Abstract
Reed-Sternberg (RS) cells, the neoplastic elements of Hodgkin’s lymphoma (HL), usually lack B-cell receptor expression. Normal germinal center B cells, with lack of or low-affinity B-cell receptor expression, are eliminated via FAS-induced apoptosis. RS cells express FAS, but are rescued from apoptosis by a transforming event. It is known that HL-derived cell lines are resistant to FAS-mediated apoptosis. To investigate potential causes for this resistance, FAS mutations and c-FLIP expression were studied in four HL-derived cell lines and 20 cases of HL. L1236 was found to have a splice donor site mutation in intron 7 that resulted in an aberrantly spliced FAS transcript. Screening of microdissected RS cells revealed loss of heterozygosity for a known exon 7 polymorphism in two of six informative cases indicating loss of one FAS allele. In one of the two cases with loss of heterozygosity a hemizygous mutation was detected in exon 9. c-FLIP expression was observed in all HL cell lines and in RS cells of all HL cases. Our data show that FAS mutations are rare and suggest that overexpression of c-FLIP, which was present in all cases, is involved in the resistance to FAS-mediated apoptosis.
A minority of clonal elements, the so-called Reed-Sternberg (RS) cells, and a majority of reactive cells characterize Hodgkin’s lymphoma (HL). RS cells generally harbor a high load of somatic mutations in the variable (V) region of immunoglobulin (Ig) gene, suggesting germinal center (GC) or post-GC B-cell origin. 1,2 These somatic mutations are often destructive and impede B-cell receptor (BcR) expression. 2 GC B cells will be driven to apoptosis, unless they are selected through high-affinity binding of antigen to the BcR. 3 It is not known which rescue mechanisms RS cell precursors use to escape from apoptosis. In a proportion of cases, Epstein-Barr virus (EBV) may play a role in this rescue. 4 However, EBV-negative cases account for >50% of HL cases in Western countries 5 and this suggests involvement of other transforming events. Although some regulators of apoptosis such as TP53, 6,7 BCL-2 8 and indirectly, IκBα 9,10 have been studied, abnormalities in these genes were only demonstrated in a minority of HL cases.
Another gene that might be involved in the rescue of RS cell precursors is the FAS gene (CD95, Apo-1, TNFRSF6) that codes for a cell surface receptor involved in death signaling. 11 In B cells, FAS is expressed during the GC phase of development and causes B cells not selected by high-affinity BcR to be eliminated through apo-ptosis. 12,13 Deletions or mutations in the FAS gene hamper the selection process and allow undesirable B cells to survive and proliferate. 14 In fact, this scenario can be observed in lpr (lymphoproliferation) FAS-deficient mice that present with lymphadenopathy, spleen and liver enlargement, a propensity to autoimmune phenomena, and the development of B-cell lymphomas. 15 In humans, germline FAS mutations result in autoimmune lymphoproliferative syndrome with variable phenotypes. 16 Some of these autoimmune lymphoproliferative syndrome patients develop solid tumors and B-cell lymphomas. 17 In autoimmune lymphoproliferative syndrome patients, the risk to develop HL is ∼50-fold higher than in the normal population. 18 HL-derived cell lines and RS cells from primary HL cases express FAS protein, 19,20 however, HL-derived cell lines are resistant to FAS ligand-mediated apoptosis. 21 This indicates a possible role for abnormalities of the FAS pathway in the escape from apoptosis by RS cells. In addition to FAS mutations, some downstream molecules in the FAS-signaling pathway may also account for the observed resistance to FAS-mediated apoptosis. In normal GC B cells and some B cell lymphomas, it has been demonstrated that FADD-like interleukin-1β-converting enzyme-inhibitory protein (c-FLIP) expression is an important mechanism to rescue these cells from apoptosis. 22-24
To gain more insight into the potential mechanisms involved in RS cell resistance to FAS-mediated apoptosis, we analyzed FAS gene mutations and c-FLIP expression in 20 HL samples and four HL-derived cell lines. To this end, RS cells were isolated by laser microdissection microscopy from the tissues involved by HL. In addition, we studied c-FLIP expression in these cases to establish a possible contribution of c-FLIP to the resistance to FAS-mediated apoptosis.
Materials and Methods
Tissues and Cell Lines
The HL-derived cell lines, L428 (EBV-negative), 25 L591 (EBV-positive), 25 and L1236 (EBV-negative) 26 were made available by Professor V. Diehl et al (Department of Internal Medicine, University of Cologne, Cologne, Germany) and the DEV cell line (EBV-negative), originally published as derived from a case of nodular scleroses (NS) HL, but this case was subsequently retyped as nodular lymphocyte predominance Hodgkin’s lymphoma (NLPHL), was established in our own laboratory. 27 Frozen tissue from 20 HL cases (16 cHL and 4 NLPHL) were retrieved from the tissue bank of the Department of Pathology, University Medical Center Groningen, and classified according to the World Health Organization classification. 28 Fifteen cases (numbers 3, 4, 12, 13, 15, 18, 23, 31, 34, 40, 44, 48, 51, 53, and 57) were previously studied for TP53 mutations and IgH rearrangements. 7 Expression analysis for c-FLIP was performed in paraffin-embedded tissues from 19 of 20 HL cases studied for FAS mutations. For case 69, no paraffin-embedded tissue was available.
