Abstract
Comprehensive expression profiling of tumors using DNA microarrays has been used recently for molecular classification and biomarker discovery, as well as a tool to identify and investigate genes involved in tumorigenesis. Application of this approach to a cohort of benign and malignant adrenocortical tissues would be potentially informative in all of these aspects. In this study, we generated transcriptional profiles of 11 adrenocortical carcinomas (ACCs), 4 adrenocortical adenomas (ACAs), 3 normal adrenal cortices (NCs), and 1 macronodular hyperplasia (MNH) using Affymetrix HG_U95Av2 oligonucleotide arrays representing ∼10,500 unique genes. The expression data set was used for unsupervised hierarchical cluster analysis as well as principal component analysis to visually represent the expression data. An analysis of variance on the three classes (NC, ACA plus MNH, and ACC) revealed 91 genes that displayed at least threefold differential expression between the ACC cohort and both the NC and ACA cohorts at a significance level of P < 0.01. Included in these 91 genes were those known to be up-regulated in adrenocortical tumors, such as insulin-like growth factor (IGF2), as well as novel differentially expressed genes such as osteopontin (SPP) and serine threonine kinase 15 (STK15). Increased expression of IGF2 was identified in 10 of 11 ACCs (90.9%) and was verified by quantitative reverse transcriptase-polymerase chain reaction. Select proliferation-related genes (TOP2A and Ki-67) were validated at the protein level using immunohistochemistry and adrenocortical tissue microarrays. Our results demonstrated significant and consistent gene expression changes in ACCs compared to benign adrenocortical lesions. Moreover, we identified several genes that represent potential diagnostic markers and may play a role in the pathogenesis of ACC.
Adrenocortical carcinoma (ACC) is a rare but highly lethal cancer with an annual incidence of 0.5 to 2 patients per million population. 1 The pathological diagnosis of ACC is straightforward in most cases, based on well-recognized features of malignancy, including large tumor size and weight, solid growth pattern, extensive tumor necrosis, fibrous bands, lipid-poor cells, abundant mitoses, atypical mitoses, nuclear pleomorphism, capsular invasion, and vascular invasion. 2 However, there are occasional adrenocortical tumors whose malignant potential is uncertain. Additionally, based on mitotic activity, it is possible to divide ACCs into prognostically significant low- and high-grade subgroups. 3 There are also undifferentiated tumors of the retroperitoneum that are not readily identified as adrenocortical in origin using routine pathological methods. Thus, additional insight into the pathology of these tumors is clearly needed.
Several genes have been reported to have diagnostic significance in adrenocortical neoplasms. Numerous studies have documented the utility of α-inhibin immunohistochemistry as a marker of adrenocortical differentiation, 4-8 with the ability to distinguish adrenocortical tumors from adrenomedullary tumors, hepatocellular carcinoma and renal tumors, 9 resulting in the acceptance of α-inhibin as a clinically useful diagnostic marker. Several studies have investigated the ability of proliferative immunohistochemical markers, such as Ki-67 and topoisomerase II α (TOP2A), to distinguish benign and malignant tumors, 10-15 resulting in a general consensus that proliferative activity as measured by these markers is significantly higher in ACCs than benign lesions. Interestingly, assessment of proliferation by proliferative cell nuclear antigen immunohistochemistry did not show a correlation with biological behavior. 10 Finally, several studies have shown that immunoreactivity to p53 is essentially restricted to ACCs. 14-16
Despite some recent advances, the molecular pathogenesis of ACC is poorly understood. Mutations of the p53 tumor suppressor gene have been implicated because of the association of ACC with Li-Fraumeni syndrome 17 and confirmed by mutational analyses. 18-20 The insulin-like growth factor II gene (IGF2) is involved in the pathogenesis of both familial ACCs, as is the case in Beckwith-Wiedemann syndrome, 21 as well as in sporadic ACCs. 22,23 Dysregulation or rearrangement at 11p15.5 results in significant up-regulation of IGF2 in ACC, resulting in an autocrine stimulatory loop. 24
Gene expression profiling provides the opportunity to assess the expression of thousands of genes simultaneously in a cohort of related tumors. Several practical applications, including but not limited to tumor classification, 25-27 biomarker discovery, 28 and prediction of therapeutic response, 29 are being developed for a variety of tumors, including those of the endocrine system. 30 Furthermore, expression profiles can provide insight into tumorigenesis and identify targets for therapeutic intervention. 31,32 Here, we report the generation of extensive gene expression profiles of a cohort of normal, hyperplastic, and neoplastic adrenocortical tissues and show that these profiles can readily distinguish benign from malignant tumors and identify tumors with unusual histopathological features. In addition, we identify numerous differentially expressed transcripts and demonstrate increased IGF2 expression as one of the dominant transcriptional changes in ACC.
