Abstract
Hormone refractory prostate cancer (PCa) is invariably lethal despite aggressive clinical treatment strategies. Detection strategies are needed to identify aggressive PCa before it becomes widely disseminated. Recently, two studies identified annexin 1 and 7 as potential biomarkers in the development of PCa progression. The annexins are a group of calcium-binding structural proteins that may play a role in the regulation of membrane trafficking, cellular adhesion, and cell signaling. Therefore the goal of this study is to simultaneously characterize the multiple members of the annexin family of genes in advanced PCa. Prostate samples from men with advanced hormone refractory PCa were compared to samples of hormone-naïve PCa and noncancerous prostate tissue. Samples from 15 patients with advanced hormone refractory PCa were used. To examine the annexin family, gene expression profiles from 21 noncancerous prostate tissues, 16 clinically localized PCas, and 20 hormone refractory PCa samples were used. By cDNA microarray analysis, annexins 1, 2, 4, 7, and 11 were significantly decreased in hormone refractory PCa when compared to localized hormone-naïve PCa with 2.2-, 1.5-, 1.3-, 1.4-, and 1.8-fold decreases, respectively (all P values <0.05). Interstudy validation of annexin family transcript expression was performed by meta-analysis of three other published prostate profiling studies. High-density tissue microarrays were used to validate a subset of annexins at the protein level by immunohistochemistry. Tissue microarray analysis revealed a significant decrease in protein expression for annexins 1, 2, 4, 7, and 11 in hormone refractory PCa as compared to localized PCa with 1.68-, 2.46-, 2.52-, and 3.01-fold decreases, respectively (Kruskal Wallis test, all P values P < 0.05). However, no significant differences were detected between the clinically localized PCa and noncancerous prostate tissues. These findings suggest that down-regulation of several members of the annexin family may contribute to PCa tumorigenesis. Annexins 1, 2, 4, 7, and 11 may play a role in tumor progression through distinct mechanisms or, alternatively, they may have redundant tumor suppressor activities. This study also suggests that a meta-analysis of existing gene expression data is useful in confirming findings from individual studies. Finally, down-regulation of several annexin family members may play a role in the development of the lethal PCa phenotype.
Prostate cancer (PCa) is the second leading cause of male cancer-related death and it affects one of nine males older than age 65. 1,2 In 2001, around 200,000 men in the United States were diagnosed with PCa and 31,500 died of the disease. The advent of prostate-specific antigen screening has led to earlier detection of clinically localized PCa. 3,4 However, to date there are no reliable predictors of PCa behavior and aggressiveness. Molecular profiling of human cancer represents a novel approach to study this multifaceted disease process. 5 Several groups including our own have adopted this high-throughput approach to evaluate the global expression of genes in PCa. 6-9 One advantage to this approach is that pathways or families of genes can be simultaneously evaluated. One difficulty is the large number of candidate genes identified using high-throughput platforms.
Annexins are a group of structurally related calcium-binding proteins that have a domain that binds to phospholipids and an amino terminal domain that determines specificity. 10-12 The annexins are involved in regulation of membrane trafficking, cellular adhesion, and possible tumorigenesis. Recent work suggests that decreased annexin 1 and 7 expression is associated with PCa progression and annexin 7 may function as a tumor suppressor. 13,14 In this study, we use cDNA microarrays to study the expression patterns of multiple annexin family members in a wide range of prostate tissue samples to determine their role in PCa progression. We use meta-analysis of gene expression data to help further validate the cDNA expression array findings. Finally, high-density tissue microarrays are used to assess annexin protein expression levels by immunohistochemistry.
Materials and Methods
Prostate Sample Collection
Prostate tissues were taken from the radical prostatectomy series and the rapid autopsy program available through the University of Michigan Prostate Cancer Specialized Program of Research Excellence Tissue Core. This program is approved by the Institutional Review Board at the University of Michigan.
Hormone-naïve, clinically localized PCa samples used for this study were taken from a cohort of men who underwent radical retropubic prostatectomy as a monotherapy (ie, no hormonal or radiation therapy) for clinically localized PCa between the years 1994 and 1998. The median age at time of surgery was 60 years (range, 39 to 74 years) with a median pretreatment prostate-specific antigen 6.2 ng/ml (range, 0.09 to 14.9 ng/ml). Gleason scores ranged from 6 to 9 with 95% having either a Gleason score of 6 or 7. Approximately 75% of cases were organ confined (pT2) and none of the tumors spread to the lymph nodes. Processing of the prostatic tissues started within 20 minutes after surgical resection. The prostates were partially sampled and ∼50% of the tissue was used for research. This protocol has been evaluated in a formal study to assure that partial sampling does not impair accurate staging and evaluation of the surgical margins. 15 The snap-frozen samples used for cDNA expression array analysis were all evaluated by one of the study pathologists (MAR). All samples were grossly trimmed to ensure that >95% of the sample represented the desired lesion. Areas of benign prostate tissue from prostates with PCa were used as normal tissue in these experiments.