Immunohistochemistry and EBV-Encoded Small RNA (EBER) in Situ Hybridization
For c-FLIP immunohistochemical analysis, a polyclonal rabbit anti-human antiserum directed against C-terminal FLIPL (Sigma-Aldrich Chemie Gmbh, Munich, Germany), dilution 1:100, was applied on paraffin sections after they were subjected to heat-induced antigen retrieval in 50 mmol/L of Tris and 2 mmol/L of ethylenediaminetetraacetic acid, pH 9.0, buffer. After appropriate washing steps, peroxidase-labeled goat anti-rabbit antibodies followed by peroxidase-labeled rabbit anti-goat antibodies (DAKO, Copenhagen, Denmark) were applied and the peroxidase enzyme was stained with diaminobenzidine and H2O2 to visualize the c-FLIPL protein-positive cells. Positive and negative controls were used to validate the assay. High expression of c-FLIPL in the dark zone of GCs and in vessel walls was used as positive controls. HL cases were semiquantitatively scored as: +, <25% of c-FLIPL-positive RS cells; ++, 25 to 75% positive RS cells; and +++, >75% of the RS cells c-FLIPL-positive. CD30 and CD20 immunostaining of RS cells and EBER in situ hybridization procedures were performed as described elsewhere. 7
Enrichment of RS Cells with Microdissection
Microdissection and DNA isolation procedures were performed as described previously. 7 Briefly, selection of RS cells was based on morphological and immunophenotypical criteria, ie, CD30- or CD20-immunolabeled RS cells were isolated, respectively, for cHL and NLPHL samples. From each case single RS cells were microdissected and 30 cells were pooled per tube containing 30 μl of polymerase chain reaction (PCR) buffer (Amersham Pharmacia Biosciences, Roosendaal, The Netherlands). For each patient a total of 10 to 15 tubes were collected, depending on the amount of RS cells present in the tissue sections. Several tubes containing ∼100 reactive cells were collected in 30 μl of PCR buffer and analyzed for every case as normal controls. For each PCR a single tube containing either 30 RS cells or 100 reactive cells were used. Control tubes, containing only 30 μl of PCR buffer, were included throughout the microdissection, DNA isolation, and PCR procedure as negative controls.
Amplification of the FAS Gene and IgH Gene Rearrangements
External primer sets were developed based on the FAS genomic and coding sequence published in the GenBank (accession numbers X63717, X81340, X81341, and X81342). Internal primer sequences used for the separate amplification of exons 1 to 9 of the FAS were derived from published data 29 (Table 1) ▶ . For HL-derived cell lines, 150 ng of DNA was amplified, using only the internal primer set. For the amplification of DNA from microdissected cells a nested PCR was performed using one tube containing 30 single RS cells. The first amplification was performed in 50 μl containing 0.2 mmol/L dNTP, 1 U Taq-polymerase (Amersham Pharmacia Biotech), the reaction buffer provided by the manufacturer and 150 ng of each PCR primer. The PCR program consisted of 33 cycles (30 seconds at 94°C, 45 seconds at 55°C, and 60 seconds at 72°C). The first denaturation step lasted for 5 minutes and the final elongation step lasted for 7 minutes (GeneAmp 9700; Perkin Elmer Applied Biosystems, Foster City, CA). The second amplification was performed with 2 μl of first-round amplification PCR product. The PCR conditions were the same as described above, except that the internal primers 29 with the GC clamps were used and that the number of cycles was decreased to 30. An aliquot of 10 μl was analyzed on a 2% agarose gel. Based on the fact that >80% of FAS mutations occur in exons 7 to 9, all cHL cases were screened for these exons. All FAS exons were studied in the HL-derived cell lines. In the NLPHL cases, only exon 9 was analyzed because of sample limitations. All analyses were performed in duplicate or triplicate using a new tube containing a total of 30 microdissected RS cells or 100 infiltrating cells. IgH gene analyses to evaluate sample enrichment and clonality of the pooled RS cells of 15 of these 20 cases were published previously. 7 The five additional samples included in this study were analyzed by the same procedures.
Table 1.
Primer Sequences for the Amplification of the FAS Gene on Genomic DNA Isolated from 30 to 100 Microdissected RS or Infiltrating Cells
| Exon | External primer set | Nested primer set 29 | PCR (bp) | ||
|---|---|---|---|---|---|
| Forward primer 5′—3′ | Reverse primer 5′—3′ | Forward primer 5′—3′ | Reverse primer 5′—3′ | ||
