Abstract
The multiprotein Mediator coactivator complex is universally required for transcription of metazoan genes. It has been proposed to function by interfacing between transcriptional activators and the RNA polymerase II machinery. However, in vitro transcription systems reconstituted from homogeneous preparations of RNA polymerase II, the general transcription initiation factors, and the cofactor PC4 display relatively robust activator (HNF-4)-dependent activity, which, nonetheless, can be further stimulated by Mediator. By contrast, an unfractionated nuclear extract-based system in which Mediator has been immunodepleted displays a near-absolute dependence on ectopic Mediator. Here, we identified and purified an activity, MSA-2, that confers extract-like Mediator responsiveness to our reconstituted system. Mass spectrometric analyses identified its two constituent polypeptides as hSpt5 and hSpt4, which also comprise the elongation factor DSIF. Mechanistically, MSA-2/DSIF acts by restricting overall transcription in the pure system, thereby imposing a strong Mediator dependence. Our data thus point to potential mechanisms for Mediator function beyond its presently believed role in promoting the initial formation of the RNA polymerase II-containing preinitiation complex.
Keywords: DRB sensitivity-inducing factor, hepatocyte nuclear factor-4, positive cofactor 2, RNA polymerase II
RNA polymerase II (Pol II) and its associated general transcription factors (GTFs: TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) are universally required at all class II genes (1–3). These factors have the innate ability to relatively efficiently assemble into a preinitiation complex (PIC) at the core promoters of these genes and lead to basal levels of transcription, at least in vitro. Gene- and cell-type-specific regulators (activators), which transduce developmental and environmental signals to target genes, are thought to function by regulating the formation and function of the Pol II PIC (4).
Gene activation also entails the action of coactivators, especially the evolutionarily conserved Mediator (5, 6). The Mediator is a multiprotein complex that is recruited to promoters via the activators and functions by modulating PIC assembly and function through its potential to interact directly with Pol II (5, 7). Importantly, in contrast to chromatin coactivators, Mediator can facilitate DNA-templated transcription, and thus acts relatively late in the overall activation pathway on natural templates (8). Several lines of evidence suggest that Mediator function is manifested both at the level of the initial establishment of the PIC as well as its postrecruitment function (5, 7, 9–15).
Despite the critical requirement of Mediator for the expression of nearly all genes in yeast (16) and, potentially, metazoan cells, it has been puzzling for some time as to why in vitro systems reconstituted with homogeneous preparations of Pol II, general transcription factors (GTFs), and positive cofactor 4 (PC4) display relatively efficient activator-dependent (and basal) transcriptional activity (13, 17). Although Mediator can further stimulate this activity, the basis of the high Mediator-independent activity has remained unclear. On the other hand, upon immunodepletion of Mediator from unfractionated nuclear extract (which presumably more closely approximates the distribution of cellular factors), both activator-dependent and basal transcription are abolished and can be effectively restored upon provision of pure Mediator (10, 12, 14). These observations suggest that in the context of the extract, and thus the cell, activity of any PICs assembled in the absence of Mediator is subject to some restriction.
Here, we have used conventional chromatographic methods to purify an activity, MSA-2, that specifically restricts PIC function in the absence of the Mediator. This function is fully restored in the presence of the Mediator, indicating that Mediator dependency in the metazoan cell is imparted, at least in part, through factors that negatively modulate unregulated PIC function. That one such activity is shown here to reside in a complex containing hSpt5 and hSpt4, which were previously identified as components of the transcription elongation factor DRB sensitivity-inducing factor (DSIF) (18, 19), further indicates a close coupling of early events in transcription (initiation and postinitiation) and the role of Mediator in the regulation thereof.
Results
Differential Mediator- and TFIIB-Responsiveness of Crude and Pure in Vitro Assay Systems.
In prior studies, we have used two in vitro assays for demonstration of Mediator functionality (17). The original system was reconstituted from homogeneous preparations of Pol II, GTFs, and PC4. Purified Mediator [either intact TRAP/SMCC (20, 21) or PC2, the form that mainly lacks the TRAP240/MED13-TRAP230/MED12-CDK8-CycC subcomplex (13, 22)] could be shown to potentiate both basal and activated transcription. Similarly, in a Mediator-depleted nuclear extract system, Mediator dependence of basal and activated transcription was readily demonstrable (10, 12, 14, 17). Thus, whereas in the purified system the Mediator boosts transcription levels that are already quite high, in the cruder system the Mediator dependence is essentially absolute. Recent studies have further demonstrated that in the unfractionated extract, the role of Mediator in supporting basal transcription could be bypassed in the presence of a molar excess of TFIIB (23).