Materials and Methods
Tumors and Histopathology
The adrenocortical tissues analyzed in this study were procured from the University of Michigan Health System between 1994 and 2001 by the Tissue Procurement Service. The transcriptional profiling and validation studies were approved by the University of Michigan Institutional Review Board (IRB-Medicine). All tissues were processed in a similar manner. Frozen tumor samples were embedded in OCT freezing media (Miles Scientific, Naperville, IL), cryotome sectioned (5 μm), and evaluated by routine hematoxylin and eosin (H&E) stains. Areas of relatively pure tumor (at least 90% tumor cells) without necrosis when present in the carcinoma samples were selected for RNA isolation. The corresponding H&E sections from original paraffin blocks and surgical pathology reports were reviewed and evaluated for various pathological features, such as tumor size, tumor weight, tumor grade, and the presence of necrosis, vascular invasion, capsular invasion, and cell type. The characteristics of the tissues used for expression profiling, along with limited clinical and laboratory information, are presented in Table 1 ▶ . Standard diagnostic criteria 2 were used to diagnose the adrenocortical tumors.
Table 1.
Pathological and Clinical Aspects of Adrenocortical Cases Used for Profiling Studies
| Designation | Tissue type | Side | Age* | Sex | Clinical/laboratory aspects | Size of 1° (cm) | Weight of 1° (gm) | Grade† | Cell type | Necrosis | Caps inv | Vasc inv | Metastatic sites |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC 4 | 1° Carcinoma | R | A | F | UN | >8.0 | UN | High | Mixed | Y | N | N | None |
| ACC 7 | 1° Carcinoma | L | A | F | UN | 17 | 2300 | High | Lipid-poor | Y | N | Y | Liver |
| ACC 8 | 1° Carcinoma | L | A | F | UN | 9 | 180 | High | Mixed | Y | Y | Y | Skull |
| ACC 11 | Metastatic carcinoma | L | A | F | Liver metastasis | UN | UN | High | Mixed | Y | NA | NA | Liver |
| ACC 13 | 1° Carcinoma | L | A | F | UN | 16 | 460 | Low | Mixed | Y | N | N | Lung, liver, bone |
| ACC 14 | 1° Carcinoma | R | A | M | R leg edema | 22 | 2000 | High | Lipid-poor | Y | Y | Y | Liver |
| ACC 15 | Metastatic carcinoma | R | A | M | Lung metastasis | UN | UN | High | Mixed | Y | NA | NA | Lung and chest wall |
| ACC 17 | 1° Carcinoma | R | A | F | Right abdominal pain | 26 | 2300 | High | Lipid-poor | Y | Y | Y | Lung and liver |
| ACC 18 | 1° Carcinoma | L | A | F | Cushing’s syndrome | 18 | 900 | High | Mixed | Y | Y | Y | None |
| ACC 19 | 1° Carcinoma | R | A | F | R Flank pain | 9 | UN | High | Lipid-poor | Y | Y | N | Liver |
| ACC 33 | 1° Carcinoma | L | P | M | Increased testosterone | 12 | 450 | High | Lipid-poor | Y | Y | N | Liver, lymph nodes |
| ACA 21 | Adenoma | R | A | F | Cushing’s syndrome | 4.5 | 65 | NA | Lipid-rich | N | NA | NA | NA |
| ACA 22 | Adenoma | L | A | F | Cushing’s syndrome | 2.5 | 12 | NA | Mixed | N | NA | NA | NA |
| ACA 28 | Adenoma | L | A | F | Cushing’s syndrome | 3.5 | 25.2 | NA | Mixed | N | NA | NA | NA |
| ACA 30 | Adenoma | R | A | F | Cushing’s syndrome | 2.5 | 16 | NA | Mixed | N | NA | NA | NA |
| MNH 29 | Macronodular hyperplasia | R | A | F | Cushing’s syndrome | 5.5 | 50 | NA | Lipid-rich | N | NA | NA | NA |
| NC 6 | Normal adrenal cortex | L | A | F | Adrenalectomy for metastatic lung carcinoma | NA | NA | NA | NA | NA | NA | NA | NA |
| NC 9 | Normal adrenal cortex | UN | A | UN | Adrenalectomy for metastatic renal cell carcinoma | NA | NA | NA | NA | NA | NA | NA | NA |
| NC 10 | Normal adrenal cortex | UN | A | UN | Adrenalectomy for metastatic gastrinoma | NA | NA | NA | NA | NA | NA | NA | NA |
RNA Isolation
Single isolates of tissue samples were homogenized in the presence of Trizol reagent (Life Technologies, Inc., Gaithersburg, MD) and total cellular RNA was purified according to manufacturer’s procedures. RNA samples were further purified using acid phenol extraction and RNeasy spin columns (Qiagen, Valencia, CA) and used to prepare cRNA probes. RNA quality was assessed by 1% agarose gel electrophoresis in the presence of ethidium bromide. Samples that did not reveal intact 18S and 28S ribosomal bands were excluded from further study.