| 1 | (40 GC)TCAGTACGGAGTTGGGGAAGC | GCCTATCCCCGGGACTAAGAC | 136 | ||
| 2 | (40 GC)ATCAATAAAATTCTCTTCATG | GACTTTCACTGTAATCTCTGG | 239 | ||
| 3 | AAACACTTGCTCCTTTTTTCC | (40 GC)TGAAATTCCAAGATTGGCC | 213 | ||
| 4 | TCCAAACTGATTTTCTAGGC | (40 GC)TCTAGTGTTTTAATCAGAGAAAGAC | 162 | ||
| 5 | (40 GC)CCAGGCTTTTGAATTTCTCC | GGGAAAGGAGGATATAACCG | 133 | ||
| 6 | TAATATGCCAATGTTCCAACC | (40 GC)CCCCAAGTTATTTCAATCTG | 173 | ||
| 7 | GGCCACTTTTAAGTTTCACTG | AGCAAGACTCCATCTCAAAC | CATGCATTCTACAAGGCTGAG | (40 GC)AGGAAGTAACAAAAAGCCAA | 254 |
| 8 | GCAACTGATTGTACTTCTTTC | TCATACGCTTCTTTCTTTCC | TCTCTGCTTCCATTTTTTGC | (40 GC)TTTACTCTGAAATTGGCCTA | 160 |
| 9.1 | TATTTCTATTTTTCAGATG | CAAACACTAATTGCATATACTC | (40 GC)TATTTTCTATTTTTCAGATG | TCATACGCTTCTTTCTTTCC | 253 |
| 9.2 | TATTTCTATTTTTCAGATG | CAAACACTAATTGCATATACTC | GTTCAACTGCTTCGTAATTG | (40 GC)GAACTGAATTTGTTGTTTT | 249 |
40 GC; GC clamp consisting of 40 nucleotides; 29 bp, base pair.
Denaturing Gradient Gel Electrophoresis (DGGE)
DGGE analysis was performed in an Ingeny PhorU-1 PCR apparatus (Ingeny, Goes, The Netherlands). The homo/heteroduplex PCR products were analyzed in a 9% polyacrylamide (acrylamide-bisacrylamide, 37.5:1) gel containing a 20 to 60% urea-formamide denaturing gradient (100% UF = 7 mol/L urea per 40% deionized formamide) parallel to the direction of electrophoresis. Electrophoresis was performed at 110 V and 59°C for 15 hours. Aberrant homoduplex bands were excised, eluted in TE (Tris ethylenediaminetetraacetic acid), and reamplified in a volume of 100 μl using PCR conditions as described above. For confirmation of the mutation, both strands were sequenced and compared to the germline sequence, as published in the GenBank. Duplex formation of FAS PCR products, purification, and sequencing of PCR products were performed as described previously. 30
Reverse Transcriptase (RT)-PCR for c-FLIP and FAS
For the HL-derived cell lines, total RNA was isolated, purified from contaminating DNA, and converted into cDNA as described previously. 31 Both long (FLIPL) and short (FLIPS) splice forms of FLIP were amplified using the following primers: FLIPL (forward, 5′-gaacatccacagaatagacc-3′; reverse, 5′-gtatctctcttcaggtatgc-3′) and FLIPS (forward, 5′-gaacatccacagaatagacc-3′; reverse, 5′-tttcagatcaggacaatggg-3′). PCR was performed on 1 μl of the cDNA synthesis reaction mix (originating from ∼100 ng of total RNA), using 1 U of Taq-polymerase (Amersham Pharmacia Biotech) and the reaction buffer provided by the manufacturer. The PCR program consisted of 30 cycles (30 seconds at 94°C, 45 seconds at 54°C, 30 seconds at 72°C). The first denaturation step lasted for 5 minutes and the final extension step lasted for 7 minutes. PCR products were analyzed on 2% agarose gels containing ethidium-bromide. For c-FLIPL, a PCR product of 262 bp was yielded whereas for c-FLIPS a product of 172 bp was obtained. A nested primer set used for FAS RT-PCR analyses was selected from the published sequence data (total coding region: forward, 5′-gaacacaccctgaggccag-3′; reverse, 5′-ccaagcagtatttacagccagc-3′; exons 6 to 8: forward, 5′-gtgcaaagaggaaggatcca-3′; reverse, 5′-(40gc)-gatatatttactcaagtcaa3′) resulting in a 1215-bp and a 248-bp PCR fragment, respectively. PCR was performed as described above (30 cycles: 30 seconds at 94°C, 45 seconds at 60°C, and 90 seconds at 72°C for total coding region and 30 seconds at 94°C, 45 seconds at 50°C, and 30 seconds at 72°C for exons 6 to 8).
Results
FAS Gene Mutation Analysis
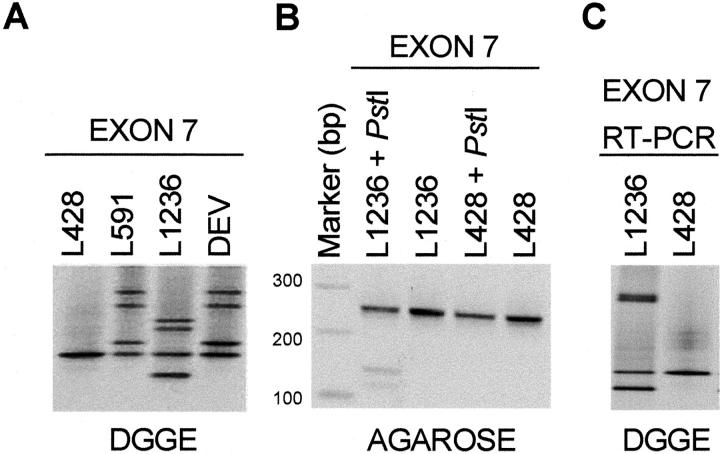
DGGE of exons 7 to 9 revealed the presence of normal bands for 16 of 20 HL cases and for the L428 cell line. FAS exon 7 analyses revealed an aberrant banding pattern (heterozygous and homozygous cases) suggesting a polymorphism. Sequence analysis of normal and aberrant bands indicated that this variation represented the previously reported polymorphism (position 836, ACC, or ACT). 32 A heterozygous pattern was observed in 4 of the 16 cases (cases 13, 15, 40, and 57) and in two of the HL cell lines (L591 and DEV). A homozygous pattern representing the ACC allele was present in cases 3, 4, 12, 18, 23, 34, 44, 53, and 68 as well as in the L428 cell line, whereas a homozygous band representing the ACT allele was present in cases 31, 48, and 51. In all 12 homozygous cases, reactive cells were also analyzed revealing a heterozygous pattern indicating loss of heterozygosity (LOH) in exon 7 for cases 4 and 23 (Figure 1A) ▶ . In L1236, a heterozygous aberrant pattern, distinct from the known polymorphism, was observed for exon 7 (Figure 2A) ▶ . Sequencing of the aberrant homoduplex band and comparison to the germ line configuration revealed a splice donor site mutation (CCTgtag→CCTgcag). This mutation results in a PstI restriction enzyme site (CTGCA!G) that is not present in the wild-type sequence. Re-amplification of exon 7 followed by a PstI restriction enzyme digestion indeed revealed a PstI restriction site, confirming the mutation (Figure 2B) ▶ . In fact, L1236 FAS RT-PCR with an exon 6 and an exon 8 primer resulted in a PCR product that appeared to be of the expected size. DGGE analysis of this RT-PCR product revealed, however, a heterozygous pattern (Figure 2C) ▶ . Sequence analysis of the abnormal homoduplex band revealed an insertion of four bases (gtag) in between exons 7 and 8, resulting in a premature termination 26 bp downstream of the insertion. The resulting truncated FAS protein lacks the death domain. Analysis of exon 7 in case 34 revealed a heterozygous pattern, different from the pattern observed for the known polymorphism, which was detected only once in triplicate experiments (Figure 1A) ▶ . Exon 9 analysis revealed aberrant patterns in two cases (cases 4 and 18) (Figure 1B) ▶ . In case 18, the RS cells had an abnormal heterozygous pattern in exon 9, which was detected only once in three different experiments. Case 4 revealed only a single aberrant band in two independent experiments with the RS microdissected cells, without identification of the normal allele. This abnormal pattern was not observed in microdissected non-RS cells, and thus exclusive for RS cells. Sequencing of the abnormal band and comparison to germ line sequence revealed a causative mutation in codon 238, GGA→GAA (Gly→Glu). Analysis of exon 7 for this case, as stated above, revealed LOH. Combination of exons 7 and 9 results for case 4 indicated that RS cells had one mutated FAS allele whereas the other FAS allele was lost.