Hormone refractory PCa samples were collected from the rapid autopsy program. 16 Snap-frozen samples were used for cDNA expression array analysis. Mirrored samples from the same lesion are placed in 10% buffered formalin. The fixed samples are embedded in paraffin. As with the prostatectomy samples, the study pathologist reviewed the glass slides, circled areas of viable PCa, avoiding areas of necrosis, and used these slides as a template for tissue microarray construction. In this study, 20 hormone refractory metastatic PCas were extracted from 15 rapid autopsy cases performed from 1997 to 2000. The patients’ ages ranged from 53 to 84 years and time from diagnosis to death ranged from 21 to 193 months. All 15 patients died with widely metastatic PCa after extensive treatment, which included anti-androgens and chemotherapy.
Prostatectomy samples were evaluated for the presence or absence of surgical margin involvement by tumor (surgical margin status), the presence of extraprostatic extension, and seminal vesicle invasion. Tumors were staged using the TNM system, which includes extraprostatic extension and seminal vesicle invasion but does not take into account surgical margin status. 17 Tumors were graded using the Gleason grading system. 18,19
cDNA Microarrays
The spotted glass cDNA microarray slides used in this study included ∼5000 known, named genes from the Research Genetics human cDNA clone set, 4400 expressed sequence tags. 8 Fluorescently labeled (Cy5) cDNA was prepared from total RNA from each of the prostate samples. The reference samples, a pool of being prostate tissue, were labeled using a second distinguishable fluorescent dye (Cy3) using a previously established protocol (www.microarrays.org). After labeling, the cDNA samples were neutralized, washed, and then applied to the microarray chips. After remainingin a hybridization water bath at 65°C overnight, the microarray slides were processed and scanned with a Genepix 4000 scanner (Axon Instruments, Union City, CA).
Primary analysis was done using the Genepix software package. Images of scanned microarrays were gridded and linked to a gene print list. Initially, data were viewed as a scatter plot of Cy3 versus Cy5 intensities. Cy3 to Cy5 ratios are determined for the individual genes along with various other quality control parameters (eg, intensity over local background). The Genepix software analysis package flags spots as absent based on spot characteristics. Furthermore, bad spots or areas of the array with obvious defects were manually flagged. Spots with small diameters (<50 μm) and spots with low-signal strengths <350 fluorescence intensity units over local background in the more intense channel were discarded. Flagged spots were not included in subsequent analyses. Data are the ratio of the fluorescent cDNA probe signal hybridized against the reference pool.
Immunohistochemistry
After paraffin removal and hydration, the tissue microarray slides were immersed in 10 mmol/L of citrate buffer placed in a pressure cooker chamber and microwaved for 10 minutes for optimal antigen retrieval. Immunostaining was performed using a DAKO autostainer (DAKO, Carpinteria, CA). The primary antibody was incubated for 45 minutes at room temperature and a secondary biotin-labeled antibody for 30 minutes. The streptavidin-LSA amplification method (DAKO K0679) was performed for 30 minutes followed by peroxidase/diaminobenzidine substrate/chromagen. The slides were counterstained with hematoxylin. Polyclonal antibodies directed against the N-terminus of annexin 1 (dilution 1:50), annexin 2 (dilution 1:100), annexin 4 (dilution 1:100), annexin 7 (dilution 1:500), and annexin 11 (dilution 1:100) were obtained from a signal source (Santa Cruz Biotechnology, Santa Cruz, CA). Cytoplasmic protein expression as determined by two pathologists (WX and MAR) using immunohistochemistry was scored as negative (score 1), weak (score 2), moderate (score 3), or strong (score 4) using a system that has been previously validated on several tissue microarray studies. 8,20
Tissue Microarray Construction, Digital Image Capture, and Analysis
Tissue microarrays were constructed as previously described to evaluate protein expression in a wide range of samples ranging from benign prostate tissue taken from the prostatectomy samples to hormone refractory PCa. 8,20 Three tissue microarrays were used for this study consisting of benign prostate, localized PCas, and hormone refractory PCa. The tissue microarrays were assembled using the manual tissue arrayer (Beecher Instruments, Silver Spring, MD) as previously described. 21,22 Tissue cores from the circled areas of interest were targeted for transfer to the recipient array blocks. The 0.6-mm-diameter tissue microarray cores were each spaced at 0.8 mm from core-center to core-center. Tissue microarray images were acquired using the BLISS Imaging System (Bacus Lab., Lombard, IL).
