Abstract
Therapies for liver diseases with stem and progenitor cells will require a detailed knowledge of the molecular mechanisms driving the in vivo differentiation process toward adult hepatic tissue. We applied quantitative gene expression methods to analyze the differentiation process of fetal liver progenitor cells after transplantation into an animal model of liver regeneration. Enhanced green fluorescent protein (EGFP)-transgenic liver progenitor cells were isolated from fetal mouse liver at stage embryonic day 13.5 and transplanted into uPA/RAG-2 mice. Two, 4, and 6 weeks after cell transplantation cryosections of liver tissue were analyzed for EGFP-positive regeneration nodules. RNA from laser-microdissected EGFP-positive tissue was isolated and used as template for quantitative real-time reverse transcriptase-polymerase chain reaction. Phenotypic differentiation was analyzed by staining of the canalicular marker enzyme dipeptidyl-peptidase IV. Proliferation in regenerative nodules and surrounding tissue was monitored with the BrdU incorporation assay. Alpha fetoprotein gene expression had already decreased 2 weeks after transplantation in EGFP-positive regeneration nodules compared to pretransplantation values and was not detectable after 4 and 6 weeks, whereas albumin slightly increased in transplanted cells indicating differentiation into a mature phenotype. The dipeptidyl-peptidase IV antigen was associated with some liver progenitor cells 2 weeks after transplantation and in virtually all cells after 4 and 6 weeks. Cell proliferation index in transplanted cells was maximally increased (4.8% BrdU-positive cells) after 2 weeks and decreased (0.4%) after 6 weeks to normal levels. Our results demonstrate that gene expression in liver progenitor cells changes from fetal to adult phenotype within 4 to 6 weeks after transplantation despite ongoing proliferation of the transplanted cells in a mouse model of liver regeneration. Quantitative gene expression profiles as shown here will have important implications in our understanding of the in vivo differentiation process of stem cells.
Cell senescence and the proliferation of hepatocytes that retain their functional repertoire are the primary limitations of the production of the substantial numbers of hepatocytes 1 necessary for liver cell therapies to become routine. 2-4 Stem cells representing a renewable resource for cell transplantation are actively studied for their ability to generate parenchymal cells and tissues and may thus open up new avenues in the treatment of liver disease. When normal reparative pathways of regeneration are impaired, the liver activates a facultative stem cell compartment to restore the liver mass and function. Like other epithelial organs, facultative liver stem cells display an extensive growth potential and are pluripotent with regard to their differentiation potential. 5-9 Recent reports suggest that stem cell tissues from outside the liver may also participate in the hepatic regeneration process. Cells derived from transplanted bone marrow differentiate into hepatocytes after liver injury and proliferate in the liver of animals with impaired hepatic regeneration. 10-12 Bone marrow-derived hepatocytes have also been detected in the livers of humans with substantially no liver damage, indicating participation of bone marrow in physiological liver regeneration. 13
Stem cells with the ability to differentiate into adult hepatocytes can also be derived from embryonic tissue. Embryonic stem cells differentiate into hepatic precursor cells in vitro, when appropriate mediators are supplied. 14 Determined liver progenitor cells can be isolated and cultured from the early embryonic liver and from fetal liver. 15 Fetal liver cells from rats and mice have been shown to differentiate into cells with biliary and hepatic phenotype after transplantation into recipient animals. 16,17
Lack of normal nontransformed liver progenitor cell lines has resulted in limited understanding of the linear relationships of different stages of liver cell differentiation. Consequently, differentiation and transdifferentiation of stem cells into fully functional hepatocytes have been observed only after transplantation experiments in vivo. Genetic information at different stages of differentiation, however, is critical for our understanding of lineage commitment and development into parenchymal cells.
In the present study we combine cell transplantation with gene expression analysis in laser-microdissected tissue samples to characterize the in vivo differentiation of transplanted fetal liver progenitor cells. We generated quantitative expression data for the developmentally regulated genes albumin and alpha fetoprotein (AFP) and followed changes in gene expression levels throughout the liver regeneration process. Our results demonstrate that only 2 weeks after transplantation into uPA/RAG2 mice, differentiation toward a mature hepatic phenotype can already be detected despite ongoing proliferation of the transplanted cells in the regenerative liver.
Materials and Methods
Animals
Enhanced green fluorescent protein (EGFP)-transgenic mice (C57BL/6-TgN(ACTbEGFP)1Osb) were purchased from The Jackson Laboratory, Bar Harbor, ME). The uPA/RAG-2 mice were generated by crossbreeding of uPA-transgenic mice originally described by Sandgren and colleagues 18 with the RAG-2 mouse as described elsewhere. 19,20 All animals were maintained and handled in accordance with institutional guidelines. For preparing fetal liver progenitor cells from EGFP embryos (embryonic day 13.5) the livers were removed under the binocular microscope. Cells were isolated by collagenase/dispase (Roche, Mannheim, Germany) digestion for 20 minutes at 37°C. The cells were washed twice in cold Dulbecco’s modified Eagle’s medium with 10% fetal calf serum, resuspended in phosphate-buffered saline (PBS) at 3 × 107 cells/ml and stored on ice. Intrasplenic transplantation of 1.5 × 106 cells in 13- to 21-day-old uPA/RAG-2 mice was performed under ketamine/rompun anesthesia. After 2, 4, and 6 weeks mice were sacrificed and the livers were removed, embedded in OCT, and snap-frozen in liquid nitrogen. One hour before sacrifice, 2 mg of BrdU solution was injected intraperitoneally for analysis of the proliferation activity.
Flow Cytometry
A 1:10 aliquot of the isolated fetal liver progenitor cells was incubated with a biotin-conjugated anti-mouse TER119 antibody (1:100 dilution) for 5 minutes on ice. Cells were washed and stained with a 1:100 dilution of a streptavidin-phycoerythrin conjugate (SAv-PE; both from BD Pharmingen, Heidelberg, Germany) for an additional 5 minutes on ice. After final washing the cells were analyzed in the presence of 1 μg/ml of propidium iodide with a FACSCalibur fluorescence flow cytometer (BD Biosciences, Heidelberg, Germany).