Figure 1.
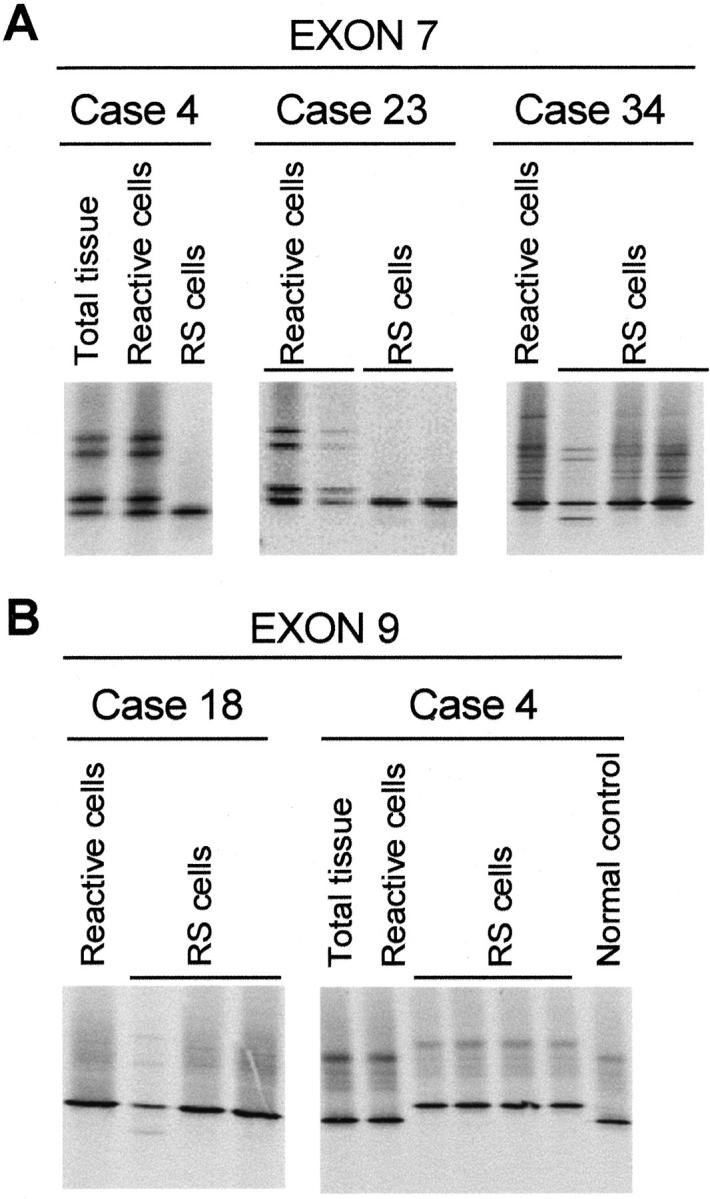
DGGE analyses of the HL samples. A: Exon 7 analysis for cases 4, 23, and 34. For cases 4 and 23 LOH can be observed on comparison of reactive and RS cells microdissected from HL-involved tissue. The duplicate analysis of case 4 was analyzed in a separate DGGE analysis (result not shown). For case 34 an aberrant banding pattern was present in RS cells in one of a duplicate experiment. B: Exon 9 analysis for cases 4 and 18. For case 18 an aberrant banding pattern was present in RS cells in one of a triplicate experiment. For case 4 an aberrant homoduplex band was detected without presence of the wild-type homoduplex band. Shown is the inverted image of an ethidium bromide-stained DGGE gel.
Figure 2.
DGGE analyses of the HL-derived cell line L1236. A: DGGE analysis of exon 7. A heterozygous banding pattern representing the known exon 7 polymorphism was detected for L591 and DEV, whereas a homozygous pattern was observed for L428. An aberrant four-banding pattern was observed for L1236. B: Reamplification and digestion with PstI revealed presence of an undigested band (255 bp) representing the normal allele and two smaller fragments (140 and 115 bp) representing the allele carrying the mutation for L1236. C: DGGE analysis of the exon 6 to 8 RT-PCR product showing an aberrant four-banded pattern for L1236 and a normal band for L428. Shown is the inverted image of ethidium bromide-stained agarose or DGGE gels.
Immunoglobulin Gene Heavy Chain Rearrangements
The five cases (cases 68 to 72) that were not included in the previous study were analyzed for IgH rearrangements to verify the efficiency of RS cell enrichment and to assess the clonality of this population. Four of the five HL cases demonstrated a single band in agarose gel and a single fluorescent peak at MEGABACE analysis (data not shown) indicating proper RS cell enrichment and monoclonal population. In case 71, no FRIII PCR product was obtained in different attempts.
Cellular FLIP (c-FLIP) Expression
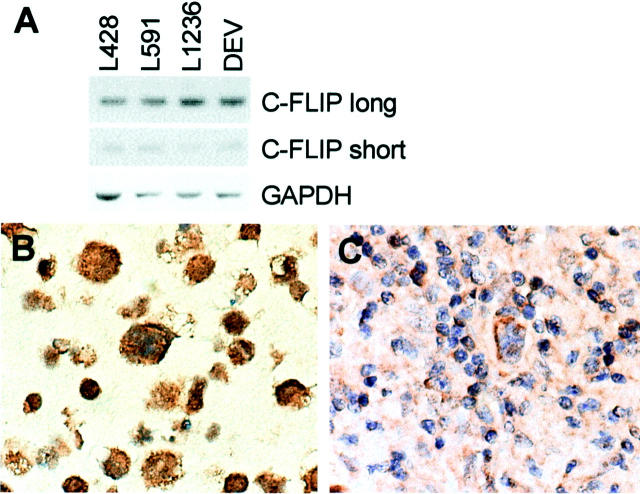
cFLIP expression was assessed by RT-PCR in the HL-derived cell lines resulting in PCR products for both long (c-FLIPL) and short (c-FLIPS) splicing forms (Figure 3A) ▶ . In addition, c-FLIPL protein expression was studied by immunohistochemistry in the HL-derived cell lines, showing high-expression levels in the cytoplasm of all cells (Figure 3B) ▶ . In the tissue sections of the 19 primary cases, RS cells from cHL as well as NLPHL stained in varying proportion and intensity for c-FLIPL (Figure 3C ▶ and Table 2 ▶ ).
