the experiment) is the simplest logical explanation for the different percentage of current inhibition caused by SMase C and SMase D (Fig. 7 A and B). This would then imply that sphingomyelin complexed to CFTR promotes gating of the channel in a way that cannot be duplicated by ceramide or ceramide-1-phosphate, with ceramide being less capable. If this is true, the fact (30, 31) that a CFTR channel reconstituted in a (nonsphingomyelin) phospholipid bilayer exhibits relatively high Po must mean either that the reconstituted CFTR channel retains its native sphingomyelin, and/or that other phospholipids with a suitable head group can also promote CFTR gating.
Many respiratory pathogens have SMase activity. For example, S. aureus is a major respiratory threat for CF patients (>50% infection rate), causing recurring infection and inflammation that significantly impair lung function (32). S. aureus also causes severe pneumonia and other types of infection in non-CF patients (33). SMase C of S. aureus was originally called β-hemolysin for its hemolytic effect on a sheep blood agar culture (34, 35). The specific role of SMase C in the pathogenesis of respiratory infections remains largely unknown. Pseudomonas aeruginosa is another major offender. It permanently colonizes the airways of virtually all late-stage CF patients and secretes a phospholipase (termed PLC-H) that can hydrolyze sphingomyelin (36).
Although CF disease undoubtedly originates from a genetic defect in CFTR, the severity of the pulmonary disease does not correlate well with genotype (37). The lung damage in CF patients is not specific, i.e., its histopathology is not so different from that seen in some other types of chronic pulmonary infection/inflammation. The fibrosis seen in CF, although very severe, is not fundamentally different from that after severe tissue damage from a variety of other causes. Significantly, aggressive antibiotic treatments and supportive measures have recently increased the median life span of CF patients from 5 to 37 years (38). These observations and arguments support the notion that CFTR defects predispose the patients to bacterial infection that in turn plays the pivotal role in pathogenesis.
Although the pathogenesis of pulmonary injuries caused by chronic bacterial infection and inflammation remains unresolved, the following facts compellingly suggest that SMases play a critical role. First, as described above, bacterial SMases C and D inhibit CFTR current, an action leading to the production of thick mucus that clogs the airways and thereby fosters further bacterial growth. Second, SMase D causes severe tissue necrosis (17, 39). Last, ceramide and ceramide-1-phosphate induce inflammation and/or trigger cell death (40–43).
Most natural strains of B. anthracis produce little SMase C, partly because of a deletion in the gene encoding a key pleiotropic regulator (44, 45). The live STI-1 (Sanitary Technical Institute) vaccine provides effective protection against these strains (46). However, once B. anthracis is engineered with genes from closely related B. cereus to express high levels of SMase C and PC-PLC, the vaccine becomes ineffective (44). Effective protection can, in principle, be achieved by new vaccines and/or antitoxins, including specific phospholipase inhibitors.
In summary, reduction of CFTR activity in CF patients leads to thick mucus that clogs airways and thus fosters infection. Infection with bacteria that produce SMase activity would further suppress CFTR activity. Inhibition probably arises because ceramide and ceramide-1-phosphate cannot substitute for sphingomyelin in some action that promotes CFTR gating, which is normally initiated primarily by phosphorylating the R domain. Independent of that, the lipid products of SMase-catalyzed sphingomyelin hydrolysis, ceramide, and ceramide-1-phosphate further aggravate the disease by triggering inflammation and cell death (40–43). These adverse effects associated with SMase activity may be important contributors to the pathogenicity of bacterial infection in both CF and non-CF patients. Although correcting the genetic defects in CFTR remains the ideal cure for CF disease, application of medicines against various bacterial virulence factors, including phospholipases and perhaps proteases, in conjunction with other effective measures, might be a viable near-term approach to improving length and quality of life for many CF patients. The same approach might also benefit patients with other types of bacterial infection.
Materials and Methods
Molecular Biology and Electrophysiological Recordings.