Statistical Analyses
To investigate the statistical significance associated with the differential expression of annexins across four independent gene expression studies, we used standard methods from meta-analysis 23 to combine the results. For each of the studies, we computed a t-statistic (with the two groups being benign tissue compared against localized PCa) and transformed the associated P values using a negative logarithmic transformation. These numbers were then doubled and added together to arrive at a summary measure of differential gene expression across the three studies. To assess the statistical significance associated with this summary measure, a permutation-based approach was adopted. 23 Namely, the tissue types were permutated within studies, and the summary measure was computed for the permutated data. A P value was computed using the permutation distribution of the summary measure. The issue then arises of whether or not the t-statistics from the three studies are comparable.
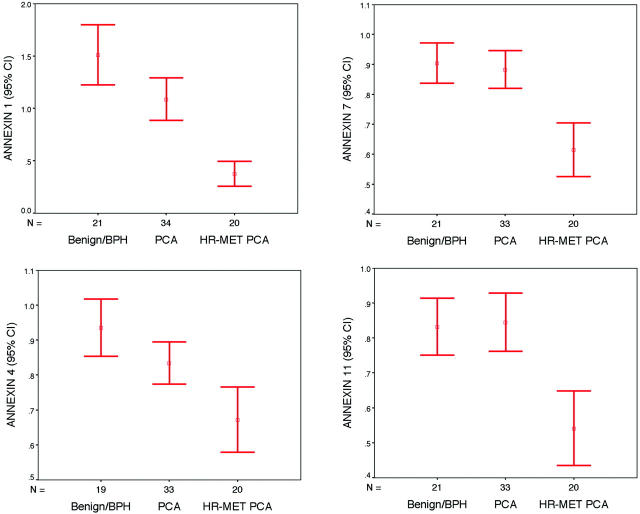
Annexin protein expressions were statistically evaluated using the mean score results from each tissue microarray sample for each prostate tissue type (ie, benign, localized PCa, and hormone refractory PCa). To determine differences between all pairs (eg, localized PCa versus benign), we performed an analysis of variance with a post hoc analysis using the Scheffé method. 24 The mean expression scores for all examined cases were presented in a graphical format by using error bars with 95% confidence intervals.
Results
Expression array analysis revealed a significant dysregulation of annexin family members with PCa progression. The cDNA expression of annexins 1, 2, 4, 7, and 11 were significantly decreased in the hormone refractory PCa samples as compared to localized hormone-sensitive PCa samples with 2.2-, 1.5-, 1.3-, 1.4-, and 1.8-fold decrease, respectively (all P values <0.01) (Table 1 ▶ and Figure 1 ▶ ). However, only annexins 1 and 4 showed significant decreases of mRNA expression in localized PCa samples as compared to the benign samples. There were no significant differences between localized hormone naive PCa and the benign samples for annexins 2, 7, and 11. No cDNA dysregulation between the tested prostate samples and annexins 8 and 13 was observed. Annexin 6 demonstrated a slight decrease in cDNA expression between localized PCa and benign samples that was not statistically significant (Table 1) ▶ .
Table 1.
Gene Expression of Select Annexins
| Annexin | Benign | BPH* | Loc-PCA† | Met-PCA‡ | Ratio PCa/Met | P Value§ | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Count | Median (n) | Count | Median (n) | Count | Median (n) | Count | Median (n) | |||
| 1 | 5 | 1.56 | 16 | 1.35 | 16 | 0.69 | 20 | 0.31 | 2.23 | < 0.001 |
| 2 | 5 | 0.79 | 16 | 0.69 | 16 | 0.74 | 20 | 0.49 | 1.51 | 0.009 |
| 4 | 5 | 0.91 | 16 | 0.97 | 16 | 0.9 | 20 | 0.69 | 1.30 | 0.001 |
| 6 | 5 | 1.2 | 16 | 1.29 | 16 | 1.05 | 20 | 1.15 | 0.91 | 0.377 |
| 7 | 5 | 0.8 | 16 | 0.88 | 16 | 0.88 | 20 | 0.62 | 1.42 | < 0.001 |
| 8 | 5 | 1.14 | 16 | 1.06 | 16 | 0.99 | 20 | 1.19 | 0.83 | 0.156 |
| 11 | 5 | 0.99 | 16 | 0.76 | 16 | 0.94 | 20 | 0.52 | 1.81 | < 0.001 |
| 13 | 5 | 1.08 | 16 | 1.35 | 16 | 1.03 | 20 | 0.94 | 1.10 | 0.393 |
*BPH, benign prostatic hyperplasia.