Staining Procedures
For hemalaun-eosin staining 5-μm cryosections were fixed with 4% p-formaldehyde (Merck, Darmstadt, Germany) in PBS for 5 minutes, washed, stained with hemalaun for 3 minutes, washed again, and stained with 0.5% eosin for an additional 3 minutes. After an additional washing step with water the slides were dehydrated in 70%, 96%, and 100% ethanol, and in xylol before they were embedded in DPX (Polysciences, Eppelheim, Germany).
Immunofluorescence staining of the dipeptidyl-peptidase (DPPIV) antigen was performed with acetone-fixed 5-μm cryosections using the anti-mouse CD26 (DPPIV) antibody (BD Pharmingen) in a 1:200 dilution. We used Cy3-labeled goat anti-rat IgG (Jackson ImmunoResearch/Dianova, Hamburg, Germany) in a 1:400 dilution as the second antibody and a 1:200 diluted AlexaFluor 488-labeled anti-EGFP antibody (A-21311; Molecular Probes, Leiden, The Netherlands) for detecting the EGFP-positive regeneration nodules, because their fluorescence is lost during acetone fixation.
For analysis of the proliferation activity the BrdU-labeling Kit I (Roche, Mannheim, Germany) was used according to the protocol given by the manufacturer with one exception: as the second antibody Cy3-conjugated goat anti-mouse IgG polyclonal antibody (Jackson ImmunoResearch) was used at a 1:200 dilution. For counterstaining of the nuclei we added 1 nmol/L of 4,6-diamidino-2-phenylindole (Sigma, Taufkirchen, Germany) and for visualization of the regeneration nodules (1:200 dilution) AlexaFluor 488-labeled anti-EGFP antibody during the incubation of the second antibody.
Laser-Manipulated Microdissection and Laser Pressure Catapulting
For laser-manipulated microdissection and laser-pressure catapulting we used the PALM Microbeam System (P.A.L.M., Bernried, Germany). The system was adapted to an inverted fluorescence microscope (Carl Zeiss, Oberkochen, Germany) and was equipped with a digital video camera and appropriate image analysis systems. Cryostat sections (10 μm) of livers from transplanted uPA-mice were mounted onto a 0.9-μm polyester membrane on thin microscope slides and kept at −20°C. Before performing laser-manipulated microdissection the specimens were dried at 37°C. EGFP-positive regeneration nodules were identified by fluorescence microscopy and areas with ∼50 cells were cut by laser-assisted microdissection. One μl of RLT lysis buffer (RNeasy kit; Qiagen, Hilden, Germany) was pipetted in a cap of a 0.5-ml Eppendorf tube, the cap was moved close to and above the specimen and the microdissected cell area was captured by laser-pressure catapulting. Afterward the cap was placed on its tube that was filled with an additional 9 μl of RLT buffer. The captured cell areas were collected by centrifugation at 8000 × g for 3 minutes.
RNA Preparation and cDNA Synthesis
The RNeasy Mini Kit (Qiagen) was used for isolation of total RNA of all samples. DNase digestion of 7 μl of total RNA was performed using 1 U DNase (Gibco Life Tech, Karlsruhe, Germany) with the supplied buffer in presence of 40 U of RNasin (Promega, Mannheim, Germany). For cDNA synthesis 10 μl of total RNA were mixed with 1 μl (100 ng) of T7-oligo dT primer 5′ GCATTAGCGGCCGCGAAATTAATACGACTCACTATAGGGAGA(T)21ACG 3′ and 9 μl of RT premix (4 μl 5× first strand buffer, 2 μl of 100 mmol/L dithiothreitol, 1 μl of 10 mmol/L dNTP, and 100 U Superscript II Polymerase (Gibco Life Tech). Samples were incubated for 45 minutes at 42°C followed by heat inactivation at 65°C for 15 minutes.
Quantitative Real-Time Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Specific mRNA were quantified using the TaqMan technique, 21-23 the qPCR Core Kit (Eurogentec, Seraing, Belgium), and the iCycler system (Bio-Rad, Munich, Germany). Primers and hybridization probes were designed with the Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) and were as follows: albumin: forward: 5′-CTCAGGTGTCAACCCCAA-3′, reverse: 5′-TCCACACAAGGCAGTCTC-3′, probe: 5′-FAM-CGTGGAGGCTGCAACAAACCTAGG-TAMRA-3′, AFP: forward: 5′-GTTTTCTGAGGGATGAAACCTATG-3′, reverse: 5′-GAAGCTCTTGTTTCATGGTCTGTA-3′, probe: 5′-FAM-CTTCCACAAGGATCTGTGCCAAGC-TAMRA-3′, GAPDH: forward: 5′AAGGAGTAAGAAACCCTGGACCAC-3′, reverse: 5′-GAAATTGTGAGGGAGATGCTCAGT-3′, probe: 5′-FAM-CACTGAGCAAGAGAGAGGCCCTATCC-TAMRA-3′, EGFP: forward: 5′-CGACGGCAACTACAAGAC-3′, reverse: 5′-TAGTTGTACTCCAGCTTGTGC-3′, probe: 5′-FAM-ACTTCAAGGAGGACGGCAACATCCT-TAMRA-3′. Ten pmol/L of the primer and hybridization probes each were used in a reaction volume of 50 μl. The amplification protocol was: 30 seconds at 95°C, 35 seconds at 55°C for 40 cycles after an initial denaturation step of 10 minutes at 95°C. All experiments were done in triplicates and threshold cycle (Ct) values are given in means ± SD.