cRNA Synthesis, Gene Expression Profiling, and Statistical Analysis
This study used commercially available high-density oligonucleotide microarrays (HG_U95Av2; Affymetrix, Santa Clara, CA). Preparation of cRNA, hybridization, scanning, and image analysis of the arrays were performed according to manufacturer’s protocols and as previously described. 25,33 The U95A arrays consist of 12,625 probe sets, each representing a transcript. Each probe set typically consists of 16 perfectly complementary 25 base long probes (PMs) as well as 16 mismatch probes (MMs) that are identical except for an altered central base. A normal adrenal cortex sample was selected as the standard and probe pairs for which PM-MM <−100 on the standard were excluded from further analysis. The average of the middle 50% of the PM-MM differences was used as the expression measure for each probe set.
A quantile normalization procedure was used to adjust for differences in the probe intensity distribution across different chips. We applied a monotone linear spline to each chip that mapped quantiles 0.01 up to 0.99 (in increments of 0.01) exactly to the corresponding quantiles of the standard. Then, the transform log[100 + max(X + 100; 0)] was applied to the data from each chip. Code to perform these computations is freely available at http://dot.ped.umich.edu:2000/ourimage/pub/index.html.
Tissue Microarrays
A tissue microarray 34 containing 4 normal adrenal cortex (NC) samples, 24 adrenocortical adenoma (ACA) samples, 12 adrenocortical hyperplasias (including both diffuse and macronodular types), 62 ACC samples, along with 3 pheochromocytomas and several various normal tissues, was constructed for immunohistochemical validation studies using the Beecher manual tissue arrayer and 0.6-mm-diameter cylindrical cores. Donor blocks were retrieved from the archives of the University of Michigan Department of Pathology and the corresponding H&E slides were reviewed and representative viable areas were chosen for sampling. Given the relatively homogeneous nature of adrenocortical tumors in general, each case was arrayed in duplicate.
Immunohistochemistry
Immunohistochemistry was performed using formalin-fixed, paraffin-embedded sections from routine paraffin blocks (mib-1) or the adrenal tissue microarray (TOP2A) using the avidin-biotin complex method. 35 The following antibodies, dilutions, and pretreatment conditions were used: anti-human Ki-67, mib-1 antibody (DAKO, Carpinteria, CA), 1:100 dilution, Tris-ethylenediaminetetraacetic acid, microwave, pH 9.0; anti-human TOP2A, clone 3F6 (Novocastra Laboratories, Newcastle, UK), 1:40 dilution, citric acid, microwave, pH 6.0; and anti-human α-inhibin (Serotec, Raleigh, NC), prediluted antibody, Tris-ethylenediaminetetraacetic acid, microwave, pH 9.0.