†Loc-PCA, localized prostate cancer.
‡Met-PCA, metastatic hormone refractory prostatic cancer. Ratio Pca/Met, ratio of expression of localized Pca over hormone refractory Pca.
§Kruskal-Wallis test.
Figure 1.
The cDNA expression of select annexin gene family members. The cDNA expression of annexins 1, 4, 7, and 11 demonstrate decreased expression going from benign prostate tissue samples (Benign/BPH), localized PCa samples (PCA), to metastatic hormone refractory PCa (Met PCA).
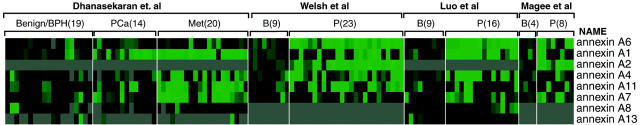
To cross validate the cDNA expression results for these annexin family members, we performed a meta-analysis of gene expression using recently described methods. 25 Annexin family members’ cDNA expression results were evaluated across publicly available data sets including our own. 6-9 The analysis evaluated annexins for each of the individual studies as well as performing a summary statistic, taking into account the significance of the gene expression across the four studies. The meta-analysis compared differences between clinically localized PCa and benign prostate tissue because not all of the studies had hormone refractory metastatic PCa. The meta-analysis (Table 2 ▶ and Figure 2 ▶ ) demonstrates that annexins 1, 2, 4, and 6 were significantly down-regulated across theses independent studies. Annexin 6 was down-regulated to a significant level in four of four studies. Annexin 1 demonstrated down-regulation in three of four studies. Annexins 2 and 4 were down-regulated in two studies and overall considered to be significantly underexpressed by the meta-analysis. Interestingly, annexin 7, which has previously been demonstrated to be down-regulated in PCa samples, 14 was not found to be significantly underexpressed in any of the four studies at the transcript level.
Table 2.
Meta-Analysis of cDNA Prostate Gene Expression Studies for Annexin Family Members
| Annexin | Dhanasekaran et al 8 | Welsh et al 6 | Luo et al 7 | Magee et al 9 | Summary P value |
|---|---|---|---|---|---|
| 6 | 0.024 | 0.0001 | 0.0001 | 0.026 | 0.0001 |
| 1 | 0.0001 | 0.031 | 0.0007 | 0.23 | 0.0001 |
| 2 | NA | 0.0001 | NA | 0.002 | 0.0001 |
| 11 | NA | 0.010 | NA | 0.6 | 0.17 |
| 7 | 0.25 | 0.48 | 0.38 | 0.088 | 0.20 |
| 4 | 0.33 | 0.023 | 0.0093 | 0.58 | 0.011 |
| 13 | 0.177 | NA | 1.00 | NA | 0.48 |
| 8 | 0.79 | NA | 0.104 | NA | 0.29 |
Figure 2.
A heat map representation of annexin family gene expression across four PCa profiling studies. Overexpression and underexpression at the transcript (ie, cDNA) level are represented by shades of red and green, respectively. Gray shading indicates that insufficient data were available. Each square represents an individual tissue sample.
Immunohistochemistry was performed to confirm these results at the protein level (Table 3 ▶ and Figure 3 ▶ ). By immunohistochemistry, a significant decrease in protein expression for annexins 1, 2, 4, 7, and 11 in hormone refractory PCa samples as compared to localized PCa samples was identified with 2.5-fold (3.8 versus 1.5 median expression), 2.4-fold (4 versus 1.7 median expression), 3.6-fold (4 versus 1.1 median expression), and 3.3-fold (4 versus 1.2 median expression) decreases, respectively (Kruskal Wallis test, all P values P < 0.05). No statistically significant differences were seen between benign and localized PCa samples in any of the annexins tested.
Table 3.
Tissue Microarray Protein Expression for Annexins by Tissue Type
| Annexin | Benign | Loc-PCA* | Met-PCA† | PCA/MET | P value‡ | |||
|---|---|---|---|---|---|---|---|---|
| Count | Median n | Count | Median n | Count | Median n | |||
| 1 | 37 | 2.59 | 360 | 2.45 | 162 | 1.46 | 1.68 | < 0.001 |
| 2 | 57 | 3.95 | 82 | 3.62 | 214 | 1.47 | 2.46 | < 0.001 |
| 4 | 23 | 3.65 | 357 | 3.96 | 141 | 1.57 | 2.52 | < 0.001 |
| 7 | 26 | 3.77 | 350 | 3.97 | 126 | 1.32 | 3.01 | < 0.001 |
| 11 | 23 | 4.00 | 360 | 3.99 | 163 | 1.30 | 3.01 | < 0.001 |
*Loc-PCA, localized prostate cancer.