Figure 3.
c-FLIP analyses in HL-derived cell lines and cases. A: RT-PCR for c-FLIP long (30 cycles) and short (35 cycles) splice forms. GAPDH housekeeping gene (18 cycles) was used as a control for RNA loading and quality. B: c-FLIP immunohistochemical staining of L428. A cytoplasmic staining can be observed in all cells. C: c-FLIP immunohistochemistry of a MCHL case. A cytoplasmic staining is present in the RS cells. Original magnifications, ×630 (B and C).
Table 2.
Overview of Four HL-Derived Cell Lines and 20 HL Cases Analyzed for FAS and TP53 Mutations and c-FLIP Expression
| Case | EBV | Subtype | TP53 mutation | FAS exon 7 polymorphism | FAS mutation | c-FLIPL expression |
|---|---|---|---|---|---|---|
| 3 | + | MCHL | − | ACC | − | + |
| 4 | + | MCHL | − | ACC/(ACT)* | + (ex 9) | + |
| 12 | + | NSHL | − | ACC | − | ++ |
| 13 | + | NSHL | − | ACC/ACT | − | ++ |
| 15 | + | NSHL | + (ex 8) | ACC/ACT | − | ++ |
| 18 | + | NSHL | − | ACC | (+/−) (ex 9) | ++ |
| 23 | + | NSHL | − | ACC/(ACT)* | − | +++ |
| 31 | − | NSHL | − | ACT | − | + |
| 34 | − | NSHL | − | ACC | (+/−) (ex 7) | + |
| 40 | − | NSHL | − | ACC/ACT | − | ++ |
| 44 | − | NSHL | + (ex 6) | ACC | − | + |
| 48 | − | NSHL | − | ACT | − | +++ |
| 51 | − | NSHL | − | ACT | − | ++ |
| 53 | − | NSHL | + (ex 7) | ACC | − | ++ |
| 57 | − | NSHL | − | ACC/ACT | − | + |
| 68 | + | MCHL | n.d. | ACC | − | ++ |
| 69 | − | NLPHL | n.d. | n.d. | − | n.d. |
| 70 | − | NLPHL | n.d. | n.d. | − | ++ |
| 71 | − | NLPHL | n.d. | n.d. | − | +++ |
| 72 | − | NLPHL | n.d. | n.d. | − | ++ |
| L428 | − | NSHL | − | ACC | − | +++ |
| L591 | + | NSHL | − | ACC/ACT | − | ++ |
| L1236 | + | MCHL | − | ACC | + (ex 7) | +++ |
| DEV | − | NLPHL | − | ACC/ACT | − | ++ |
HL, Hodgkin’s lymphoma; +, positive; −, negative; (+/−), abnormality observed once in triplicate experiments;
*, the allele lost in the RS cells is shown in between brackets;
MCHL, mixed cellularity HL; NSHL, nodular sclerosis HL; NLPHL, nodular lymphocyte predominance HL; ex, exon; n.d., not done; c-FLIPL expression: + with less than 25% of c-FLIPL-positive RS cells, ++ with 25 to 75% positive RS cells, and +++ with more than 75% of the RS cells were c-FLIPL-positive.
Discussion
RS cells are considered to be derived from GC B cells in the vast majority of HL cases. 1,2 Physiologically, GC B cells with low-affinity BcR are eliminated via FAS signaling pathway. 13,33 Because RS precursor cells are considered to be GC B cells without BcR expression 2,34 that have escaped from apoptosis, we decided to study FAS gene mutations, which may confer apoptosis resistance to RS cells.
We analyzed 20 primary HL cases and four HL-derived cell lines for FAS mutations in the hotspot region (exons 7 to 9), where >80% of FAS mutations occurs. 29,33 We detected a heterozygous pattern for the exon 7 polymorphism in six HL cases and two HL-derived cell lines, whereas homozygous patterns were detected for the 10 remaining cases and for two HL cell lines. In these samples, the frequency of the ACC allele was 0.65 (26 times) and the frequency of the ACT allele was 0.35 (14 times). This frequency is in agreement with the normal population, in which the frequency of the two alleles is estimated to be ∼0.66 and 0.33 for ACC and ACT, respectively. 32
LOH of the 10q24 region including the FAS locus was not previously described in HL, but we demonstrated LOH in two HL cases (4 and 23). In a recent study, single RS cells were analyzed for FAS mutations, and both FAS alleles could be amplified in five informative cases for two polymorphisms in the 5′ untranslated region of FAS 35 . This finding excluded LOH in these HL samples. In addition, no deletions in the FAS gene region have been described by cytogenetic studies in HL cases and cell lines. 36-38 However, classical cytogenetic, fluorescence in situ hybridization, and comparative genomic hybridization, approaches could easily have missed small deletions in this region because of sensitivity limitations. Overall the frequency of LOH at the FAS locus appears to be low in HL.