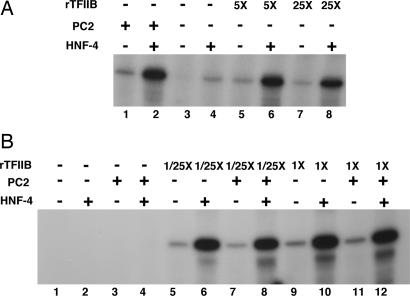
Toward identifying the basis of the differential Mediator requirements in the two assay systems, we first ascertained whether excess TFIIB also allows a Mediator requirement to be bypassed for activator-dependent transcription in the nuclear extract-based assay. For this purpose, Mediator-depleted HeLa cell nuclear extract was used to monitor transcription from a template that is responsive to the orphan nuclear receptor hepatocyte nuclear factor-4 (HNF-4) (14, 17). As described, both basal and HNF-4-driven transcription was strongly Mediator (PC2)-dependent (Fig. 1A, lanes 1–4). Supplementation with excess (5-fold) TFIIB (Fig. 1A, lanes 5–8), but not with TFIIE or TFIIF (data not shown), was almost as effective as PC2 in restoring both basal (Fig. 1A, lanes 7 and 5 vs. lane 1) and HNF-4-driven transcription (Fig. 1A, lanes 6 and 8 vs. lane 2). Thus, in a crude extract, excess TFIIB allows a Mediator requirement for activated transcription to be bypassed just as for basal transcription (23).
Fig. 1.
Dependence of transcription on Mediator and TFIIB. (A) In vitro transcription reactions contained Mediator-depleted nuclear extract. Both basal (odd numbered lanes) and HNF-4-driven (even numbered lanes) transcription from a cognate template was monitored. Reactions were supplemented with PC2 (lanes 1 and 2) or increasing amounts of recombinant TFIIB (rTFIIB; 50 ng, lanes 5 and 6; 250 ng, lanes 7 and 8). Control reactions showing residual transcription in Mediator-depleted extract to which neither Mediator nor TFIIB have been added were also included (lanes 3 and 4). (B) In vitro transcription reactions were reconstituted with recombinant TFIIA, TFIIE, TFIIF, and PC4, and affinity-purified TFIID, TFIIH, and Pol II; HNF-4 and PC2 were added as indicated. In addition, reactions contained either no TFIIB (lanes 1–4), 0.4 ng TFIIB (lanes 5–8), or 10 ng TFIIB (lanes 9–12).
To eliminate the possibility that inclusion of excess TFIIB in our reactions accounts for the absence of a strong Mediator dependency in our pure reconstituted system, we titrated down the amounts of TFIIB in our standard reactions (Fig. 1B). For this assay, we reconstituted reactions with pure preparations of Pol II, the GTFs (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH), and PC4 and evaluated basal and HNF-4 driven transcription as a function of PC2. Reactions were carried out with our usual complement of TFIIB (Fig. 1B, lanes 9–12) or with a 25-fold lower amount of this factor (Fig. 1B, lanes 5–8). As expected, the standard system supported both low level basal and strong HNF-4-dependent transcription (Fig. 1B, lane 10 vs. lane 9). However there was no additional stimulation by Mediator of either basal (Fig. 1B, lane 11 vs. lane 9) or HNF-4-dependent (Fig. 1B, lane 12 vs. lane 10) transcription under these conditions. Importantly, lowering the TFIIB concentration did not confer Mediator dependence for basal (Fig. 1B, lane 7 vs. lane 5) or HNF-4-driven (Fig. 1B, lane 8 vs. lane 6) transcription. Control experiments established that the assay system is nevertheless critically dependent on TFIIB (Fig. 1B, lanes 1–4). These results confirm that the purified system, despite being competent for activator-dependent transcription, may be deficient in some respects.
Identification of an Activity That Confers Mediator Dependency.
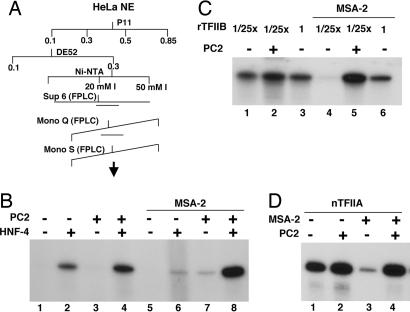
We tested various chromatographic fractions derived from HeLa nuclear extract for their ability to confer Mediator-responsiveness to our pure assay system (Fig. 2A). In a preliminary screen, we found that the phosphocellulose (P11) 0.1 M KCl flow-through fraction of the extract conferred significant Mediator-responsiveness to HNF-4-dependent transcription in our reconstituted system (data not shown). Subsequent fractionation over DE52 and Ni-NTA agarose (Fig. 2A) pointed to at least two chromatographically separable activities. One activity was largely stimulatory for both Mediator-independent and Mediator-dependent transcription and has not been characterized further. The other activity, designated MSA-2, most faithfully recapitulated the Mediator responsiveness that is observed in the context of the unfractionated extract (Fig. 2B). Thus, in its presence, Mediator-independent activation by HNF-4 was suppressed (Fig. 2B, lane 6 vs. lane 2) whereas with PC2 in the reaction, transcription activity was fully recovered (Fig. 2B, lane 8 vs. lane 6 and lane 4) and closely paralleled what was seen in the context of the crude extract.