†Met-PCA, metastatic hormone refractory prostatic cancer.
‡Kruskal Wallis Test.
Figure 3.
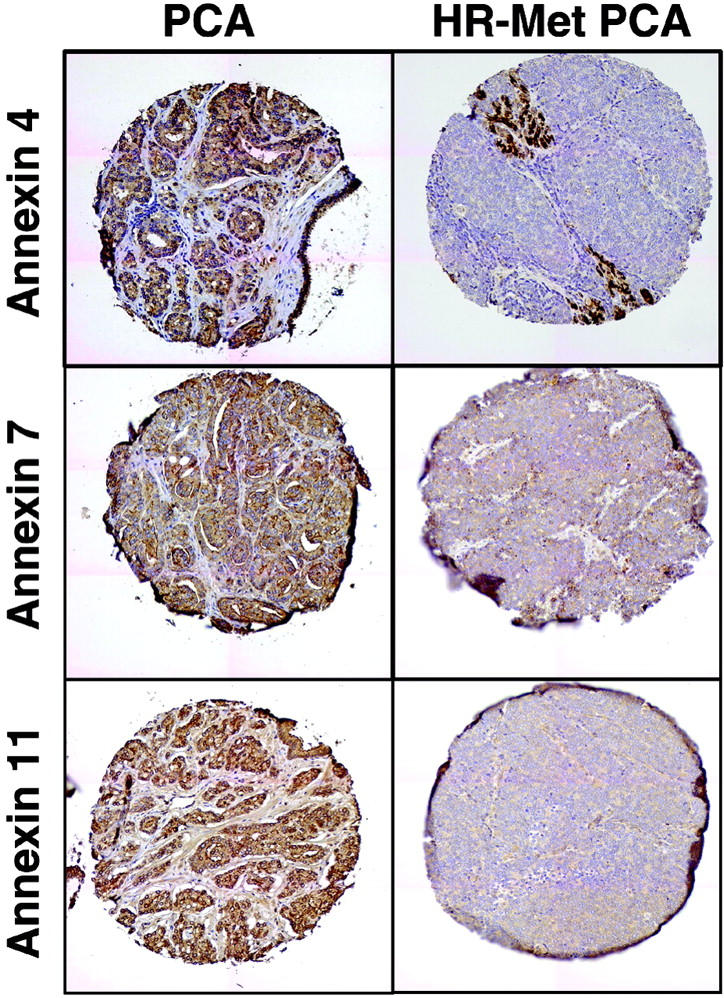
Protein expression of annexin for select annexin family members. Strong protein expression was observed in clinically localized PCa samples for annexins 4, 7, and 11. A significant decrease in protein expression was observed in hormone refractory in benign prostatic tissue, localized PCas, and metastatic hormone refractory PCas. Original magnifications, ×200.
Discussion
The dysregulation of annexin family members has been reported in multiple neoplasms. Overexpression ofannexin VIII was identified in acute promyelocytic leukemia. 26 Annexin II was reported as overexpressed in drug-resistant small-cell lung cancer cell lines, 27 pancreatic tumor cell lines and primary pancreatic tumors, 28 and astrocytic brain tumors. 29 Annexin VI has been demonstrated to have tumor suppressor activity in squamous tumor cell lines. 30 Three isoforms of annexin I have recently been seen to be down-regulated in esophageal cancers. 31 This current study focuses on the expression of multiple annexin family members in PCa.
Eight annexins were evaluated for their mRNA expression levels in benign prostatic tissue, localized hormone naïve PCa, and metastatic hormone refractory PCa samples. Five annexins (1, 2, 4, 7, and 11) demonstrated a progressive down-regulation at the transcript level going from benign prostatic tissue to localized PCa to hormone refractory PCa. To validate the cDNA expression array finding of these five annexin family members, we performed a meta-analysis, which confirmed that when looking across four studies in which at least two studies reported results, annexins 1, 2, 4, and 6 were significantly down-regulated in localized PCa samples when compared to benign prostatic tissue. Therefore the meta-analysis confirmed results on annexins 1, 2, and 4. In these examples, summary statistics across all data sets found these annexins to be significantly down-regulated at the cDNA level. However, not all of the four studies had significant down-regulation. Annexin 4, for example, was significantly down-regulated in two of four studies but the resultant summary statistic, which also takes into account the number of samples evaluated, was statistically significant. Mixed results and missing cases also demonstrates limitations of meta-analysis, as seen in the example of annexin 11. Although annexin 11 was significantly down-regulated in one microarray study, it was not present on two other studies and not significantly dysregulated in the fourth study. This would suggest that further data are probably required. Annexins 7, 8, and 13 were not found to be significantly underexpressed. The results for annexin 7 are particularly interesting as this annexin has been implicated as a putative tumor suppressor gene. 14 However, previous work on annexin 7 suggested that the greatest area of down-regulation was in the hormone refractory metastatic PCa samples. Only our study had a significant number of metastatic samples to evaluate. As demonstrated in Figure 1 ▶ , annexin 7 does decrease significantly when comparing localized PCa and metastatic PCa.