Results
Characterization of Isolated Fetal Liver Cells
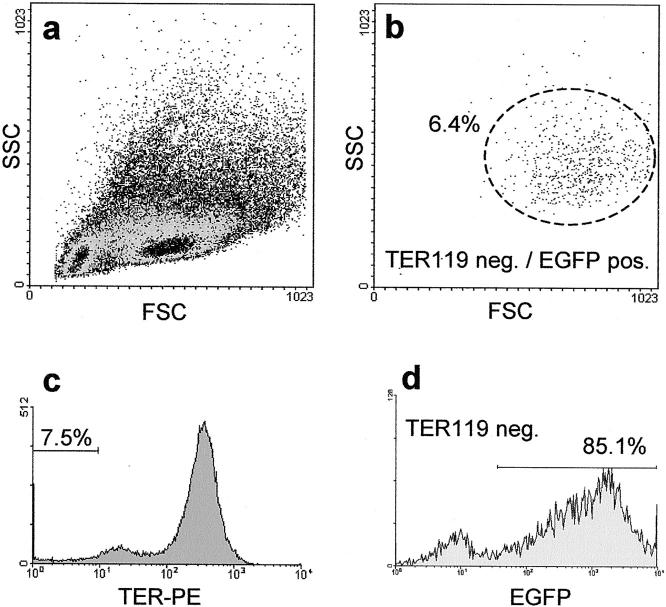
EGFP-transgenic breeder pairs were phenotyped for green fluorescence of their naked skin and eyes using a standard blacklight lamp. Because EGFP homozygous mice were reported to be lethal, 24 heterozygous mice were used with the consequence that some prepared fetal livers did not contain EGFP-positive cells. Nucleated cells express EGFP, but not mature red blood cells, which are the most common cells in fetal liver. Therefore, freshly prepared cells from fetal livers were characterized by flow cytometry (Figure 1, a and b) ▶ . Of all isolated vital cells 92.5% showed TER119-positive staining (Figure 1c) ▶ and were EGFP-negative. From the remaining 7.5% TER-negative cells the majority (85.1%, representing 6.4% of all cells) was EGFP-positive (Figure 1d) ▶ , illustrating only a small proportion of EGFP-negative fetal livers in the initial tissue preparation procedure. The propidium iodide exclusion test revealed an overall vitality of 97.5%.
Figure 1.
FACS analysis of day 13.5 fetal liver progenitor cells of EGFP-C57BL/6 mice. For fluorescent cell cytometry analysis of cell suspension derived from fetal liver (embryonic day 13.5) staining with the erythrocyte marker TER119 was performed and vitality was determined by propidium iodide (PI) exclusion. Cells (n = 100,000) were collected and PI-negative cells (97.5%, data not shown) were analyzed by the foreword/sideward scatter (a). Most of the cells in fetal livers were of hematopoietic origin with a high percentage of erythrocytes (92.5% TER-positive) that did not express EGFP (c). Because heterozygous mice were used for the mating, there are some embryos with EGFP-negative liver progenitor cells. From the remaining 7.5% TER-negative cells the majority was EGFP-positive (85.1%), demonstrating a small number of cells derived from EGFP-negative fetal livers only (d). The overall percentage of TER119-negative EGFP-positive cells was 6.4%. These cells contained the fraction of liver progenitor cells and were distributed typically in the FSC/SSC blot (b).
Table 1 ▶ gives a summary of the mouse transplantation procedures. As recipients we used immunodeficient mice that express the uroplasminogen activator in the liver, controlled by the albumin promotor (uPA/RAG2-mice). 11,12 Because for each transplantation 1.5 × 106 total fetal liver cells were used we estimated to have transplanted ∼1 × 105 EGFP-positive/TER-negative cells (6.4% of total cells). From the analysis of at least six different specimens per liver we calculated a total of 100 to 150 EGFP-positive regeneration nodules representing 1 to 2% of the regenerated liver mass.
Table 1.
Experimental Groups of uPA/RAG2 Mice Transplanted with EGFP-Positive Fetal Liver Progenitor Cells
| Mouse no. | No. of transplanted cells | Time of sacrifice | Proliferation activity |
|---|---|---|---|
| 1245 | 1.5 × 106 cells | 2 weeks after transplantation | (64/1342) |
| 1246 | 4.77% | ||
| 1258 | |||
| 1255 | 1.5 × 106 cells | 4 weeks after transplantation | (27/1625) |
| 1272 | 1.66% | ||
| 1303 | 1.5 × 106 cells | 6 weeks after transplantation | (6/1453) |
| 1305 | 0.41% | ||
| 1308 |
Experimental groups of uPA/RAG2 mice transplanted with EGFP-transgenic fetal liver progenitor cells. The proliferation activity was calculated by the number of BrdU-incorporating versus the total number of DAPI-stained nuclei in 10 fetal liver progenitor cell-derived regeneration nodules.
Analysis of EGFP-Positive Regeneration Nodules
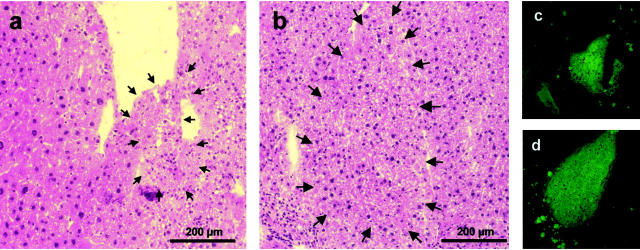
Two weeks after fetal liver progenitor cell transplantation, small EGFP-positive regeneration nodules were detectable by immunofluorescence in the uPA/RAG2 mouse liver. Hemalaun-eosin staining showed distinct regeneration nodules surrounded by diseased liver tissue (Figure 2, a and c) ▶ as well as larger EGFP-negative parenchyma of endogenous regenerated liver cells. Four weeks (Figure 2, b and d) ▶ and 6 weeks after the cell transplantation, the EGFP-positive nodules became larger and were more and more integrated in the EGFP-negative regenerated parenchyma, although some demarcation was visible most likely because of freezing and staining procedures.
Figure 2.
Regeneration nodules after transplantation of EGFP-positive fetal liver progenitor cells in uPA/RAG-2 mice. Five-μm cryosections of uPA/RAG-2 mouse livers were stained with hemalaun-eosin and consecutive slides were analyzed for EGFP fluorescence after EGFP-positive fetal liver progenitor cell transplantation. Two weeks (a and c) after transplantation, small EGFP-positive regeneration nodules (arrows) are detectable between normal endogenously regenerated (a, left) and uPA-damaged liver tissue (a, right). After 4 weeks (b and d) the damaged liver tissue nearly was diminished and the nodules have become larger, whereby the liver morphology in the H&E staining developed a normal pattern (b). On the right part of this nodule there is a demarcation line, which is probably a result of the freezing, fixation, and staining procedure.
For further analysis of these EGFP-positive regeneration nodules (Figure 3; a, d, and g) ▶ immunofluorescence staining of the ecto-ATPase DPPIV was performed. DPPIV is typically located on the apical membrane of hepatocytes resulting in a canalicular-staining pattern (Figure 3; b, e, and h) ▶ . The overlay of the corresponding EGFP and DPPIV fluorescence proved the presence of DPPIV in fetal liver cell-derived regeneration nodules 2, 4, and 6 weeks after transplantation (Figure 3; c, f, and i) ▶ . Additionally these nodules were increasingly well integrated in the surrounding tissue throughout time (Figure 3f) ▶ , which suggests functionality of the transplanted cells.