Fig. 2.
Description of MSA-2 as a distinct activity. (A) Fractionation scheme showing chromatographic steps that defined MSA-2 as a distinct activity. (B) In vitro transcription reactions were reconstituted with recombinant TFIIA, TFIIB (0.4 ng), TFIIE, TFIIF, and PC4, and affinity-purified TFIID, TFIIH, and Pol II. HNF-4 and PC2 were added as indicated. Reactions were also supplemented with the MSA-2 Ni-NTA agarose fraction. (C) In vitro transcription reactions were reconstituted with recombinant TFIIA, TFIIE, TFIIF, and PC4, and affinity-purified TFIID, TFIIH, and Pol II. PC2 and variable amounts of TFIIB (0.4 ng, lanes 1, 2, 4, and 5; 10 ng, lanes 3 and 6) were added as indicated. All reactions contained HNF-4. MSA-2 Ni-NTA agarose fraction was added to reactions in lanes 4–6. (D) In vitro transcription reactions were reconstituted as in B, except that pure natural TFIIA was used in place of the recombinant preparation. PC2 and MSA-2 fractions were added as indicated. All reactions contained HNF-4.
To confirm that MSA-2 recreates extract-like conditions, we retested whether high TFIIB concentrations now allow Mediator requirements to be bypassed in its presence (Fig. 2C). Transcription reactions were reconstituted with either our normal TFIIB concentration or a 25-fold lower concentration. In the absence of MSA-2, lowering the TFIIB concentration had essentially no effect on the already high level of Mediator-independent transcription (Fig. 2C, lane 3 vs. lane 1). However, in the presence of MSA-2 and absence of Mediator, little or no HNF-4-driven transcription was observed at the lower TFIIB concentration in accord with the ability of MSA-2 to suppress transcription (Fig. 2C, lane 4). By contrast, elevating the TFIIB concentration restored a high level of transcription (Fig. 2C, lane 6 vs. lane 4) that approached that seen with PC2 at the low TFIIB concentration (Fig. 2C, lane 6 vs. lane 5). These data demonstrate that in the presence of MSA-2 the Mediator requirement for transcription is in large part bypassed by excess TFIIB, as was seen for the crude extract.
We also analyzed the MSA-2 fraction for the presence of any known GTFs and cofactors by immunoblot analysis using available antibodies. TFIIA was the only GTF to be found in any significant amounts (data not shown). Further experiments showed that although we had used recombinant TFIIA in the preceding assays, the mild stimulatory component of MSA-2 could be accounted for by coeluted TFIIA. Thus, the use of highly purified natural TFIIA in our assay system almost totally amortized this stimulation (Fig. 2D, lane 4 vs. lane 2). Yet, both the MSA-2 mediated repression of PC2-independent transcriptional activity (Fig. 2D, lane 3 vs. lane 1) and the reversal of the inhibition by PC2 (Fig. 2D, lane 4 vs. lane 3) were still allowed. Therefore, only natural TFIIA was used in subsequent assays.
Identification of MSA-2 as the DSIF Complex.
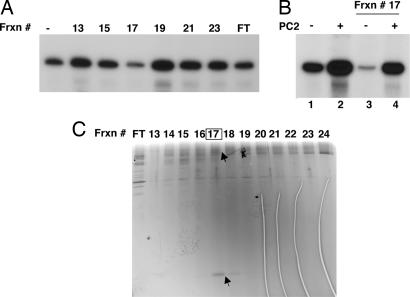
To identify the polypeptides that comprise MSA-2, we used a series of conventional columns (Fig. 2A). The analysis of the final Mono S column fractions is shown in Fig. 3 where peak inhibitory activity (assayed in the absence of Mediator) was seen in fraction 17. Fraction 17 not only suppressed Mediator-independent activity much more efficiently than other Mono S fractions (Fig. 3A), but also exhibited authentic MSA-2 activity with respect to the conferral of Mediator-responsiveness (Fig. 3B, lane 4 vs. lane 3). Furthermore, silver staining of the fractions revealed a strong correlation between two polypeptides of ≈160 kDa and 16 kDa and the MSA-2 activity (Fig. 3C).
Fig. 3.
Purification of MSA-2 to near homogeneity. (A) Transcriptional assay of the Mono S fractions. In vitro transcription reactions were reconstituted as described in the legend to Fig. 2D. All reactions contained HNF-4 but no Mediator was added. (B) The peak fraction (#17) was rigorously assayed for MSA-2 activity in the presence or absence of PC2 with reactions reconstituted as in A. (C) Silver staining of an SDS/PAGE analysis of fractions eluting from the final Mono S column. Candidate bands that correlated with activity throughout the various chromatography steps are marked with arrowheads.