The mib-1 and TOP2A immunostains were evaluated by counting the number of mib-1 or TOP2A immunoreactive tumor nuclei and the total number of tumor nuclei. The results for both immunostains were recorded as the percentage of immunoreactive tumor nuclei/total tumor nuclei. Tonsil with germinal centers was used as a positive control and negative controls were performed with no primary antibody.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (Q-RT-PCR)
cDNA was synthesized from 1 μg of total RNA using a first strand synthesis kit for RT-PCR (Retroscript; Ambion, Austin, TX) and poly(dT) primers. The relative abundance of IGF2 transcripts was assessed using the 5′ fluorogenic nuclease assay to perform real-time Q-PCR. 36 IGF2 primers (forward 5″-CCGTGCTTCCGGACAACT-3, reverse 5′-GGACTGCTTCCAGGTGTCATATT-3′) and a fluorogenic probe (5′-CCCCAGATACCCCGTGGGCAA-3′) were designed using the Primer Express software package (Applied Biosystems, Foster City, CA) and obtained from Applied Biosystems. The sequence of the IGF2 amplicon was compared to reported genomic sequences using the BLAST program to assure the amplicon was unique. The primers and probe set were optimized with respect to MgCl2 concentration and time and temperature of the hybridization step. Multiplex Q-PCR using a SmartCycler (Cepheid, Sunnyvale, CA) was performed in 30-μl reactions consisting of 1× Q-PCR Supermix-UDG reaction mix (Life Technologies, Inc., Gaithersburg, MD) supplemented with the appropriate MgCl2 concentration. Relative expression of mRNA for IGF2 was calculated using the comparative CT method as described 37 using the CT of GAPDH as the reference.
Results
Transcriptional Profiles Distinguish Benign and Malignant Adrenocortical Tumors, Reflect Morphology, and Identify Differentially Expressed Transcripts
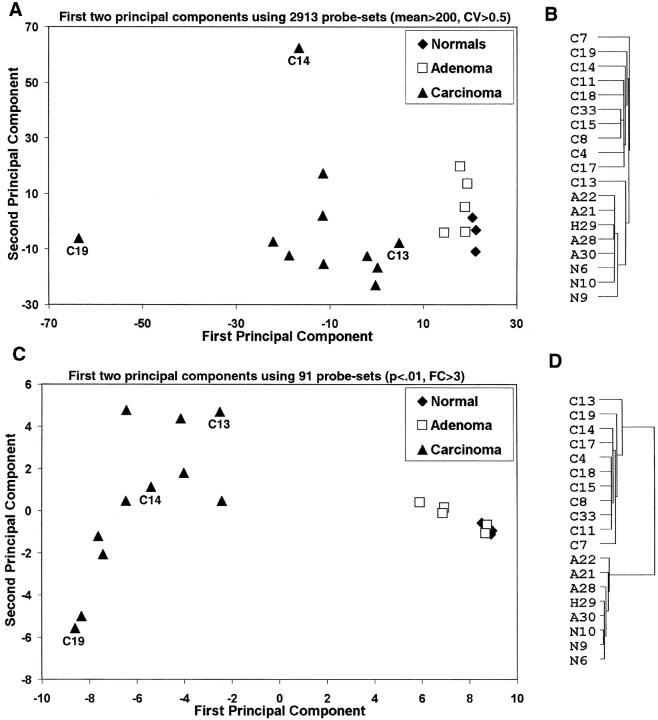
Transcriptional profiles of 11 ACCs (nine primary and two metastatic), 4 ACAs, 3 NCs, and 1 macronodular hyperplasia (MNH) were generated using oligonucleotide arrays with 12,625 probe sets interrogating ∼10,500 genes. To provide visual representations of the tissues and tumors based on gene expression, we used principal component analysis (PCA) to locate the two-dimensional views that capture the greatest amount of variability in the data, using several thousand of the most variable probe sets. The resulting PCA view showed a clear separation between the ACC and benign cohorts (Figure 1A) ▶ . The greatest variability was seen within the ACC cohort, with two tumors, ACC19 and ACC14, located far from the remaining ACCs. One of these tumors (ACC19) was a high-grade ACC with little morphological (Figure 2) ▶ or transcriptional evidence of adrenocortical differentiation, although the tumor was focally and weakly positive for α-inhibin (not shown). The other outlying tumor, ACC14, was a myxoid variant of ACC, a rare but recognized variant 38 (Figure 2) ▶ . Interestingly, the ACC closest to the NC/ACA cohort, ACC13, was a low-grade ACC 2 that shared some histological features with the benign cohort (Figure 2) ▶ . A typical high-grade ACC (ACC17) is also shown for comparison in Figure 2 ▶ .The PCA view showed little differences between the NC and the ACA/MNH cohorts (Figure 1A) ▶ , a not entirely unexpected finding based on their similar morphologies.
Figure 1.