Figure 3.
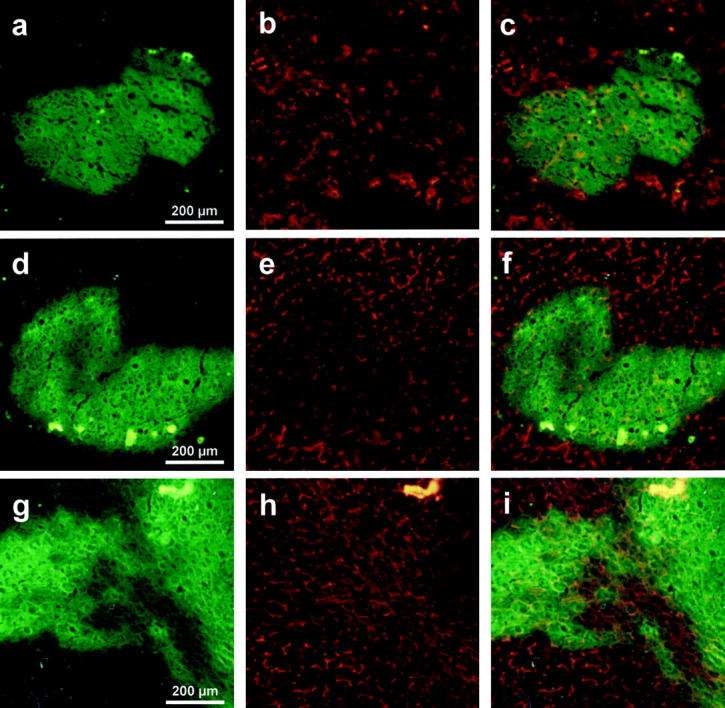
Immunofluorescence analysis of the DPPIV antigen in EGFP-positive regeneration nodules. Five-μm cryosections from uPA/RAG2-mouse livers were analyzed 2 weeks (no. 1246, a–c), 4 weeks (no. 1255, d–f), and 6 weeks (no. 1308, g–i) after transplantation of EGFP fetal liver cells. For analysis of the fluorescent (green) regeneration nodules co-staining of the hepatic marker enzyme DPPIV was performed (red fluorescence), illustrating the canalicular membranes of polarized hepatocytes (b, e, h). The overlay of the green fluorescent regeneration nodules with the red DPPIV signal shows the presence of few polarized hepatocytes 2 weeks after transplantation (c), much more after 4 weeks (f), and a normal pattern after 6 weeks (i), demonstrating the developing liver architecture and the integration of the regenerative tissue.
During the regeneration process cell proliferation is needed to restore the cell mass of the diseased liver, which was investigated by BrdU incorporation. Ten areas of fetal liver cell-derived green fluorescent regeneration nodules were analyzed 2, 4, and 6 weeks after cell transplantation by counting BrdU-incorporating versus 4,6-diamidino-2-phenylindole-stained nuclei. Two weeks after transplantation ∼4.8% of the fetal liver cells showed proliferation activity (Table 1 ▶ ; Figure 4, a and b ▶ ), decreasing to 1.7% (Figure 4, c and d) ▶ and 0.4% 4 and 6 weeks after transplantation, respectively. Most of the proliferating cells were observed in the periphery, which suggests expansion of the regeneration nodules. As expected, 2 weeks after transplantation endogenous proliferating nodules were also detectable (∼1.9% BrdU-positive nuclei), but 4 weeks and 6 weeks after transplantation the surrounding tissue shows no increased proliferation activity (Figure 4, e and f) ▶ .
Figure 4.
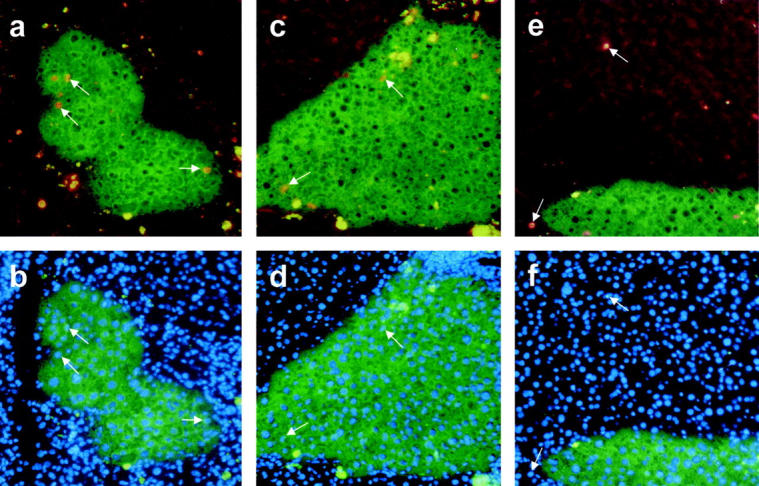
Proliferation activity index in EGFP-positive regeneration nodules. Specimens of fetal liver progenitor cell-derived regeneration nodules (green fluorescence) in uPA/RAG2-mouse liver were analyzed 2 weeks (no. 1246, a and b) and 4 weeks (no. 1255, c–f) after transplantation for BrdU incorporation (red nuclear fluorescence, arrows in a, c, e). The ratio of the number of BrdU versus the total number of 4,6-diamidino-2-phenylindole staining of all nuclei (b, d, f) was calculated, as demonstrated by the corresponding arrows. Some unspecific autofluorescence in the diseased liver tissue appears in the green and red channel resulting in yellow spots around the regeneration nodules. Increased proliferation activity is shown in regeneration nodules at 2 weeks (a and b) and at a lower degree also at 4 weeks (c and d) after fetal liver progenitor cell transplantation, whereas in the surrounding endogenously regenerated liver tissue (e and f) only few proliferating cells are detectable (see also Table 1 ▶ ).