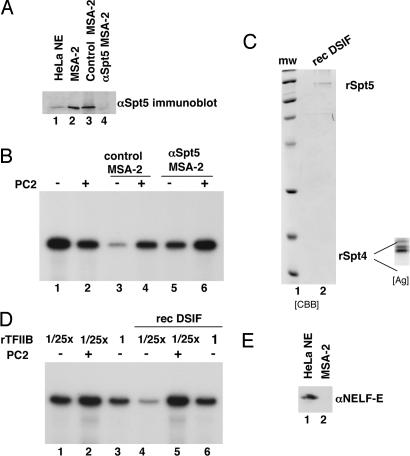
Mass spectrometric analysis of p160 and p16 identified them as hSpt5 and hSpt4, respectively. These polypeptides, which form a tight complex, have been identified as components of the transcriptional elongation factor DSIF (19). To evaluate their relevance to the MSA-2 activity, we first used anti-Spt5 antibodies to deplete the MSA-2 (Ni-NTA agarose) fraction of this complex and tested it for its ability to suppress Mediator-independent transcription and confer Mediator responsiveness (Fig. 4A). As before, PC2 had little effect on HNF-4-driven transcription (Fig. 4B, lane 2 vs. lane 1). (In this particular experiment, PC2 reduced transcription by a half.) A control MSA-2 fraction that had been mock-depleted displayed typical MSA-2 characteristics in that Mediator-independent transcription was reduced 9-fold but was reversed in the presence of PC2 for a net Mediator effect of ≈4-fold. By contrast, Spt5-depleted MSA-2 was compromised in this ability because Mediator-independent activity in its presence remained at levels that were only reduced by 2.8-fold; conversely, in the presence of PC2, transcription went up by no more than 2-fold.
Fig. 4.
MSA-2 activity of DSIF. (A) The MSA-2 Ni-NTA agarose fraction was immunodepleted over an anti-Spt5-protein A Sepharose resin and analyzed by immunoblotting using anti-Spt5 antibodies (lane 4). A mock-depleted fraction (control MSA-2, lane 3) was also analyzed. (B) Transcriptional activity of anti-Spt5-depleted MSA-2 Ni-NTA agarose fraction. Mock (lanes 3 and 4) and anti-Spt5-depleted MSA-2 fractions (lanes 5 and 6) were analyzed for PC2 responsiveness in in vitro transcription reactions reconstituted as in Fig. 2D. HNF-4 was present in all reactions. RNA products were quantified by phosphorimaging (see Results). (C) DSIF was reconstituted from the individual recombinant proteins, which were corenatured. The resulting heterodimer was further purified by gel filtration and analyzed by SDS/PAGE and Coomassie staining (CBB). The bottom part of the gel was also stained with silver (Ag; Inset) to better visualize the smaller hSpt4 subunit, which was not easily seen with Coomassie. (D) Transcriptional assay of recombinant DSIF. In vitro transcription reactions were reconstituted as in legend to Fig. 2D except that variable amounts of TFIIB were used: 0.4 ng in lanes 1, 2, 4, and 5; 10 ng in lanes 3 and 6. PC2 and recombinant DSIF (rec DSIF) were added as indicated. (E) Immunoblot analysis to show absence of NELF (using antibodies against the NELF-E subunit) in the MSA-2 Ni-NTA agarose fraction. Lane 1, HeLa nuclear extract; lane 2, MSA-2 fraction.
To prove that the hSpt5-hSpt4/DSIF complex alone accounts for bulk of the MSA-2 activity, these subunits were expressed in bacteria, purified, and after heterodimer assembly (Fig. 4C) tested in our reconstituted assay. The results (Fig. 4D) show that recombinant DSIF fully displayed the following MSA-2 activities: (i) it suppressed transcription in the absence of Mediator (Fig. 4D, lane 4 vs. lane 1); (2) it allowed restoration of transcription by addition of Mediator i.e., it imposed Mediator-responsiveness (Fig. 4D, lane 5 vs. lane 4); and (3) it allowed restoration of transcription by excess TFIIB in the absence of Mediator (Fig. 4D, lane 6 vs. lane 4). Furthermore, these results point to a distinct mode of DSIF function that is independent of the well characterized effect through negative elongation factor (NELF), which is not present in our system (Fig. 4E).
Functional Interplay Between DSIF and Mediator.
Given that earlier work had implicated the Mediator in PIC assembly and transcription initiation on the one hand and DSIF in elongation on the other, we investigated possible mechanisms that could provide linkage between the two apparently disparate phenomena.