The protein expression levels of all above five annexins tested were statistically significantly decreased in hormone refractory PCa samples when compared to either localized PCa or benign prostate tissue. Four of five annexins also demonstrated a decrease in protein expression in clinically localized PCa as compared to benign prostate tissue. However, in none of these cases was the protein expression found to be significantly decreased. This second validation method at the protein level confirmed the cDNA expression array data for annexins 1, 2, 4, 7, and 11. Interestingly, our protein expression array data supports down-regulation of annexins 7 and 11. One of the limitations of the meta-analysis was that hormone refractory metastatic PCa samples were not present on all of the data sets and therefore could not be evaluated. However, by immunohistochemistry, down-regulation was confirmed for five of the five annexins tested.
Based on our expression array data, localized PCa cells down-regulate their mRNA levels of annexins but maintained the corresponding protein expression levels. Posttranslational alteration may compensate for decreased mRNA, producing enough protein to maintain levels seen with benign samples. Because annexins play an important role in maintaining cellular adhesion, 10 once the cells eventually lose this ability, tumor progression may occur. Therefore, as one might anticipate, annexin expression levels decreased significantly in the advanced hormone refractory PCa samples. This was confirmed at the protein level by significant decreases as demonstrated by immunohistochemistry.
We observed a sequential down-regulation of annexins in both transcriptional and translational levels in metastatic PCa samples. Annexin I, also called lipocortin, has been described as a phospholipase A2 inhibitor, and served as a substrate of epidermal growth factor receptor. 32,33 The significant reduction of protein level has been shown in esophageal and prostate tumor cells, in which the exact mechanism is not yet known. 13 Annexin 2, also called p36, appears an efficient substrate of protein kinase C and Src pp60. 34 Annexin 4, called endonexin, regulates Cl− flux by mediating calmodulin kinase II (CaMKII) activity. 35 Annexin 7, synexin, is involved in Duchenne’s muscular dystrophy. 36 Its gene is located on human chromosome 10q21, and its protein expression was decreased in hormone refractory tumor cells. It might function as a tumor suppressor gene in PCa progression. 14
Srivastava and colleagues 14 examined the role of annexin 7 as a putative tumor suppressor gene. They observed growth suppression of the prostate tumor cell lines DU145 and LNCaP by transfecting these cell lines with annexin 7. They also used high-density tissue microarrays to evaluate annexin 7 protein expression in a wide range of prostate tissue samples. Annexin expression was observed to be lost in a significant percentage of the metastatic prostate tumors (57% loss) and in the local recurrences of hormone refractory PCa (63% loss). Annexin 7 was strongly expressed in ∼90% of benign prostatic tissues, high-grade prostatic intraepithelial neoplasia, and localized PCa. Therefore the findings in the current study support a down-regulation of annexin 7 at the transcript and protein level in hormone refractory metastatic PCa. Our own cDNA data and the meta-analysis, which includes three other studies, did not detect significant differences between benign samples and localized PCa in their annexin 7 expression. However, when we compare the cDNA expression difference between localized and metastatic PCa samples from our current study, we note a significant difference. Our tissue microarray data confirms these differences.
One potential limitation to this study is that the specificity of antibodies used to determine protein expression. Therefore some cross reactivity between annexin family members may exist. This study and others using antibodies to characterize annexin family expression may best be viewed as confirming the down-regulation of annexins in advanced PCa as a group of genes.
In conclusion, down-regulation of several annexin family members may play a role in the development of the lethal PCa phenotype. This study further demonstrates how combining cDNA and tissue microarray technology can be used to characterize families of genes.
Acknowledgments
We thank Michelle LeBlanc and Nancy McAnsh (Comprehensive Cancer Center Immunohistochemistry/Histology Laboratory) for slide preparation.
Footnotes
Address reprint requests to Mark A. Rubin, M.D., Department of Pathology, Brigham and Women’s Hospital, 75 Francis Street, Boston, MA 02115-6110. E-mail: marubin@partners.org.