Probe sets (n = 2913) with mean expression greater than 200 and SD divided by the mean >0.5 for the entire data set were selected. A: First two principal components for these probe sets. Probe sets were standardized by subtracting the mean and dividing by the SD. B: Dendrogram from average clustering 62 using these probe sets. C and D: Similar principal components and dendrograms using only 91 probe sets with a fold change greater than three between the carcinoma and normal/adenoma cohorts with a P < 0.01 in both cases.
Figure 2.
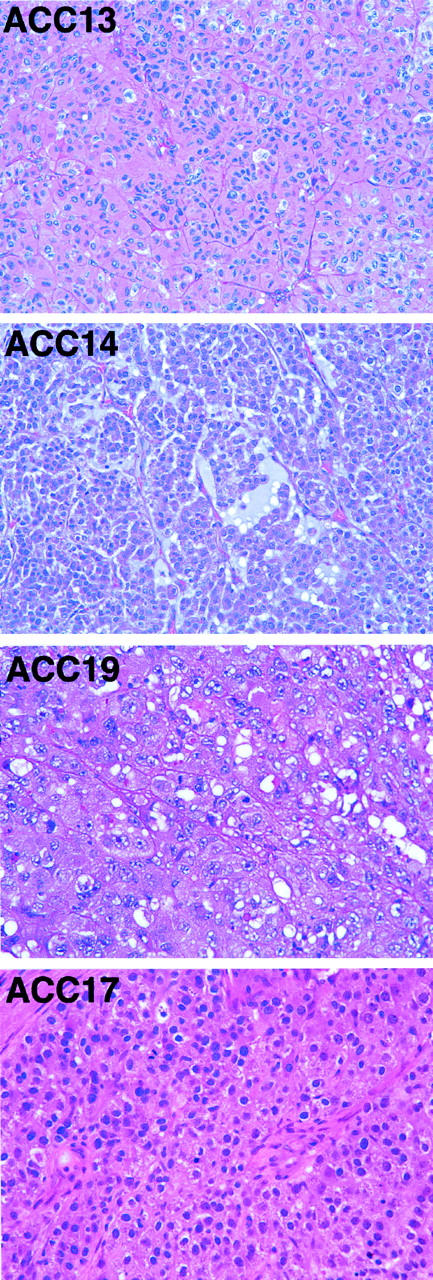
Histology of typical ACC and the three outlying tumors identified by PCA. ACC13 is a well-differentiated or low-grade ACC. ACC14 is a myxoid variant of ACC. ACC19 is a poorly differentiated ACC. ACC17 is a typical high-grade ACC. H&E; original magnification, ×200.
In addition to PCA, we used hierarchical clustering to generate another visual relationship between the adrenocortical tissues and tumors. The resulting dendrogram (Figure 1B) ▶ placed 10 of 11 ACCs on the same branch, with the low-grade tumor ACC13 on the branch with the NCs, ACAs, and MNH, which likely reflects its well-differentiated nature. However, the dendrogram clearly showed a difference between ACC13 and the NC/ACA cohort. With respect to this cohort, the hierarchical clustering was in agreement with the PCA analysis (Figure 1A) ▶ , as the dendrogram (Figure 1B) ▶ showed minimal differences within the NC/ACA cohort.
One of the main interests in this study was to identify transcripts of greater abundance in ACC samples than in ACA/MNH and NC samples. A simple one-way analysis of variance using the entire data set of 12,625 probe sets on these three groups of samples obtained 677 probe sets that gave P < 0.01 when comparing ACCs with ACAs, and 473 when ACCs were compared with NCs. These numbers of probe sets are several times larger than the number expected by chance alone. To highlight those differences of potentially greatest biological interest, as well as to reduce the fraction of false-positive findings, we further required that expression values in the ACCs be at least twofold different from the average value in the other group, and that the probe sets meet these requirements in both comparisons (ACC versus NC, ACC versus ACA). One hundred fifty-eight probe sets met these criteria, and for 91 probe sets the fold changes were greater than three for both comparisons. Randomly permuting the sample labels 10,000 times, on average less than one probe set met the final criterion of P < 0.01 and fold change larger than three for both comparisons, and no permutations gave more than the observed number of 91 probe sets. This permutation result shows that very few of the selected 91 genes have been identified because of chance variation alone.
A PCA view of the data for these 91 probe sets (Figure 1C) ▶ , similar to the previous PCA view based on thousands of variable probe sets (Figure 1A) ▶ , showed a greater relative distance between the ACC and benign cohorts, as expected because of the method of probe set selection. Interestingly, there was persistent intragroup variability within the ACC group, with a trend toward displaying the tumors along a differentiation spectrum.