One of the 20 cases showed a causative hemizygous FAS mutation in exon 9, whereas the other FAS allele based on exon 7 analysis was lost, a combination (mutation and LOH) not described previously. It can be speculated that LOH alone does not affect the FAS function and may have anteceded the FAS mutation. In the majority of the published cases, a heterozygous FAS mutation is sufficient to hamper FAS signaling, probably because of improper trimerization of FAS molecules. 33 It is not clear whether or which biological advantages LOH would add to RS cell tumorigenesis. Solid tumors and B cell-derived lymphomas may have LOH of the FAS genomic region. 33 Müschen and colleagues 39 suggests that this phenomenon occurs invariably in association with a mutation in the first eight FAS exons, indicating that, in these exons, LOH alone is not sufficient for malignant transformation and progression. Overall, our study is in line with the findings of Müschen and colleagues, 35 who also described low frequency of FAS mutations in the death domain (1 of 10 cHL cases). In this case a nonclonal FAS mutation was identified in exon 9 (two different mutations in 12 single cells analyzed). Therefore, FAS mutations play a role in only a minority of HL cases (2 of 30). In cases 18 and 34, in which abnormalities were present only once in triplicate experiments, our approach does not permit distinction between an aberration present in a minority of RS cells and an artifact generated by PCR because of Taq-polymerase errors. Although we cannot prove the origin of these aberrant bands we believe that lack of consistency of the results in these two cases suggests that these findings are most likely caused by Taq-polymerase errors. Therefore, we did not proceed to a detailed analysis. We observed a splice donor site mutation in the L1236 HL cell line on one allele, and confirmed this by restriction site and RT-PCR analyses. This mutation leads to a four-base insertion resulting in a truncated FAS protein that lacks the death domain. This mutation was also confirmed by Re and colleagues 21 who had previously reported on an exclusive expression of wild-type FAS. Moreover, analysis of bone marrow cells of the respective patient from whom the L1236 cell line was established revealed germ line sequence at the corresponding region (D. Re, A. Staratschek-Jox, personal communications).
FAS somatic mutations are also present in other lymphomas. 29 In general, GC and post-GC B-cell-derived lymphomas have a higher frequency (∼20%) than pre-GC lymphomas (∼2.5%). 33 Lymphomas with FAS mutations often have extranodal presentation and are associated with autoimmune phenomena. 29 It was recently demonstrated that normal GC B cells harbor, at a low frequency, FAS mutations. 14 Therefore, the higher frequency of mutations in antigen-experienced B-cell-derived lymphomas may merely reflect mutations acquired during GC reaction. FAS mutations have also been demonstrated in T-cell adult leukemia 40,41 and solid tumors, 42,43 indicating that mechanisms other than somatic hypermutations may also be involved in the generation of FAS mutations in neoplastic cells.
The low frequency of FAS mutations and the fact that RS cells are resistant to FAS-induced apoptosis, suggest a role for FAS downstream molecules in conferring FAS apoptosis resistance. In recent years, up-regulation of c-FLIP was shown to exert a protective effect from apoptosis in GC B cells and lymphoma cells. 22-24 Cellular FLIP expression impedes recruitment of procaspase-8 to the death-inducing signaling complex (DISC). 44,45 As a result, cleavage of procaspase-8 is not achieved, and activation of downstream caspases and apoptosis are prevented. 44,45 A recent study demonstrated c-FLIP expression in HL-derived cell lines and in primary HL cases. 46 Our results are in agreement with these findings, suggesting that c-Flip overexpression rather than FAS mutations may confer resistance to FAS-mediated apoptosis in RS cells. C-FLIPL staining scores were not associated with subtype, EBV status, and presence of mutations in either FAS or TP53 genes (Table 1) ▶ . The exact mechanisms of c-FLIP up-regulation in HL are unclear. High-affinity BcR expression is one of the possible mechanisms for c-FLIP up-regulation, 22 however BcR is in general not expressed in HL. 2 CD40/CD40L interaction is a possible mechanism to up-regulate c-FLIP expression, 47 however, this up-regulation lasts only for 24 hours, whereas the RS cells in HL have constant c-FLIP expression. 46 Moreover, c-FLIP expression is observed in HL cell lines, a system in which CD40L is not present. Therefore, other mechanisms activating the CD40 pathway may be involved and it will be of interest to determine which regulatory mechanisms participate in c-FLIP expression in HL.
TP53 mutations are one of the mechanisms involved in deregulation of FAS expression. 48 Down-regulation of FAS expression has been described as a mechanism for malignant progression. Intron 1 of the FAS gene harbors a p53-responsive element and binding of wild-type p53 protein results in up-regulation of FAS expression. 48 In addition to up-regulation of FAS, wild-type p53 collaborates with transportation of FAS to the cell membrane where it exerts its functions. 49 The low frequency of TP53 mutations 6,7 in HL and frequent FAS expression in RS cells 19,20 suggests that FAS down-regulation is not essential for tumor progression in HL.
In conclusion, the results demonstrate that the resistance to FAS-mediated apoptosis as observed in RS cells is rarely associated with FAS mutations, but may well be because of c-FLIP overexpression.
Acknowledgments
We thank Ms. Inge Platteel for her assistance in the development and optimization of the FAS primer sets.
Footnotes
Address reprint requests to Sibrand Poppema, M.D., Ph.D., Department of Pathology and Laboratory Medicine, University Medical Center Groningen, Hanzeplein 1, PO Box 30.001, 9700RB Groningen, The Netherlands. E-mail: s.poppema@med.rug.nl.
Supported by the Dutch Cancer Society (grant RUG 97-1580).