The CFTR, CTFR-ΔR (9), and PKA-C cDNAs were subcloned in the pGEMHE plasmid (47) (see acknowledgments for the source of all cDNA constructs). The ΔF508 and R117H mutant CFTR cDNAs were obtained through PCR-based mutagenesis and confirmed by DNA sequencing. The cRNAs were synthesized with T3 or T7 polymerase using the corresponding linearized cDNAs as templates. Channel currents were recorded from whole oocytes previously injected with the corresponding cRNAs and stored at 18°C, using a two-electrode voltage–clamp amplifier (Warner OC-725C). Background leak currents were not corrected for. The resistance of electrodes filled with 3 M KCl was 0.2–0.3 MΩ. Unless specified otherwise, the bath solution contained: 95 mM Na+ (Cl− + OH−), 5 mM KCl, 0.3 mM CaCl2, 1 mM MgCl2 and 10 mM Hepes; pH was adjusted to 7.6 with NaOH. SMases (2–5 μl) were manually added to the recording chamber (100 μl). B. cereus PC-PLC was purchased from Sigma (St. Louis, MO).
Identification and Purification of Spider SMase D.
For purification, L. reclusa venom samples purchased from Spider Pharm (Yarnell, AZ) were loaded onto a size-exclusion FPLC column (Superdex G200, Amersham Biosciences), where the running buffer contained 50 mM NaCl and 5 mM Tris chloride (pH 6.4). The most abundant active fraction was subsequently loaded onto a reverse-phase HPLC column (C18, Beckman) and eluted with a water-acetonitrile gradient (1% per minute). Aqueous and organic mobile phases contained 0.1% and 0.07% trifluoroacetic acid, respectively. SMase D activity was drastically reduced after HPLC purification because of exposure to low pH. The sample corresponding to the large peak on the HPLC chromatogram contained the activity and was subsequently analyzed with MALDI-TOF MS for mass identification and also run on SDS/PAGE for further purification. The single visible Coomassie blue-stained band on SDS/PAGE was excised and digested with trypsin. The digestion products were then analyzed with liquid chromatography tandem MS, yielding 9 and 12 partial peptide sequences corresponding to Lr1 and Lr2 isoforms of SMase D (SI Fig. 8).
Molecular Cloning of SMase C.
Full length and “truncated” cDNAs of SMase C were produced with PCR, primed with a pair of oligonucleotides corresponding to the 5′ or 3′ translated regions against the genomic DNA isolated from B. anthracis (Sterne strain) and S. aureus ATCC 29213, respectively. The truncated cDNA of S. aureus was further extended to full length (22) with PCR.
Production of Recombinant SMases.
To produce the mature recombinant wild-type and mutant SMases, E. coli BL21 (DE3) cells were transformed with the respective cDNAs cloned into pET30 vector (Novagen, San Diego, CA), grown in LB medium to ≈0.6 OD at 600 nm, and induced with 1 mM isopropyl β-D-thiogalactoside for 2 h. The bacteria were harvested, resuspended, and sonicated. The resulting samples were loaded onto a cobalt affinity column and eluted by stepping the imidazole concentration from 50 to 500 mM (all SMase proteins contain N- and C-terminal His tags). The imidazole was later removed by dialysis.
Supplementary Material
Acknowledgments
We thank D. Gadsby (Rockefeller University, New York) for CFTR and CFTR-ΔR cDNAs in the pGEMHE vector; S. Billington (University of Arizona, Tucson) for CpSMase D cDNA; K. Lynch (University of Virginia, Charlottesville) for Lr2SMase D cDNA; H. Goldfine and Z. Wei for genomic DNA of B. anthracis and discussion; J. Weiser, A. Ratner, and M. Shchepetov (University of Pennsylvania, Philadelphia) for S. aureus and the genomic DNA isolation method; S. Taylor (University of California, San Diego) for PKA-C cDNA; C.-X. Yuan (University of Pennsylvania, Phildelphia) for liquid chromatography tandem MS sequencing; and C. Armstrong and P. De Weer (University of Pennsylvania, Philadelphia) for discussion and improving our manuscript. This study was supported by a grant from National Institute of General Medical Sciences (to Z.L.).
Abbreviations
- CF
cystic fibrosis
- CFTR
CF transmembrane conductance regulator
- SMase
sphingomyelinase
- R domain
regulatory domain
- PLC
phospholipase C
- PC
phosphatidylcholine
- IBMX
isobutylmethylxanthine
- PKA-C
catalytic subunit of PKA
- I–V
current–voltage.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701354104/DC1.