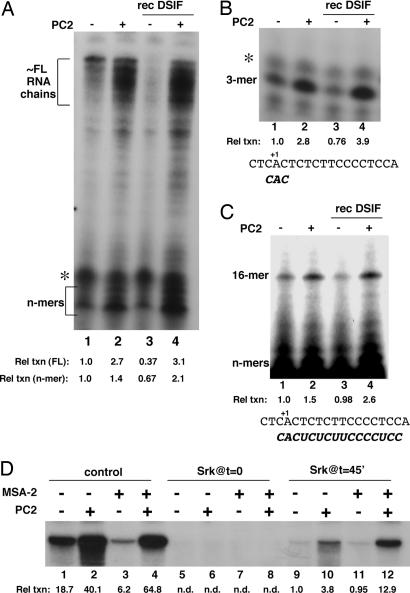
Because no evidence for a direct physical interaction between Mediator and DSIF was found [supporting information (SI) Fig. 6] we focused on functional links. Given a role in transcriptional elongation, we also tested whether DSIF acted to limit RNA synthesis by inducing transcriptional pausing in our system (19). For this purpose, we analyzed the earliest nascent RNAs as well as longer chains on high-resolution gels (Fig. 5A). We could detect both a cluster of long RNA products that corresponded to completed or nearly completed RNA chains and a significant accumulation of early RNA intermediates that were present in stoichiometric excess and represented the abortively synthesized 4- to 8-mers (“n-mers”; Fig. 5A, lane 1). When PC2 was added to the reactions, synthesis of the n-mer cluster was stimulated by 1.4-fold and of the longer RNA cluster by 2.3-fold (Fig. 5A, lane 2 vs. lane 1). Addition of recombinant DSIF markedly reduced the synthesis of long RNAs by 2.7-fold (Fig. 5A, lane 3 vs. lane 1). The inhibitory effect of DSIF on the synthesis of the n-mers was somewhat less (1.5-fold). When PC2 was now added, transcription levels more than recovered with a net Mediator effect in the presence of DSIF being 8.2-fold on long RNAs but only 3.2-fold on the n-mers (Fig. 5A, lane 4 vs. lane 3). No discrete paused transcripts (especially at ≈+25) that could be attributed to DSIF were detectable throughout the length of the template.
Fig. 5.
Mechanistic analysis of MSA-2/DSIF function. (A) Transcription reactions were reconstituted as in Fig. 2D. HNF-4 was present in all reactions. PC2 and DSIF were added as indicated. RNA products were analyzed on a composite 4%/25% polyacrylamide gel and quantitated by phosphorimaging. Clusters of very short (“n-mers”) and nearly full-length (FL) products are bracketed. The relative transcription levels (Rel txn) for each cluster (normalized to the corresponding products in reaction 1) are indicated. Asterisk marks a nonspecific band seen in all lanes. (B) Transcription reactions as in A, except that CpA and [α-32P]CTP were used to generate a trimer (3-mer). Relative transcription levels for each reaction are shown. The DNA sequence around the start site and the expected product are also shown. (C) Transcription reactions as in B, except that CpA, UTP, and [α-32P]CTP were used. Note that the gel was overexposed to visualize the 16-mer. Relative transcription levels are shown as are the DNA sequence around the start site and the expected product. (D) Transcription reactions were reconstituted as described for Fig. 2D. HNF-4 was present in all reactions. PC2 and MSA-2 fraction were added at time 0, where indicated. PICs were formed in the absence of NTPs for 45 min; transcription was then allowed to initiate by addition of a radiolabeled NTP mix. Sarkosyl (Cf = 0.02%) was added 45 s later to reactions in lanes 9–12. Reactions in lanes 5–8 received sarkosyl at time 0. Reactions in lanes 1–4 (“control”) were untreated. Relative transcription levels for each reaction are shown.
Although the above analysis indicated a predominant negative effect of DSIF on longer RNAs, there also was a significant effect on nascent chains. We therefore followed synthesis of discrete small RNAs to further analyze the functional interplay of DSIF and Mediator on nascent transcription. To synthesize a trimer, which reflected the formation of the first phosphodiester bond, reactions included only a CpA dinucleotide primer and CTP (Fig. 5B). As for the RNAs shown in Fig. 5A, the synthesis of this trimer in the absence of Mediator also was minimally reduced (1.3-fold) by DSIF (Fig. 5A, lane 3 vs. lane 1). Furthermore, the induction by PC2 was 2.8–fold in the absence of DSIF (Fig. 5A, lane 2 vs. lane 1) and 5.1–fold in its presence (Fig. 5A, lane 4 vs. lane 3). Also, the synthesis of a 16-mer (Fig. 5C), which is the expected product when CpA, CTP and UTP are included in the reactions and likely originates from a Pol II molecule that has just left the promoter, also followed a similar pattern with PC2 induction (Fig. 5C, lane 4 vs. lane 3) being 2.7-fold in the presence of DSIF and 1.5-fold in its absence (Fig. 5C, lane 2 vs. lane 1).