References
- 1.Kanzler H, Küppers R, Hansmann ML, Rajewsky K: Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med 1996, 184:1495-1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Küppers R, Rajewsky K: The origin of Hodgkin and Reed Sternberg cells in Hodgkin’s disease. Annu Rev Immunol 1998, 16:471-493 [DOI] [PubMed] [Google Scholar]
- 3.Rajewsky K: Clonal selection and learning in the antibody system. Nature 1996, 381:751-758 [DOI] [PubMed] [Google Scholar]
- 4.Chapman ALN, Rickinson AB: Epstein-Barr virus in Hodgkin’s disease. Ann Oncol 1995, 9:5-16 [DOI] [PubMed] [Google Scholar]
- 5.Jarrett RF, MacKenzie J: Epstein-Barr virus and other candidate viruses in the pathogenesis of Hodgkin’s disease. Semin Hematol 1999, 36:260-269 [PubMed] [Google Scholar]
- 6.Montesinos-Rongen M, Roers A, Küppers R, Rajewsky K, Hansmann ML: Mutation of the p53 gene is not a typical feature of Hodgkin and Reed-Sternberg cells in Hodgkin’s disease. Blood 1999, 94:1755-1760 [PubMed] [Google Scholar]
- 7.Maggio EM, Stekelenburg E, van den Berg A, Poppema S: TP53 gene mutations in Hodgkin lymphoma are infrequent and not associated with absence of Epstein-Barr virus. Int J Cancer 2001, 94:60-66 [DOI] [PubMed] [Google Scholar]
- 8.Gravel S, Delsol G, Al Saati T: Single cell analysis of the t(14;18)(q31,q21) chromosomal translocation in Hodgkin’s disease demonstrates the absence of this rearrangement in neoplastic Hodgkin and Reed-Sternberg cells. Blood 1998, 91:2866-2874 [PubMed] [Google Scholar]
- 9.Emmerich F, Meiser M, Hummel M, Demel G, Foss HD, Jundt F, Mathas S, Krappmann D, Scheidereit C, Stein H, Dörken B: Overexpression of I kappa B alpha without inhibition of NF-κB activity and mutations in the I kappa B alpha gene in Reed-Sternberg cells. Blood 1999, 94:3129-3134 [PubMed] [Google Scholar]
- 10.Jungnickel B, Staratschek-Jox A, Bräuninger A, Spieker T, Wolf J, Diehl V, Hansmann ML, Rajewsky K, Küppers R: Clonal deleterious mutations in the IκBα gene in the malignant cells in HL. J Exp Med 2000, 91:395-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagata S, Golstein P: The Fas death factor. Science 1995, 267:1449-1456 [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Valdez H, Guret C, de Bouteiller O, Fugier I, Banchereau J, Liu YJ: Human germinal-center B cells express the apoptosis inducing genes Fas, c-myc, p53 and Bax but not the survival gene, bcl-2. J Exp Med 1996, 183:971-977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Eijk M, Defrance T, Hennino A, Groot C: Death-receptor contribution to the germinal center reaction. Trends Immunol 2001, 22:677-682 [DOI] [PubMed] [Google Scholar]
- 14.Müschen M, Re D, Jurgnickel B, Diehl V, Rajewsky K, Küppers R: Somatic mutation of the CD95 gene in human B cells as a side-effect of the germinal center reaction. J Exp Med 2000, 192:1833-1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe-Fukunaga R, Brannan PE, Copeland NG, Jenkins N, Nagata S: Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediate apoptosis. Nature 1992, 356:314-317 [DOI] [PubMed] [Google Scholar]
- 16.Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, Strober W, Lenardo MJ: Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell 1995, 81:935-946 [DOI] [PubMed] [Google Scholar]
- 17.Lim MS, Straus SE, Dale JK, Fleischer TA, Steler-Stevenson M, Strober W, Sneller MC, Puck JM, Lenardo MJ, Elenitoba-Johnson KS, Lin AY, Raffeld M, Jaffe ES: Pathological findings in a human autoimmune lymphoproliferative syndrome. Am J Pathol 1998, 153:1541-1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Straus SE, Jaffe ES, Puck JM, Dale JK, Elkon KB, Rosen-Wolff A, Peters AM, Sneller MC, Hallahan CW, Wang J, Fischer RE, Jackson CM, Lin AY, Baumler C, Siegert E, Marx A, Vaishnaw AK, Grodzicky T, Fleisher TA, Lenardo MJ: The development of lymphomas in families with autoimmune lymphoproliferative syndrome with germline Fas mutations and defective lymphocyte apoptosis. Blood 2001, 98:194-200 [DOI] [PubMed] [Google Scholar]
- 19.Xerri L, Carbuccia N, Parc P, Hassoun J, Birg F: Frequent expression of Fas/Apo-1 in Hodgkin’s disease and anaplastic large cell lymphomas. Histopathology 1995, 27:235-241 [DOI] [PubMed] [Google Scholar]
- 20.Metkar SS, Naresh KN, Redkar AA, Soman CS, Advani SH, Nadkarni JJ: Expression of Fas and Fas ligand in Hodgkin’s disease. Leuk Lymphoma 1999, 33:521-530 [DOI] [PubMed] [Google Scholar]
- 21.Re D, Hofmann A, Wolf J, Diehl V, Staratschek-Jox A: Cultivated H-RS cells are resistant to CD95L-mediated apoptosis despite expression of wild-type CD95. Exp Hematol 2000, 28:31-35 [DOI] [PubMed] [Google Scholar]
- 22.Hennino A, Bérard M, Krammer PH, Defrance T: FLICE-inhibitory Protein is a key regulator of germinal center B cell apoptosis. J Exp Med 2001, 193:447-458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djerbi M, Screpanti V, Catrina AI, Bogen B, Biberfeld P, Grandieri A: The inhibitor of death receptor signaling, FLICE-inhibitory protein defines a new class of tumor progression factors. J Exp Med 1999, 190:1025-1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irisarri M, Plumas J, Bonnefoix T, Jacob MC, Roucard, Pasquier MA, Sotto JJ, Lajmanovich A: Resistance to CD95-mediatyed apoptosis through constitutive c-FLIP expression in a non-Hodgkin lymphoma B cell line. Leukemia 2000, 14:2149-2158 [DOI] [PubMed] [Google Scholar]
- 25.Diehl V, Kirchner HH, Burrichter H, Stein H, Fonatsch C, Gerdes J, Schaadt M, Heit W, Uchanska-Ziegler B, Ziegler A, Heintz F, Sueno K: Characteristics of Hodgkin’s disease derived cell lines. Cancer Treat Rep 1982, 66:615-632 [PubMed] [Google Scholar]