As above, hierarchical clustering was applied to the data set with the 91 differentially expressed genes. As expected, the resulting dendrogram (Figure 1D) ▶ showed a significant difference between the ACC and NC/ACA cohorts. Tumor ACC13, while now included within the ACC branch, showed the least amount of relatedness within the ACC cohort, again reflecting its well-differentiated nature.
The probe set designation, identity of the individual genes, as well as the fold change of the mean expression level relative to mean expression level of NC for the 91 differentially expressed probe sets described above are shown in Figure 3 ▶ .
Figure 3.
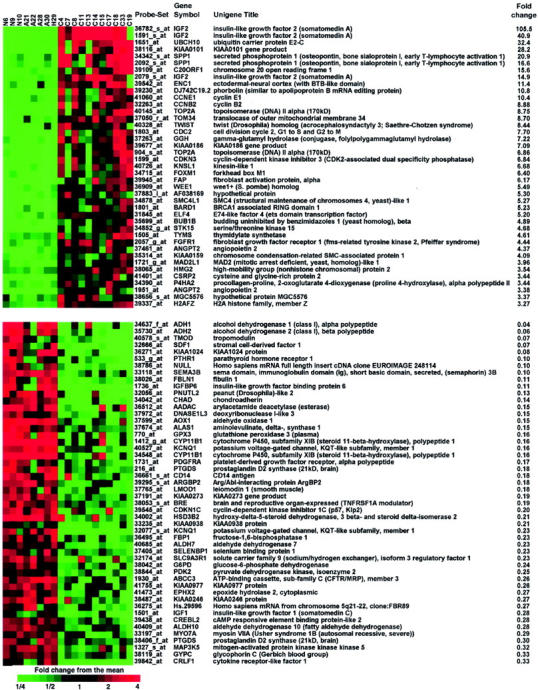
Ninety-one probe sets that gave P < 0.01 for F-tests of both carcinoma versus normal and carcinoma versus adenoma that also had both fold changes larger than three, or smaller than 0.333 (see text). A central value for each probe set was determined by averaging log-transformed data, and taking the anti-logarithm. Colors represent the fold change for each sample from this value. The heat map was made using Treeview. 65 The probe set designation, the gene symbol, and the unigene title, as well as the fold change in transcript level, are shown. Samples are labeled as follows: N = NC, A = ACA, H = MNH, and C = ACC.
Few transcriptional changes were found between the NC and ACA cohorts because just 58 probe sets with a fold change greater than 1.5 in either direction were identified at a significance level of P < 0.01. Because the chip contains 12,625 probe sets, 126 probe sets would be expected by chance alone at P < 0.01, suggesting that many of these 58 are likely to be false-positives. Examination of the expression data for the three NC samples strongly suggests that some normal adrenal medulla contaminated two of the specimens, a not unexpected finding given the adjacent and often interdigitating relationship between adrenal cortex and medulla. These two samples, NC9 and NC10, showed increased expression levels for several probable medulla-related genes, such as SCG2 (secretogranin II), TH (tyrosine hydroxylase), PENK (proenkephalin), DBH (dopamine B-hydroxylase), DDC (dopa-decarboxylase), SST (somatostatin), and CART (cocaine- and amphetamine-regulated transcript). However, most of the remaining candidate differentially expressed genes were not the result of contamination based on consistent expression across all three NC samples (Figure 3) ▶ and some, such as SGK (serum/glucocorticoid regulated kinase), likely represent true positives.