Gene Expression Profiles in Fetal Liver Cell-Derived Tissue
Gene expression profiling is a suitable tool for staging differentiating processes in the liver because transcripts of the fetal liver cell protein AFP are hardly detectable in adult tissue, whereas the RNA level of the major secretion protein albumin rises constantly during liver development. Regeneration nodules derived from the transplanted cells were identified by fluorescence microscopy and isolated by laser-mediated microdissection. In preliminary examinations a pool of five microdissected areas, each of ∼50 cells, has been proven to result in sufficient RNA quantities for 12 analyses, which are needed for expression profiling of four genes in triplicates. This amount of RNA gave Ct values in the same order of magnitude as obtained with 2 ng of standard cDNA. Therefore, the complete RNA of five pooled areas was transcribed into cDNA (20 μl) and diluted 1:3 for 12 × 5-μl sample volumes.
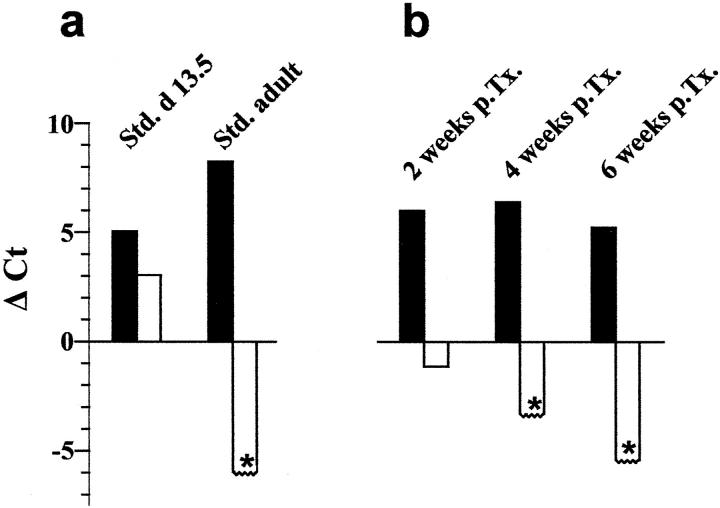
For the analysis of the eight different mouse liver samples, two iCycler runs had to be performed, including nontemplate controls and standard cDNA of adult and fetal (day 13.5) mouse liver (Table 2) ▶ . Quantitative RT-PCR of 2 ng of fetal liver cDNA gave Ct values of 26.7 and 25.8 for albumin, of 29.3 and 27.2 for AFP and of 32.3 and 30.3 for the housekeeping gene GAPDH (Table 2) ▶ , resulting in a difference of albumin and AFP expression of ∼1.4 to 2.6 cycles. For comparison, in 2 ng of adult liver cDNA Ct values of 24.1 and 23.5 were measured for albumin and Ct values of 32.6 and 31.5 for GAPDH, whereas the AFP expression was not detectable until cycle 38 in these samples. So, the albumin/AFP difference rises to >13.9 in adult mouse liver. No EGFP expression was found in our standard cDNA of fetal and adult liver from wild-type mice. These data were additionally shown in Figure 5a ▶ , by normalizing the albumin and AFP expression as differences to the GAPDH Ct values.
Table 2.
Gene Expression Profile of Regeneration Nodules 2, 4, and 6 Weeks after Transplantation of Day 13.5 Fetal Liver Progenitor Cells in uPA/RAG2 Mice
| Sample | Albumin | AFP | GAPDH | EGFP | Δ (Alb-AFP) |
|---|---|---|---|---|---|
| Standard, day 13.5 2 ng | 26.7 ± 0.38 | 29.3 ± 0.35 | 32.3 ± 0.59 | n.d. (>38) | 2.6 |
| Standard, day 13.5 2 ng | 25.8 ± 0.40 | 27.2 ± 0.26 | 30.3 ± 0.49 | n.d. (>38) | 1.4 |
| Standard, adult 2 ng | 24.1 ± 0.31 | n.d. (>38) | 32.6 ± 0.58 | n.d. (>38) | >13.9 |
| Standard, adult 2 ng | 23.5 ± 0.06 | n.d. (>38) | 31.5 ± 0.31 | n.d. (>38) | >14.5 |
| No. 1245, 2 weeks p.Tx. | 27.3 ± 0.35 | 37.5 ± 0.42 | 35.7 ± 0.28 | 37.4 ± 0.78 | 10.2 |
| No. 1246, 2 weeks p.Tx. | 32.7 ± 0.10 | 37.6 ± 0.40 | 35.3 ± 0.55 | 37.7 ± 0.44 | 4.9 |
| No. 1258, 2 weeks p.Tx. | 30.2 ± 0.52 | 36.5 ± 0.78 | 37.2 ± 1.36 | 36.6 ± 0.75 | 6.3 |
| No. 1255, 4 weeks p.Tx. | 27.8 ± 0.84 | n.d. (>38) | 34.5 ± 0.44 | 34.0 ± 0.42 | >10.2 |
| No. 1272, 4 weeks p.Tx. | 28.8 ± 0.42 | n.d. (>38) | 34.9 ± 0.97 | 35.6 ± 0.42 | >9.2 |
| No. 1303, 6 weeks p.Tx. | 27.5 ± 0.17 | n.d. (>38) | 32.3 ± 0.12 | 32.3 ± 1.05 | >10.5 |
| No. 1305, 6 weeks p.Tx. | 26.7 ± 0.61 | n.d. (>38) | 32.3 ± 0.51 | 31.3 ± 0.38 | >11.3 |
| No. 1308, 6 weeks p.Tx. | 27.9 ± 0.25 | n.d. (>38) | 33.2 ± 0.25 | 33.2 ± 0.75 | >10.1 |
p.Tx.: after transplantation.
Samples of EGFP-positive regeneration nodules (2, 4, and 6 weeks after EGFP-transgenic fetal liver cell transplantation) were isolated by laser-assisted microdissection. Five areas were pooled for each sample before RNA extraction and cDNA synthesis was performed. For quantitative RT-PCR analysis of albumin, AFP, GAPDH, and EGFP expression the Taqman method was used. The resulting Ct values are given and show a distinct expression profile for adult and fetal (day 13.5) liver standard cDNA, respectively. Ct values over 38 were out of the detection range and marked as n.d. (not detectable). The increasing albumin/AFP difference indicates the differentiation of the transplanted fetal liver cells towards mature hepatocytes.
Figure 5.