References
- 1.Knowles MR, Stutts MJ, Spock A, Fischer N, Gatzy JT, Boucher RC. Science. 1983;221:1067–1070. doi: 10.1126/science.6308769. [DOI] [PubMed] [Google Scholar]
- 2.Quinton PM. Nature. 1983;301:421–422. doi: 10.1038/301421a0. [DOI] [PubMed] [Google Scholar]
- 3.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 4.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 5.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, et al. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 6.Tabcharani JA, Chang XB, Riordan JR, Hanrahan JW. Nature. 1991;352:628–631. doi: 10.1038/352628a0. [DOI] [PubMed] [Google Scholar]
- 7.Cheng SH, Rich DP, Marshall J, Gregory RJ, Welsh MJ, Smith AE. Cell. 1991;66:1027–1036. doi: 10.1016/0092-8674(91)90446-6. [DOI] [PubMed] [Google Scholar]
- 8.Rich DP, Gregory RJ, Anderson MP, Manavalan P, Smith AE, Welsh MJ. Science. 1991;253:205–207. doi: 10.1126/science.1712985. [DOI] [PubMed] [Google Scholar]
- 9.Csanady L, Chan KW, Seto-Young D, Kopsco DC, Nairn AC, Gadsby DC. J Gen Physiol. 2000;116:477–500. doi: 10.1085/jgp.116.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bompadre SG, Ai T, Cho JH, Wang X, Sohma Y, Li M, Hwang TC. J Gen Physiol. 2005;125:361–375. doi: 10.1085/jgp.200409227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramu Y, Xu Y, Lu Z. Nature. 2006;442:696–699. doi: 10.1038/nature04880. [DOI] [PubMed] [Google Scholar]
- 12.Ramos-Cerrillo B, Olvera A, Odell GV, Zamudio F, Paniagua-Solis J, Alagon A, Stock RP. Toxicon. 2004;44:507–514. doi: 10.1016/j.toxicon.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Lynch KR. Biochem J. 2005;391:317–323. doi: 10.1042/BJ20050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Andrade SA, Murakami MT, Cavalcante DP, Arni RK, Tambourgi DV. Toxicon. 2006;47:380–386. doi: 10.1016/j.toxicon.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Murakami MT, Fernandes-Pedrosa MF, Tambourgi DV, Arni RK. J Biol Chem. 2005;280:13658–13664. doi: 10.1074/jbc.M412437200. [DOI] [PubMed] [Google Scholar]
- 16.Barksdale L, Linder R, Sulea IT, Pollice M. J Clin Microbiol. 1981;13:335–343. doi: 10.1128/jcm.13.2.335-343.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurpiewski G, Forrester LJ, Barrett JT, Campbell BJ. Biochim Biophys Acta. 1981;678:467–476. doi: 10.1016/0304-4165(81)90128-8. [DOI] [PubMed] [Google Scholar]
- 18.Bernheimer AW, Campbell BJ, Forrester LJ. Science. 1985;228:590–591. doi: 10.1126/science.3983643. [DOI] [PubMed] [Google Scholar]
- 19.McNamara PJ, Cuevas WA, Songer JG. Gene. 1995;156:113–118. doi: 10.1016/0378-1119(95)00002-n. [DOI] [PubMed] [Google Scholar]
- 20.Lipsky BA, Goldberger AC, Tompkins LS, Plorde JJ. Rev Infect Dis. 1982;4:1220–1235. doi: 10.1093/clinids/4.6.1220. [DOI] [PubMed] [Google Scholar]
- 21.Linder R. Emerg Infect Dis. 1997;3:145–153. doi: 10.3201/eid0302.970207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Projan SJ, Kornblum J, Kreiswirth B, Moghazeh SL, Eisner W, Novick RP. Nucleic Acids Res. 1989;17:3305. doi: 10.1093/nar/17.8.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, et al. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 25.Doery HM, Magnusson BJ, Cheyne IM, Sulasekharam J. Nature. 1963;198:1091–1092. doi: 10.1038/1981091a0. [DOI] [PubMed] [Google Scholar]