We also considered the possibility that DSIF effects could also be exerted at the level of reinitiation. We therefore gauged the extent of recycling in our standard system by using sarkosyl to limit transcription to a single round (Fig. 5D). In this experiment, transcription reactions were reconstituted either in the presence or absence of the MSA-2 fraction and the PICs were allowed to form for 45 min in the absence of NTPs. Transcription was allowed to proceed by addition of NTPs, and sarkosyl was added soon thereafter to restrict further cycling of the transcriptional machinery (Fig. 5D, lanes 9–12). Assuming that the lowest detectable transcription levels obtained in the presence of sarkosyl (Fig. 5D, lanes 9 and 11) represent a single round of HNF-4-driven transcription, our measurements show that in the absence of either Mediator or MSA-2 up to 20 rounds of transcription may be taking place from a given committed template (Fig. 5D, lane 9 vs. lane 1). Most importantly, in the presence of MSA-2/DSIF, where transcription levels are low (Fig. 5D, lane 3 vs. lane 1), the down-regulation is primarily evident as a restriction of transcription to significantly fewer cycles (≈3; Fig. 5D, lane 3 vs. lane 11). Our data further show that upon addition of Mediator, which restores transcription to higher levels (Fig. 5D, lane 4 vs. lane 3), the effect is manifested both at the level of increased PIC formation (Fig. 5D, lane 12 vs. lane 11) as well as increased reinitiation (Fig. 5D, lane 12 vs. lane 4).
Discussion
In this article, we identify hSpt4-hSpt5/DSIF as an accessory factor for optimal Mediator function. DSIF is a transcription elongation factor with a general role in surveillance through key checkpoints. Thus, our results reveal a potential mechanism whereby Mediator control of gene expression may be imposed by coupling initial PIC formation to subsequent steps of the transcriptional cycle.
Although it is generally believed that Mediator functions by promoting PIC assembly by interfacing between the activator and Pol II (5), it has been suspected that additional mechanisms are also at work. For example, Mediator is a component of the scaffold, which remains once the Pol II from the pioneer PIC has progressed into the elongation mode (24), and likely plays a role in facilitating subsequent cycles of reinitiation. Potentially related, there also is strong evidence for a postrecruitment function for the Mediator (14, 15). Further complicating straightforward conclusions regarding Mediator mechanisms are newer observations, which have been described here and also previously published (23), that excess TFIIB allows the Mediator requirement to be bypassed.
Our present identification of DSIF, a well established transcription elongation factor (18, 19, 25), as one factor that allows Mediator function to be fully realized also suggests additional mechanisms whereby Mediator effects may be exerted. Although DSIF acts with NELF (26) to couple transcription to RNA processing, especially capping (25), our failure both to observe DSIF-induced pausing and the apparent lack of requirement for NELF point to a rather different mode of action in the present context. Consistent with the notion of DSIF function at a more fundamental level that is distinct from its previously disclosed properties, we note also that whereas both Spt4 and Spt5 are fairly well conserved within eukaryotes, NELF may be restricted to metazoans (27). Indeed, Spt5 and Spt4 orthologs are found in both eubacteria and archaea (19, 28, 29).
In our assays, DSIF suppresses overall transcription when Mediator is not present. As discussed below, DSIF seems to target multiple steps. Thus, in as much as the Mediator (and TFIIB) can reverse each of those DSIF effects, one can infer that they likely also control these various steps. Importantly, together with a recent report identifying Gdown1 as another factor that determines Mediator dependencies (30), it seems that sensing of total “transcriptional flux” may be generally important in the specification of a Mediator requirement. Thus when transcriptional flux is high there would neither be any need nor an opportunity for Mediator function. Conversely, restricting the flux via DSIF could impose such a requirement and concurrently confer the advantage of a regulatory capability built into the Mediator.
At this time, our data do not allow us to unambiguously conclude as to which of the steps of the transcription process is determinative for the DSIF-Mediator interplay that we describe here. We find that DSIF has effects on initiation of the first-round of transcription, on reinitiation, and potentially on promoter clearance. Thus DSIF slightly, but reproducibly, reduces both the formation of a 16-mer, which reflects single-round transcription by Pol II molecules that have just cleared the promoter and formation of a trimer, which reflects reiterative abortive transcription by promoter-bound Pol II molecules. Under steady-state conditions, it preferentially affects the synthesis of longer RNAs relative to abortive products. Moreover, sarkosyl restriction suggests that DSIF also limits Pol II recycling in our system. We also note that the absolute levels of Mediator-supported transcription are actually higher in the presence of DSIF than its absence.