Supported by the Specialized Program in Research Excellence in Prostate Cancer (P50 CA69568), the National Cancer Institute (grant P50CA69568 to A. M. C. and M. A. R.), and the University of Michigan Bioinformatics Program (pilot grant 379206).
References
- 1.Ruijter E, van de Kaa C, Miller G, Ruiter D, Debruyne F, Schalken J: Molecular genetics and epidemiology of prostate carcinoma. Endocr Rev 1999, 20:22-45 [DOI] [PubMed] [Google Scholar]
- 2.Abate-Shen C, Shen MM: Molecular genetics of PCa. Genes Dev 2000, 14:2410-2434 [DOI] [PubMed] [Google Scholar]
- 3.Catalona WJ, Richie JP, Ahmann FR, Hudson MA, Scardino PT, Flanigan RC, deKernion JB, Ratliff TL, Kavoussi LR, Dalkin BL, et al: Comparison of digital rectal examination and serum prostate specific antigen in the early detection of PCa: results of a multicenter clinical trial of 6,630 men. J Urol 1994, 151:1283-1290 [DOI] [PubMed] [Google Scholar]
- 4.Etzioni R, Legler JM, Feuer EJ, Merrill RM, Cronin KA, Hankey BF: Cancer surveillance series: interpreting trends in PCa—part III: quantifying the link between population prostate-specific antigen testing and recent declines in PCa mortality. J Natl Cancer Inst 1999, 91:1033-1039 [DOI] [PubMed] [Google Scholar]
- 5.Liotta L, Petricoin E: Molecular profiling of human cancer. Nat Rev Genet 2000, 1:48-56 [DOI] [PubMed] [Google Scholar]
- 6.Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Jr, Hampton GM: Analysis of gene expression identifies candidate markers and pharmacological targets in PCa. Cancer Res 2001, 61:5974-5978 [PubMed] [Google Scholar]
- 7.Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB: Human PCa and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res 2001, 61:4683-4688 [PubMed] [Google Scholar]
- 8.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM: Delineation of prognostic biomarkers in PCa. Nature 2001, 412:822-826 [DOI] [PubMed] [Google Scholar]
- 9.Magee JA, Araki T, Patil S, Ehrig T, True L, Humphrey PA, Catalona WJ, Watson MA, Milbrandt J: Expression profiling reveals hepsin overexpression in PCa. Cancer Res 2001, 61:5692-5696 [PubMed] [Google Scholar]
- 10.Smith PD, Moss SE: Structural evolution of the annexin supergene family. Trends Genet 1994, 10:241-246 [DOI] [PubMed] [Google Scholar]
- 11.Mailliard WS, Haigler HT, Schlaepfer DD: Calcium-dependent binding of S100C to the N-terminal domain of annexin I. J Biol Chem 1996, 271:719-725 [DOI] [PubMed] [Google Scholar]
- 12.Waisman DM: Annexin II tetramer: structure and function. Mol Cell Biochem 1995, 149–150:301-322 [DOI] [PubMed] [Google Scholar]
- 13.Paweletz CP, Ornstein DK, Roth MJ, Bichsel VE, Gillespie JW, Calvert VS, Vocke CD, Hewitt SM, Duray PH, Herring J, Wang QH, Hu N, Linehan WM, Taylor PR, Liotta LA, Emmert-Buck MR, Petricoin III EF: Loss of annexin 1 correlates with early onset of tumorigenesis in esophageal and prostate carcinoma. Cancer Res 2000, 60:6293-6297 [PubMed] [Google Scholar]
- 14.Srivastava M, Bubendorf L, Srikantan V, Fossom L, Nolan L, Glasman M, Leighton X, Fehrle W, Pittaluga S, Raffeld M, Koivisto P, Willi N, Gasser TC, Kononen J, Sauter G, Kallioniemi OP, Srivastava S, Pollard HB: ANX7, a candidate tumor suppressor gene for PCa. Proc Natl Acad Sci USA 2001, 98:4575-4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollenbeck BK, Bassily N, Wei JT, Montie JE, Hayasaka S, Taylor JM, Rubin MA: Whole mounted radical prostatectomy specimens do not increase detection of adverse pathological features. J Urol 2000, 164:1583-1586 [PubMed] [Google Scholar]
- 16.Rubin MA, Putzi M, Mucci N, Smith DC, Wojno K, Korenchuk S, Pienta KJ: Rapid (“warm”) autopsy study for procurement of metastatic PCa. Clin Cancer Res 2000, 6:1038-1045 [PubMed] [Google Scholar]