Increased IGF2 Expression Is One of the Dominant Transcriptional Events in ACC
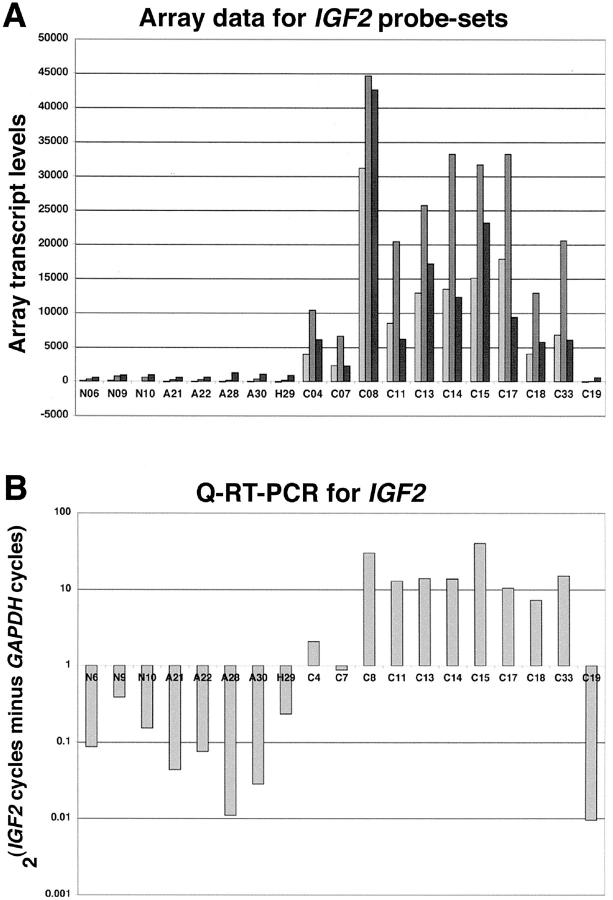
Three distinct probe sets for the IGF2 gene were present on the U95A chip, thereby providing triplicate measures for each of the tissues. All three probe sets showed substantially increased expression of IGF2 in 10 of 11 (90.9%) ACCs compared to the mean of the NC/ACA cohort (Figure 4A) ▶ . One carcinoma (ACC19) did not show increased IGF2 expression for the three probe sets and was one of the two ACCs identified by PCA as an outlier (see above). The fold changes of the mean expression of the ACC cohort compared to the mean of the NC/ACA cohort were 105.5, 40.9, and 14.9 for probe sets 36782_s_at, 1591_s_at, and 2079_s_at, respectively (Figure 3) ▶ . Q-RT-PCR for IGF2 and GAPDH transcripts confirmed the pattern of expression for the arrayed tissues (Figure 4B) ▶ . Significant IGF2 transcripts were present in the same 10 of 11 ACCs, whereas ACC19 contained few IGF2 transcripts relative to GAPDH transcripts.
Figure 4.
Up-regulation of IGF2 in ACC. A: Expression of IGF2, represented by three probe sets (light gray = 36782_s_at, medium gray = 1591_s_at, and dark gray = 2079_s_at), in the adrenocortical samples. B: Q-RT-PCR for IGF2 and GAPDH in the adrenocortical samples. Samples are labeled as follows: N = NC, A = ACA, H = MNH, and C = ACC.
Relative Absence of Growth Factor Receptor Expression
Given the recent development of therapies targeted to growth factor receptors (eg, trastuzumab and ERB-B2), the transcript levels of several receptor tyrosine kinases were assessed. Such genes represented on the U95A chip included EGFR, ERB-B2, HER3, PDGFRA, PDGFRB, KIT, FGFR1, FGFR4, IGF1R, IGFR2, INSR, ESR1, ESR2, and PGR, among others. The only receptor tyrosine kinase that displayed differential transcript levels was FGFR1, which was represented four times on the array and showed 5.6-, 2.1-, and 2.8-fold increased expression in the ACCs compared to the ACAs in three of the probe sets with statistical significance (2057_g_at, P = 0.00009; 2056_at, P = 0.0003; and 424_s_at, P = 0.003, respectively). Importantly, most of these other genes showed absolute transcript levels indicative of either no expression (eg, KIT) or low expression (eg, PDGFRA and PDGFRB).