Whereas ongoing studies will elucidate the underlying mechanisms more precisely, several possibilities emerge from our present analyses: First, a relatively straightforward possibility, especially in light of its effects on the synthesis of the abortive trimer and on reinitiation, is that DSIF interferes both with the initial establishment of the PIC for the pioneer round of transcription as well with its efficient reassembly for subsequent rounds. Correspondingly, given the roles of Mediator and TFIIB in PIC assembly and the likely role of the Mediator-containing scaffold in promoting reinitiation, in part by facilitating TFIIB recruitment, either the Mediator or TFIIB are able to facilitate PIC assembly in the face of this interference by DSIF. With respect to the ability of TFIIB in reversing DSIF effects, it may be that they result from a competition for binding to a common site on Pol II somewhat analogous to the mutually exclusive binding of NusA and sigma factors to, respectively, the elongating and initiating form of prokaryotic RNA polymerase (31).
If our data that there is preferential diminution of longer RNAs relative to smaller transcripts in the presence of DSIF are taken at face value, an alternative possibility is that a step closely associated with promoter clearance may be a critical checkpoint. Mediator could then help in overcoming the resulting postinitiation arrest of the PIC potentially by means of its ability to stimulate the CTD kinase activity of TFIIH and concomitant promoter clearance (9, 32). This primary effect of DSIF may be amplified in steady-state transcription conditions where committed templates would be prevented from freely reinitiating perhaps owing to the promoter-proximally stalled Pol II. Potentially related, and in view of the possibility that TFIIB likely has multiple roles in the transcription cycle (3, 33), the ability of TFIIB to bypass the Mediator requirement may also reflect additional functions at the postinitiation level.
Other possibilities include DSIF action at more than one point or action per se at one step but its manifestation at multiple points because of as yet uncharacterized interconnectivity between them. Either way, overall, our data would point to a cumulative effect of DSIF both on early transcription and on reinitiation and of the Mediator in its counteraction at each of those levels. These conclusions are consistent with emerging ideas that the role of the Mediator is not limited to initial formation of the PIC.
Finally, we should note that our synthesis here of the effects of Pol II, TFIIB, Mediator, and hSpt4-hSpt5/DSIF is strongly supported by yeast genetics. Thus the MED31/SOH1 subunit of the Mediator displays genetic interactions not only with Pol II (34), but also with TFIIB (34) and several transcription elongation factors that include SII (35), the last in turn displaying close relationships with hSpt4-hSpt5/DSIF (36–39). Furthermore, consistent with its role in early elongation, TFIIB has also been connected to diverse elongation and processing factors (40).
Materials and Methods
Transcription Factor Preparations and in Vitro Transcription.
RNA polymerase II, TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and PC4 were purified to homogeneity and used in reconstituted transcription reactions as described (17). TFIIB amounts were variable and are noted in figure legends. Standard steady-state in vitro transcription reactions using the HNF-4 responsive G-less template pA4XMLΔ53 giving rise to a circa 300 nucleotide product were performed as described (17). Briefly, reaction mixtures containing the various factors, template and NTPs (ATP, UTP, and [α-32P]CTP) were incubated at 30°C for 1 h; reactions were stopped, and processed by phenol-chloroform extraction and ethanol precipitation. Electrophoresis was on 4% polyacrylamide gels containing 7 M urea except for the products shown in Fig. 5A (see legend). CpA-primed reactions contained the same basic components of the reactions as for standard reactions; NTP additions were as indicated in figure legends. After trimer or 16-mer synthesis, reaction products were treated with calf intestinal phosphatase and analyzed by electrophoresis on 25% polyacrylamide gels containing 7 M urea as described (41). All transcription gels were autographed; selected experiments were quantified by using phosphorimaging. Natural TFIIA was purified to near homogeneity from HeLa nuclear extract after chromatography on P11, DE52, Ni-NTA-agarose, and Mono S. Depletion of Mediator from HeLa nuclear extract using anti-NUT2 antibodies has been published (17).
Purification of Recombinant DSIF.
His-tagged recombinant hSpt4 and Spt5 were separately expressed in Escherichia coli from corresponding vectors [gift of Drs. T. Wada and H. Handa (19)]. The insoluble proteins were denatured in 8 M urea and purified over Ni-NTA-agarose. The proteins were then corenatured by slow dialysis and finally purified over Superose 6.
Purification and Polypeptide Identification of MSA-2.
MSA-2 was purified from standard HeLa cell nuclear extract. The phosphocellulose P11 flow-through fraction (0.1 M KCl, F1) was bound to DE52 and eluted with 0.3 M KCl (F3) and loaded directly on a Ni-NTA-agarose column. The column was step-eluted with imidazole. The 20 mM imidazole eluate (F5) was purified over Superose 6 (AKTA-FPLC) from which MSA-2 activity eluted with an apparent molecular mass of 300 kDa (at 0.1 M KCl). Further purification was over Mono Q and Mono S columns, both of which were eluted with linear salt gradients (AKTA-FPLC). All chromatography buffers were of the “BC” type (17).