- 26.Kanzler H, Hansmann ML, Kapp U, Wolf J, Diehl V, Rajewsky K, Kuppers R: Molecular single cell analysis demonstrates the derivation of a peripheral blood derived cell line (L1236) from the Hodgkin/Reed-Sternberg cells of a HL patient. Blood 1996, 87:3429-3436 [PubMed] [Google Scholar]
- 27.Poppema S, De Jong B, Atmosoerodjo J, Idenburg V, Visser L, De Ley L: Morphologic, immunologic, enzyme histochemical and chromosomal analysis of a cell line derived from Hodgkin’s disease. Evidence for a B-cell origin of Reed-Sternberg cells. Cancer 1985, 55:683-690 [DOI] [PubMed] [Google Scholar]
- 28.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD: The World Health Organization classification of neoplastic diseases of the haematopoietic and lymphoid tissues. Report of the Clinical Advisory Committee meeting, Airlie House, Virginia, November, 1997. Ann Oncol 1999, 10:1419-1432 [DOI] [PubMed] [Google Scholar]
- 29.Grønbaek K, Straten PT, Ralfkiaer E, Ahrenkiel V, Andersen MK, Hansen NE, Zeuthen J, Hou-Jensen K, Guldberg P: Somatic Fas mutations in non-Hodgkin’s lymphoma: association with extranodal disease and autoimmunity. Blood 1998, 91:3018-3024 [PubMed] [Google Scholar]
- 30.Van den Berg A, Maggio E, Diepstra A, Van Krieken J, Poppema S: Germ line FAS gene mutation in a case of ALPS and NLP Hodgkin lymphoma. Blood 2002, 99:1492-1494 [DOI] [PubMed] [Google Scholar]
- 31.Maggio EM, Van den Berg A, Visser L, Diepstra A, Kluiver J, Emmens R, Poppema S: Common and differential chemokine expression patterns in RS cells of NLP, EBV positive and negative classical Hodgkin lymphomas. Int J Cancer 2002, 99:665-672 [DOI] [PubMed] [Google Scholar]
- 32.Fiucci G, Ruberti G: Detection of polymorphisms within the Fas cDNA gene sequence by GC-clamp denaturing gradient gel electrophoresis. Immunogenetics 1994, 39:437-439 [DOI] [PubMed] [Google Scholar]
- 33.Müschen M, Warskulat U, Beckmann MW: Defining CD95 as a tumor suppressor gene. J Mol Med 2000, 78:312-325 [DOI] [PubMed] [Google Scholar]
- 34.Marafioti T, Hummel M, Foss HD, Laumen H, Korbjuhn P, Anagnostopoulos I, Lammert H, Demel G, Theil J, Wirth T, Stein H: Hodgkin and Reed-Sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood 2000, 95:1443-1450 [PubMed] [Google Scholar]
- 35.Müschen M, Re D, Brauninger A, Wolf J, Hansmann ML, Diehl V, Kuppers R, Rajewsky K: Somatic mutations of the CD95 gene in Hodgkin and Reed-Sternberg cells. Cancer Res 2000, 60:5640-5643 [PubMed] [Google Scholar]
- 36.Diehl V, Kirchner HH, Schaadt M, Fonatsch C, Stein H, Gerdes J, Boie C: Hodgkin’s disease: establishment and characterization of four in vitro cell lines. J Cancer Res Clin Oncol 1981, 101:111-124 [DOI] [PubMed] [Google Scholar]
- 37.Joos S, Kupper M, Ohl S, von Bonin F, Mechtersheimer G, Bentz M, Marynen P, Moller P, Pfreundschuh M, Trumper L, Lichter P: Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res 2000, 60:549-552 [PubMed] [Google Scholar]
- 38.Joos S, Menz CK, Wrobel G, Siebert R, Gesk S, Ohl S, Mechtersheimer G, Trumper L, Moller P, Lichter P, Barth TF: Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood 2002, 99:1381-1387 [DOI] [PubMed] [Google Scholar]
- 39.Müschen M, Rajewsky K, Kronke M, Kuppers R: The origin of CD95-gene mutations in B-cell lymphoma. Trends Immunol 2002, 23:75-80 [DOI] [PubMed] [Google Scholar]
- 40.Tamiya S, Etoh K, Suzushima H, Takatsuki K, Matsuoka M: Mutation of CD95 (Fas/Apo-1) gene in adult T-cell leukemia cells. Blood 1998, 91:3935-3942 [PubMed] [Google Scholar]
- 41.Beltinger C, Kurz E, Bohler T, Schrappe M, Ludwig WD, Debatin KM: CD95 (APO-1/Fas) mutations in childhood T-lineage acute lymphoblastic leukemia. Blood 1998, 91:3943-3951 [PubMed] [Google Scholar]
- 42.Lee SH, Shin MS, Park WS, Kim SY, Dong SM, Pi JH, Lee HK, Kim HS, Jang JJ, Kim CS, Kim SH, Lee JY, Yoo NJ: Alterations of Fas (Apo-1/CD95) gene in transitional cell carcinomas of urinary bladder. Cancer Res 1999, 59:3068-3072 [PubMed] [Google Scholar]
- 43.Lee SH, Shin MS, Park WS, Kim SY, Dong SM, Pi JH, Lee HK, Kim HS, Jang JJ, Kim CS, Kim SH, Lee JY, Yoo NJ: Alterations of Fas (Apo-1/CD95) gene in non-small cell lung cancer. Oncogene 1999, 18:3754-3760 [DOI] [PubMed] [Google Scholar]
- 44.Muzio M, Chinnaiyan AM, Kischkel FC, O′Rourke K, Shevchenko ANJ, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM: FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95(Fas/Apo-1) death-inducing signaling complex. Cell 1996, 85:817-827 [DOI] [PubMed] [Google Scholar]
- 45.Medema JP, Scaffidi C, Kischkel FC, Shevchenko ANJ, Mann M, Krammer PH, Peter ME: FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J 1997, 16:2794-2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas RK, Kallenborn A, Wickenhauser C, Schultze JL, Draube A, Vockerodt M, Re D, Diehl V, Wolf J: Constitutive expression of c-FLIP in Hodgkin and Reed-Sternberg cells. Am J Pathol 2002, 160:1521-1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hennino A, Berard M, Casamayor-Palleja M, Krammer PH, Defrance T: Regulation of the Fas death pathway by FLICE-inhibitory protein in primary human B cells. J Immunol 2000, 165:3023-3030 [DOI] [PubMed] [Google Scholar]
- 48.Müller M, Wilder S, Bannasch D, Israeli D, Lehlback K, Li-Weber M, Friedmann SL, Galle PR, Stremmel W, Oren M, Krammer PH: p53 activates the CD95 (Apo-1/Fas) gene in response to DNA damage by anticancer drugs. J Exp Med 1998, 188:2033-2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett M, Macdonald K, Chan SW, Luzio JP, Simari R, Weissberg P: Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science 1998, 282:290-293 [DOI] [PubMed] [Google Scholar]