Expression profiles of albumin and AFP in liver progenitor cell differentiation. The albumin (black bars) and AFP expression (white bars) was normalized to GAPDH by subtracting their Ct values from the GAPDH Ct value (see Table 2 ▶ ). For calculating GAPDH-AFP Ct differences the detection limit of AFP expression was set at a value of 38 (asterisks). For each experimental group the means were calculated and shown in this figure. In the control experiments (a) the standard cDNA of day 13.5 fetal liver and adult liver showed an albumin expression of +5.05 and +8.25, respectively. In contrast the AFP expression was +3.05 in the fetal liver cDNA and under the detection limit (<−5.95) in adult liver cDNA. Two, 4, and 6 weeks after transplantation (b) the expression profiles of the transplanted liver progenitor cells altered toward mature hepatocytes. The albumin expression remained constant (+6.00, +6.40, +5.23), whereas the AFP-expression decreased gradually (−1.13 after 2 weeks) under the detection limit (<−3.30 and <−5.4 after 4 and 6 weeks, respectively).
For staging of the gene expression profile at different time points after transplantation of EGFP-positive fetal liver cells in uPA/RAG2 mice these control data were used. Two weeks after cell transplantation only weak AFP expression was detectable with Ct values ranging from 36.5 to 37.5 (Table 2) ▶ . The resulting albumin/AFP differences were 4.9 (mouse no. 1246), 6.3 (mouse no. 1258), and 10.2 (mouse no. 1245) indicating a more differentiated stage of the transplanted fetal liver cells.
These differences became even larger in regeneration nodules analyzed after 4 and 6 weeks. The AFP expression level was decreased under the detection limit (Ct > 38), whereby the albumin expression remained in the same range compared to GAPDH and EGFP expression. Analysis of the albumin/AFP differences (>10.2, >9.2, >10.5, >11.3, and >10.1; see mice nos. 1255 to 1308 in Table 2 ▶ ) and of the normalized expression levels (Figure 5b) ▶ demonstrates an even more differentiated gene expression profile, suggesting a terminal differentiation into mature hepatocytes of the transplanted fetal liver cells.
Discussion
The early fetal liver contains progenitor cells, which have the ability to differentiate either into hepatocytes or biliary epithelial cells. A small fraction of cells even exhibit bipotential activity and generate both epithelial cell types. 15,17 Fetal liver progenitor cells proliferate extensively in vitro 16 and differentiate into adult parenchymal phenotypes after transplantation into a host animal. 17,25 In the present study we used EGFP-transgenic mice as donors for fetal liver progenitor cells. In contrast to other transplantation systems such as the DPPIV rat model, 26 which relies on staining of a developmentally regulated membrane enzyme, all nucleated cells in EGFP mice starting from oocytes constitutively express the fluorogenic transgene. 24 Furthermore, cells transplanted into the liver can be identified in native or fixed tissue regardless of the phenotype and differentiation status. Transplantation of the fetal liver cell suspension from embryonic day 13.5 embryos into the liver regeneration model uPA/RAG-2 resulted in fluorescent cell clusters within a period of 6 weeks after transplantation morphologically resembling hepatic cord structures. The fetal cell suspensions used in the transplantation experiments most likely also contained hematopoietic progenitor cells, because the liver is the predominant site of hematopoiesis during this phase of development. 27,28 In cell suspensions derived from fetal liver only a small fraction (7.5%) was negative for the surface marker TER 199, which identifies erythroid cells from the early erythroblast to the mature erythrocyte in flow cytometric analysis. 29 Despite strong hematopoietic activity and the presence of hematopoietic stem cells in the fetal liver, however, we did not observe hematopoiesis originating from transplanted fetal cells either in the liver or in the bone marrow.
The alb-uPA mice used as recipient animals for cell transplants express the uro-plasminogen protein exclusively in hepatocytes. The transgene destroys the hepatocytes after birth and induces a subacute liver failure. In the heterozygous state some host cells can delete the transgene and form large regeneration nodules. 19,20 The transplanted EGFP-transgenic liver progenitor cells participated in tissue regeneration and formed cohesive cell clusters within the host liver of the uPA mice. The high frequency of small- and medium-sized EGFP-positive regeneration nodules in our experiments suggests efficient initial integration of fetal liver progenitor cells in the host liver and subsequent clonal expansion. The regeneration originating from the transplanted cells always competes with endogenous regeneration and is terminated with restoration of the normal liver mass. In contrast, repopulation models based on hematopoietic stem cell transdifferentiation have resulted only in a few, but large cell clones after a long period of positive/negative selection pressure. 12
Identification of transplanted cells in the liver without tissue staining allowed us to follow and analyze the differentiation process during the early phase of liver regeneration. Recently, a contamination-free laser-assisted microdissection procedure was developed that is useful to isolate intact mRNA of frozen tissue samples. 30-32 In dried cryosections we were able to identify the EGFP-positive cell clusters by fluorescence microscopy, precisely isolate the target cells by laser microdissection and generate RNA from the tissue samples. Normalized for the GAPDH and EGFP mRNA content we quantified mRNAs for the hepatocyte-specific proteins AFP and albumin. For the first time this experimental approach provides quantitative data of the in vivo differentiation process at the transcriptional level. In a previous study from our group albumin has been shown to be expressed at developmental stage embryonic day 11.5 in mice at a level comparable to AFP. At later stages the albumin mRNA level constantly increases and reaches a maximum in the adult liver (A. Jochheim et al, manuscript in preparation). AFP mRNA expression also rises during fetal development, although at overall lower levels compared to albumin, and drops to very low levels immediately after birth. The quantitative mRNA levels of both genes thus represent excellent markers of liver maturation and may characterize the transition of fetal liver progenitor cells into mature hepatocytes. The data show that 2 weeks after transplantation the differentiation process in the transplanted cells is already initiated. The (AFP-ALB) Ct value substantially increased compared to values obtained from the primary fetal tissue, but did not reach the level obtained for adult liver tissue. After 4 and 6 weeks AFP mRNA was not detectable anymore, whereas the albumin/GAPDH ratio reached levels comparable to adult liver. These data strongly suggest terminal differentiation of the transplanted fetal liver progenitor cells into mature hepatocytes after 4 to 6 weeks. The staining pattern of the DPPIV antigen, which is associated with the mature parenchymal cells in the liver, 26 in EGFP-positive regeneration nodules also supports the obtained gene expression data. In 2- and 4-week-old cell clusters the DPPIV antigen staining appeared scattered and faint, whereas in 6-week-old nodules continuous networks with strong DPPIV staining had already formed.