A fraction from the final Mono S column that contained peak MSA-2 activity was concentrated by trichloroacetic acid precipitation and resolved by SDS/PAGE. Bands containing polypeptides of 160 kDa and 16 kDa were excised and subjected to mass spectrometric analysis.
Supplementary Material
Acknowledgments
We thank Dr. R. G. Roeder for constant support and encouragement, Drs. T. Wada and H. Handa for the hSpt4 and hSpt5 expression vectors, and Joe Fernandez of the Proteomics Core Facility at The Rockefeller University for mass spectrometric analysis. This work was supported by National Institutes of Health Grant RO1 DK060764 (to S.M.) and a Rockefeller University Women and Science fellowship (to M.J.B.).
Abbreviations
- Pol II
RNA polymerase II
- GTF
general transcription factor
- PIC
preinitiation complex
- HNF-4
hepatocyte nuclear factor-4
- PC
positive cofactor
- DSIF
DRB sensitivity-inducing factor
- NELF
negative elongation factor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608717104/DC1.
References
- 1.Malik S, Roeder RG. In: Handbook of Cell Signaling. Bradshaw RA, Dennis EA, editors. Vol 3. New York: Academic; 2003. pp. 11–19. [Google Scholar]
- 2.Hahn S. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woychik NA, Hampsey M. Cell. 2002;108:453–463. doi: 10.1016/s0092-8674(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 4.Roeder RG. CSH Symp Quant Biol. 1998;68:201–218. doi: 10.1101/sqb.1998.63.201. [DOI] [PubMed] [Google Scholar]
- 5.Malik S, Roeder RG. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Kornberg RD. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Chadick JZ, Asturias FJ. Trends Biochem Sci. 2005;30:264–271. doi: 10.1016/j.tibs.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Roeder RG. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 10.Baek HJ, Malik S, Qin J, Roeder RG. Mol Cell Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malik S, Guermah M, Yuan CX, Wu W, Yamamura S, Roeder RG. Mol Cell Biol. 2004;24:8244–8254. doi: 10.1128/MCB.24.18.8244-8254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittler G, Kremmer E, Timmers HT, Meisterernst M. EMBO Rep. 2001;2:808–813. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik S, Baek HJ, Wu W, Roeder RG. Mol Cell Biol. 2005;25:2117–2129. doi: 10.1128/MCB.25.6.2117-2129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik S, Wallberg AE, Kang YK, Roeder RG. Mol Cell Biol. 2002;22:5626–5637. doi: 10.1128/MCB.22.15.5626-5637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 17.Malik S, Roeder RG. Methods Enzymol. 2003;364:257–284. doi: 10.1016/s0076-6879(03)64015-2. [DOI] [PubMed] [Google Scholar]
- 18.Hartzog GA, Wada T, Handa H, Winston F. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 21.Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 22.Malik S, Gu W, Wu W, Qin J, Roeder RG. Mol Cell. 2000;5:753–760. doi: 10.1016/s1097-2765(00)80254-3. [DOI] [PubMed] [Google Scholar]
- 23.Baek HJ, Kang YK, Roeder RG. J Biol Chem. 2006;281:15172–15181. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- 24.Yudkovsky N, Ranish JA, Hahn S. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 25.Sims RJ, III, Belotserkovskaya R, Reinberg D. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 26.Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, et al. Mol Cell Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterlin BM, Price DH. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Steiner T, Kaiser JT, Marinkovic S, Huber R, Wahl MC. EMBO J. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponting CP. Nucleic Acids Res. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu X, Malik S, Negroiu CC, Hubbard K, Velalar CN, Hampton B, Grosu D, Catalano J, Roeder RG, Gnatt A. Proc Natl Acad Sci USA. 2006;103:9506–9511. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mooney RA, Darst SA, Landick R. Mol Cell. 2005;20:335–345. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 32.Takagi Y, Calero G, Komori H, Brown JA, Ehrensberger AH, Hudmon A, Asturias F, Kornberg RD. Mol Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Pal M, Ponticelli AS, Luse DS. Mol Cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 34.Fan HY, Cheng KK, Klein HL. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malagon F, Tong AH, Shafer BK, Strathern JN. Genetics. 2004;166:1215–1227. doi: 10.1534/genetics.166.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui Y, Denis CL. Mol Cell Biol. 2003;23:7887–7901. doi: 10.1128/MCB.23.21.7887-7901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindstrom DL, Hartzog GA. Genetics. 2001;159:487–497. doi: 10.1093/genetics/159.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, III, Hartzog GA. Mol Cell Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palangat M, Renner DB, Price DH, Landick R. Proc Natl Acad Sci USA. 2005;102:15036–15041. doi: 10.1073/pnas.0409405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ansari A, Hampsey M. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malik S, Guermah M, Roeder RG. Proc Natl Acad Sci USA. 1998;95:2192–2197. doi: 10.1073/pnas.95.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.