- 17.Bostwick DG, Foster CS: Predictive factors in PCa: current concepts from the 1999 College of American Pathologists Conference on Solid Tumor Prognostic Factors and the 1999 World Health Organization Second International Consultation on Prostate Cancer. Semin Urol Oncol 1999, 17:222-272 [PubMed] [Google Scholar]
- 18.Gleason DF: Classification of prostatic carcinomas. Cancer Chemother Rep 1966, 50:125-128 [PubMed] [Google Scholar]
- 19.Tannenbaum MP: Urologic Pathology: The Prostate. 1977:pp xii and 419 Lea and Febiger, Philadelphia
- 20.Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM: Apha-methylacyl coenzyme A racemase as a tissue biomarker for PCa. JAMA 2002, 287:1662-1670 [DOI] [PubMed] [Google Scholar]
- 21.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP: Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998, 4:844-847 [DOI] [PubMed] [Google Scholar]
- 22.Perrone EE, Theoharis C, Mucci NR, Hayasaka S, Taylor JM, Cooney KA, Rubin MA: Tissue microarray assessment of PCa tumor proliferation in African-American and white men. J Natl Cancer Inst 2000, 92:937-939 [DOI] [PubMed] [Google Scholar]
- 23.Hedges LV, Olkin I: Statistical Methods for Meta-Analysis. 1985:pp xxii and 369 Academic Press, Orlando
- 24.Scheffé H: The Analysis of Variance. 1959:p 477 Wiley, New York
- 25.Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM: Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res 2002, 62:4427-4433 [PubMed] [Google Scholar]
- 26.Chang KS, Wang G, Freireich EJ, Daly M, Naylor SL, Trujillo JM, Stass SA: Specific expression of the annexin VIII gene in acute promyelocytic leukemia. Blood 1992, 79:1802-1810 [PubMed] [Google Scholar]
- 27.Cole SP, Pinkoski MJ, Bhardwaj G, Deeley RG: Elevated expression of annexin II (lipocortin II, p36) in a multidrug resistant small cell lung cancer cell line. Br J Cancer 1992, 65:498-502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vishwanatha JK, Chiang Y, Kumble KD, Hollingsworth MA, Pour PM: Enhanced expression of annexin II in human pancreatic carcinoma cells and primary pancreatic cancers. Carcinogenesis 1993, 14:2575-2579 [DOI] [PubMed] [Google Scholar]
- 29.Roseman BJ, Bollen A, Hsu J, Lamborn K, Israel MA: Annexin II marks astrocytic brain tumors of high histologic grade. Oncol Res 1994, 6:561-567 [PubMed] [Google Scholar]
- 30.Theobald J, Hanby A, Patel K, Moss SE: Annexin VI has tumour-suppressor activity in human A431 squamous epithelial carcinoma cells. Br J Cancer 1995, 71:786-788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia SH, Hu LP, Hu H, Ying WT, Xu X, Cai Y, Han YL, Chen BS, Wei F, Qian XH, Cai YY, Shen Y, Wu M, Wang MR: Three isoforms of annexin I are preferentially expressed in normal esophageal epithelia but down-regulated in esophageal squamous cell carcinomas. Oncogene 2002, 21:6641-6648 [DOI] [PubMed] [Google Scholar]
- 32.Pepinsky RB, Sinclair LK: Epidermal growth factor-dependent phosphorylation of lipocortin. Nature 1986, 321:81-84 [DOI] [PubMed] [Google Scholar]
- 33.Wallner BP, Mattaliano RJ, Hession C, Cate RL, Tizard R, Sinclair LK, Foeller C, Chow EP, Browing JL, Ramachandran KL, et al: Cloning and expression of human lipocortin, a phospholipase A2 inhibitor with potential anti-inflammatory activity. Nature 1986, 320:77-81 [DOI] [PubMed] [Google Scholar]
- 34.Hubaishy I, Jones PG, Bjorge J, Bellagamba C, Fitzpatrick S, Fujita DJ, Waisman DM: Modulation of annexin II tetramer by tyrosine phosphorylation. Biochemistry 1995, 34:14527-14534 [DOI] [PubMed] [Google Scholar]
- 35.Chan HC, Kaetzel MA, Gotter AL, Dedman JR, Nelson DJ: Annexin IV inhibits calmodulin-dependent protein kinase II-activated chloride conductance.A novel mechanism for ion channel regulation. J Biol Chem 1994, 269:32464-32468 [PubMed] [Google Scholar]
- 36.Selbert S, Fischer P, Menke A, Jockusch H, Pongratz D, Noegel AA: Annexin VII relocalization as a result of dystrophin deficiency. Exp Cell Res 1996, 222:199-208 [DOI] [PubMed] [Google Scholar]