It is notable that differentiation of fetal liver progenitor cells into mature hepatocytes occurred despite ongoing cell proliferation. BrdU pulse labeling showed a substantially increased proliferation index compared to normal liver tissue in 2-week-old regeneration nodules. After 4 weeks the index decreased but was still higher than in control tissue and dropped to levels comparable to normal liver tissue after 6 weeks. The BrdU-labeled nuclei were located predominantly in the periphery of regeneration nodules clearly indicating a growth of the cell clusters rather than increased turnover of cells caused by acute damage and apoptosis. Our data suggest that the differentiation pathway is distinct from the process leading to reactivation of AFP expression in carcinoma cells. 33 In fact, Dabeva and colleagues 34 have shown that cell division of hepatocytes is not associated with dedifferentiation and reactivation of AFP expression indicating dissociation of proliferation from AFP expression in hepatocytes.
Sandhu and colleagues 35 reported that embryonic day 14 fetal liver progenitor cells derived from DPPIV+ rats showed a higher proliferation capacity compared to adult hepatocytes and proliferated for up to 6 month after transplantation into a DPPIV− host liver and subsequent partial hepatectomy. In our model we could not observe a further increase in the size of regeneration nodules derived from liver progenitor cells beyond the 6 week period. Similar results have also been observed by transplanting adult hepatocytes, because the hepatocyte proliferative stimulus lasts in uPA/RAG-2 mice ∼8 to 10 weeks after birth, until the transplanted hepatocyte mass becomes comparable to the liver mass of nontransgenic normal mice.
Quantitative RT-PCR has been performed with RNA from <50 microdissected cells or <2 ng of total RNA. In a previous study we were able to show, that combination of laser-mediated microdissection with a linear RNA amplification protocol 36 makes even low abundance genes such as transcription factors accessible for molecular analysis (T. Cantz et al, manuscript in preparation). Our quantitative molecular approach may thus be applicable in the analysis of in situ differentiation involving scattered adult hepatic and extrahepatic stem cells. Recently, differentiation of adult stem cells into mature hepatocytes has been demonstrated in murine animal models as well as in humans. Transdifferentiation of bone marrow cells into liver progenitor cells and hepatocytes has been described. 10-12 Generation of hepatocytes from bone marrow cells also occurs after allergenic liver transplantation in humans and is associated with the frequency and severity of rejection episodes. 37 The molecular events, however, inducing differentiation and transdifferentiation in adult and embryonic/fetal stem cells are primarily unknown. Stage- and differentiation-specific gene expression patterns with quantitative PCR and genome-wide technologies may be helpful in unraveling the underlying molecular mechanisms and will be crucial for developing in vitro differentiation protocols for stem cell populations.
Acknowledgments
We thank Andrea Jochheim, Jennifer Scharf, and Tina Hillemann for expert help in generating the quantitative RT-PCR data.
Footnotes
Address reprint requests to Michael Ott, M.D., Department of Gastroenterology, Hepatology, and Endocrinology, Hanover Medical School, Carl-Neuberg-Straβe 1, D-30625 Hanover, Germany. E-mail: ott-mhh@gmx.de.
Supported in part by grants of the Deutsche Forschungsgemeinschaft (OT 131/4-1 and PE 608/2-3).
References
- 1.Cascio SM: Novel strategies for immortalization of human hepatocytes. Artif Organs 2001, 25:529-538 [DOI] [PubMed] [Google Scholar]
- 2.Fox IJ, Chowdhury JR, Kaufman SS, Goertzen TC, Chowdhury NR, Warkentin PI, Dorko K, Sauter BV, Strom SC: Treatment of the Crigler-Najjar syndrome type I with hepatocyte transplantation. N Engl J Med 1998, 338:1422-1426 [DOI] [PubMed] [Google Scholar]
- 3.Bilir BM, Guinette D, Karrer F, Kumpe DA, Krysl J, Stephens J, McGavran L, Ostrowska A, Durham J: Hepatocyte transplantation in acute liver failure. Liver Transpl 2000, 6:32-40 [DOI] [PubMed] [Google Scholar]
- 4.De Vree JM, Ottenhoff R, Bosma PJ, Smith AJ, Aten J, Oude Elferink RP: Correction of liver disease by hepatocyte transplantation in a mouse model of progressive familial intrahepatic cholestasis. Gastroenterology 2000, 119:1720-1730 [DOI] [PubMed] [Google Scholar]
- 5.Overturf K, Al Dhalimy M, Ou CN, Finegold M, Grompe M: Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol 1997, 151:1273-1280 [PMC free article] [PubMed] [Google Scholar]
- 6.Alison M, Sarraf C: Hepatic stem cells. J Hepatol 1998, 29:676-682 [DOI] [PubMed] [Google Scholar]
- 7.Yasui O, Miura N, Terada K, Kawarada Y, Koyama K, Sugiyama T: Isolation of oval cells from Long-Evans Cinnamon rats and their transformation into hepatocytes in vivo in the rat liver. Hepatology 1997, 25:329-334 [DOI] [PubMed] [Google Scholar]
- 8.Brill S, Holst P, Sigal S, Zvibel I, Fiorino A, Ochs A, Somasundaran U, Reid LM: Hepatic progenitor populations in embryonic, neonatal, and adult liver. Proc Soc Exp Biol Med 1993, 204:261-269 [DOI] [PubMed] [Google Scholar]
- 9.Thorgeirsson SS: Hepatic stem cells in liver regeneration. EMBO J 1996, 10:1249-1256 [PubMed] [Google Scholar]
- 10.Theise ND, Badve S, Saxena R, Henegariu O, Sell S, Crawford JM, Krause DS: Derivation of hepatocytes from bone marrow cells in mice after radiation-induced myeloablation. Hepatology 2000, 31:235-240 [DOI] [PubMed] [Google Scholar]
- 11.Petersen BE, Bowen WC, Patrene KD, Mars WM, Sullivan AK, Murase N, Boggs SS, Greenberger JS, Goff JP: Bone marrow as a potential source of hepatic oval cells. Science 1999, 284:1168-1170 [DOI] [PubMed] [Google Scholar]