- 26.Titball RW. Microbiol Rev. 1993;57:347–366. doi: 10.1128/mr.57.2.347-366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ago H, Oda M, Takahashi M, Tsuge H, Ochi S, Katunuma N, Miyano M, Sakurai J. J Biol Chem. 2006;281:16157–16167. doi: 10.1074/jbc.M601089200. [DOI] [PubMed] [Google Scholar]
- 28.Drumm ML, Wilkinson DJ, Smit LS, Worrell RT, Strong TV, Frizzell RA, Dawson DC, Collins FS. Science. 1991;254:1797–1799. doi: 10.1126/science.1722350. [DOI] [PubMed] [Google Scholar]
- 29.Dean M, White MB, Amos J, Gerrard B, Stewart C, Khaw KT, Leppert M. Cell. 1990;61:863–870. doi: 10.1016/0092-8674(90)90196-l. [DOI] [PubMed] [Google Scholar]
- 30.Bear CE, Li CH, Kartner N, Bridges RJ, Jensen TJ, Ramjeesingh M, Riordan JR. Cell. 1992;68:809–818. doi: 10.1016/0092-8674(92)90155-6. [DOI] [PubMed] [Google Scholar]
- 31.Gunderson KL, Kopito RR. J Biol Chem. 1994;269:19349–19353. [PubMed] [Google Scholar]
- 32.Patient Registry Annual Data Report. Bethesda, MD: Cystic Fibrosis Foundation; 2004. [Google Scholar]
- 33.Kasper DL, Braunwald E, Fauci A, Hauser S, Longo D, Jameson JL. Harrison's Principles of Internal Medicine. New York: McGraw–Hill; 2004. [Google Scholar]
- 34.Glenny AT, Stevens NF. J Pathol Bacteriol. 1935;40:201–210. [Google Scholar]
- 35.Dinges MM, Orwin PM, Schlievert PM. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stonehouse MJ, Cota-Gomez A, Parker SK, Martin WE, Hankin JA, Murphy RC, Chen W, Lim KB, Hackett M, Vasil AI, et al. Mol Microbiol. 2002;46:661–676. doi: 10.1046/j.1365-2958.2002.03194.x. [DOI] [PubMed] [Google Scholar]
- 37.Pilewski JM, Frizzell RA. Physiol Rev. 1999;79:S215–S255. doi: 10.1152/physrev.1999.79.1.S215. [DOI] [PubMed] [Google Scholar]
- 38.Annual Report. Bethesda, MD: Cystic Fibrosis Foundation; 2005. [Google Scholar]
- 39.Rees RS, Nanney LB, Yates RA, King LE., Jr J Invest Dermatol. 1984;83:270–275. doi: 10.1111/1523-1747.ep12340340. [DOI] [PubMed] [Google Scholar]
- 40.Chalfant CE, Spiegel S. J Cell Sci. 2005;118:4605–4612. doi: 10.1242/jcs.02637. [DOI] [PubMed] [Google Scholar]
- 41.Chan C, Goldkorn T. Am J Respir Cell Mol Biol. 2000;22:460–468. doi: 10.1165/ajrcmb.22.4.3376. [DOI] [PubMed] [Google Scholar]
- 42.Thevissen K, François IE, Winderickx J, Pannecouque C, Cammue BP. Mini Rev Med Chem. 2006;6:699–709. doi: 10.2174/138955706777435643. [DOI] [PubMed] [Google Scholar]
- 43.Thon L, Mohlig H, Mathieu S, Lange A, Bulanova E, Winoto-Morbach S, Schutze S, Bulfone-Paus S, Adam D. FASEB J. 2005;19:1945–1956. doi: 10.1096/fj.05-3726com. [DOI] [PubMed] [Google Scholar]
- 44.Pomerantsev AP, Staritsin NA, Mockov Y, Marinin LI. Vaccine. 1997;15:1846–1850. doi: 10.1016/s0264-410x(97)00132-1. [DOI] [PubMed] [Google Scholar]
- 45.Slamti L, Perchat S, Gominet M, Vilas-Boas G, Fouet A, Mock M, Sanchis V, Chaufaux J, Gohar M, Lereclus D. J Bacteriol. 2004;186:3531–3538. doi: 10.1128/JB.186.11.3531-3538.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shlyakhov EN, Rubinstein E. Vaccine. 1994;12:727–730. doi: 10.1016/0264-410x(94)90223-2. [DOI] [PubMed] [Google Scholar]
- 47.Liman ER, Tytgat J, Hess P. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.