- 12.Lagasse E, Connors H, Al Dhalimy M, Reitsma M, Dohse M, Osborne L, Wang X, Finegold M, Weissman IL, Grompe M: Purified hematopoietic stem cells can differentiate into hepatocytes in vivo. Nat Med 2000, 6:1229-1234 [DOI] [PubMed] [Google Scholar]
- 13.Theise ND, Nimmakayalu M, Gardner R, Illei PB, Morgan G, Teperman L, Henegariu O, Krause DS: Liver from bone marrow in humans. Hepatology 2000, 32:11-16 [DOI] [PubMed] [Google Scholar]
- 14.Hamazaki T, Iiboshi Y, Oka M, Papst PJ, Meacham AM, Zon LI, Terada N: Hepatic maturation in differentiating embryonic stem cells in vitro. FEBS Lett 2001, 497:15-19 [DOI] [PubMed] [Google Scholar]
- 15.Rogler LE: Selective bipotential differentiation of mouse embryonic hepatoblasts in vitro. Am J Pathol 1997, 150:591-602 [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki A, Zheng Y, Kondo R, Kusakabe M, Takada Y, Fukao K, Nakauchi H, Taniguchi H: Flow-cytometric separation and enrichment of hepatic progenitor cells in the developing mouse liver. Hepatology 2000, 32:1230-1239 [DOI] [PubMed] [Google Scholar]
- 17.Dabeva MD, Petkov PM, Sandhu J, Oren R, Laconi E, Hurston E, Shafritz DA: Proliferation and differentiation of fetal liver epithelial progenitor cells after transplantation into adult rat liver. Am J Pathol 2000, 156:2017-2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandgren EP, Palmiter RD, Heckel JL, Daugherty CC, Brinster RL, Degen JL: Complete hepatic regeneration after somatic deletion of an albumin-plasminogen activator transgene. Cell 1991, 66:245-256 [DOI] [PubMed] [Google Scholar]
- 19.Petersen J, Dandri M, Gupta S, Rogler CE: Liver repopulation with xenogenic hepatocytes in B and T cell-deficient mice leads to chronic hepadnavirus infection and clonal growth of hepatocellular carcinoma. Proc Natl Acad Sci USA 1998, 95:310-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dandri M, Burda MR, Torok E, Pollok JM, Iwanska A, Sommer G, Rogiers X, Rogler CE, Gupta S, Will H, Greten H, Petersen J: Repopulation of mouse liver with human hepatocytes and in vivo infection with hepatitis B virus. Hepatology 2001, 33:981-988 [DOI] [PubMed] [Google Scholar]
- 21.Holland PM, Abramson RD, Watson R, Gelfand DH: Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 1991, 88:7276-7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson UE, Heid CA, Williams PM: A novel method for real time quantitative RT-PCR. Genome Res 1996, 6:995-1001 [DOI] [PubMed] [Google Scholar]
- 23.Heid CA, Stevens J, Livak KJ, Williams PM: Real time quantitative PCR. Genome Res 1996, 6:986-994 [DOI] [PubMed] [Google Scholar]
- 24.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y: ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett 1997, 407:313-319 [DOI] [PubMed] [Google Scholar]
- 25.Lilja H, Arkadopoulos N, Blanc P, Eguchi S, Middleton Y, Meurling S, Demetriou AA, Rozga J: Fetal rat hepatocytes: isolation, characterization, and transplantation in the Nagase analbuminemic rats. Transplantation 1997, 64:1240-1248 [DOI] [PubMed] [Google Scholar]
- 26.Gupta S, Rajvanshi P, Lee CD: Integration of transplanted hepatocytes into host liver plates demonstrated with dipeptidyl peptidase IV-deficient rats. Proc Natl Acad Sci USA 1995, 92:5860-5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamiya A, Kinoshita T, Ito Y, Matsui T, Morikawa Y, Senba E, Nakashima K, Taga T, Yoshida K, Kishimoto T, Miyajima A: Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J 1999, 18:2127-2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita T, Sekiguchi T, Xu MJ, Ito Y, Kamiya A, Tsuji K, Nakahata T, Miyajima A: Hepatic differentiation induced by oncostatin M attenuates fetal liver hematopoiesis. Proc Natl Acad Sci USA 1999, 96:7265-7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikuta K, Kina T, MacNeil I, Uchida N, Peault B, Chien YH, Weissman IL: A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell 1990, 62:863-874 [DOI] [PubMed] [Google Scholar]
- 30.Schutze K, Posl H, Lahr G: Laser micromanipulation systems as universal tools in cellular and molecular biology and in medicine. Cell Mol Biol 1998, 44:735-746 [PubMed] [Google Scholar]
- 31.Schermelleh L, Thalhammer S, Heckl W, Posl H, Cremer T, Schutze K, Cremer M: Laser microdissection and laser pressure catapulting for the generation of chromosome-specific paint probes. Biotechniques 1999, 27:362-367 [DOI] [PubMed] [Google Scholar]
- 32.Fink L, Seeger W, Ermert L, Hanze J, Stahl U, Grimminger F, Kummer W, Bohle RM: Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med 1998, 4:1329-1333 [DOI] [PubMed] [Google Scholar]
- 33.Spear BT: Alpha-fetoprotein gene regulation: lessons from transgenic mice. Semin Cancer Biol 1999, 9:109-116 [DOI] [PubMed] [Google Scholar]
- 34.Dabeva MD, Laconi E, Oren R, Petkov PM, Hurston E, Shafritz DA: Liver regeneration and alpha-fetoprotein messenger RNA expression in the retrorsine model for hepatocyte transplantation. Cancer Res 1998, 58:5825-5834 [PubMed] [Google Scholar]
- 35.Sandhu JS, Petkov PM, Dabeva MD, Shafritz DA: Stem cell properties and repopulation of the rat liver by fetal liver epithelial progenitor cells. Am J Pathol 2001, 159:1323-1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baugh LR, Hill AA, Brown EL, Hunter CP: Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res 2001, 29:E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleeberger W, Rothamel T, Glockner S, Flemming P, Lehmann U, Kreipe H: High frequency of epithelial chimerism in liver transplants demonstrated by microdissection and STR-analysis. Hepatology 2002, 35:110-116 [DOI] [PubMed] [Google Scholar]