Immunohistochemical Protein Validation Studies
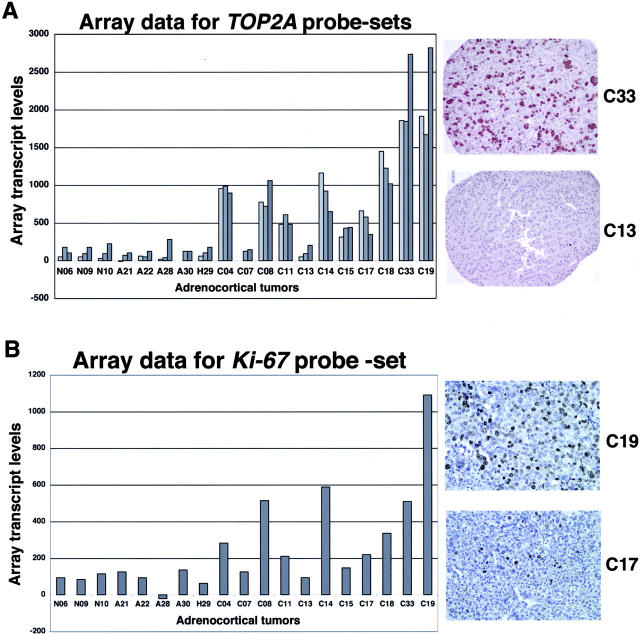
To validate differential expression at the protein level, immunohistochemistry was performed for select genes for which specific literature regarding adrenocortical tumors already existed. Accordingly, two proliferation-related markers, Ki-67 and TOP2A, were chosen for validation. Although KI-67 was not on the most differentially expressed gene list, it was up-regulated in the ACC cohort compared to the ACA (3.7-fold change, P < 0.006) and NC cohorts (3.7-fold change, P < 0.036). Comparison of the gene expression data with the corresponding immunostains of the same tumors showed strong correlation between transcript levels and tumoral immunoreactivity (Figure 5 ▶ and Table 2 ▶ ). Specifically, TOP2A, represented by three distinct probe sets, showed significantly increased expression in the ACC cohort compared to the ACA/MNH cohort, as expected (Figure 5A) ▶ . Tumors with relatively high transcript levels of TOP2A (eg, ACC33) showed the highest percentage of immunoreactive cells, whereas the tumors with low transcript levels (eg, ACC13) showed only rare immunoreactive cells (Figure 5A) ▶ . Likewise, tumors with relatively high transcript levels of MKI67 showed a high percentage of Ki-67 (mib-1) immunoreactive cells (Figure 5B) ▶ . Transcript levels and the percentage of immunoreactive tumor nuclei showed a high level of correlation for both proliferation markers (Table 2) ▶ .
Figure 5.
Array transcript data and protein validation studies of select proliferation markers. A: Up-regulation of TOP2A in ACC. Transcript data for three probe sets (light gray = 40145_at, medium gray = 904_s_at, and dark gray = 1592_at) the adrenocortical cohort and two representative tissue cores that show high- and low-level immunoreactivity for TOP2A. B: Up-regulation of Ki-67 in ACC. Transcript data (419_at) for the adrenocortical cohort and two representative tissue cores that show high and moderate levels of mib-1 immunoreactivity.
Table 2.
Correlation between Transcript Levels and IHC for Ki-67 (mib-1) and TOP2A
| Probe set name | Gene symbol | ACA 21 | ACA 22 | ACA 28 | ACA 30 | MNH 29 | ACC 4 | ACC 7 | ACC 8 | ACC 11 | ACC 13 | ACC 14 | ACC 15 | ACC 17 | ACC 18 | ACC 19 | ACC 33 | Transcript versus IHC correlation (r value) | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 40145_at | TOP2A | −10 | 63 | 21 | 0 | 63 | 955 | 0 | 776 | 483 | 52 | 1161.9 | 314 | 661 | 1448 | 1912.3 | 1857 | 0.7238989 | 0.002 |
| 904_s_at | TOP2A | 73 | 52 | 42 | 126 | 105 | 986.5 | 126 | 720.6 | 608.3 | 94 | 921.7 | 431 | 577.7 | 1225.2 | 1668 | 1847 | 0.7536438 | 0.001 |
| 1592_at | TOP2A | 105 | 126 | 283 | 126 | 178 | 895.9 | 147 | 1060 | 483 | 204.2 | 651 | 441 | 347 | 1018 | 2819 | 2733 | 0.8540047 | <0.0001 |
| Top2A IHC | 1.4 | 0 | NP | 4.3 | 2.3 | 0 | 0.9 | 2.9 | 5.8 | 0 | 7.2 | 4.4 | 17.3 | 0 | 33 | 38.2 | |||
| 419_at | MKI67 | 126 | 94 | −21 | 136 | 63 | 283 | 126 | 514 | 210 | 94 | 588 | 147 | 220 | 336 | 1090.4 | 509 | 0.7450237 | 0.0009 |
| mib-1 IHC | 0 | 0 | 0 | 3.8 | 0.5 | 7.3 | 0 | 3.5 | 6.7 | 1.5 | 5.5 | 0 | 17.5 | 10.7 | 29 | 29.6 |
NP, not performed.
Discussion
DNA microarray technology allows the opportunity to comprehensively examine the transcriptional profile of tumors. It is rapidly being deployed to address a variety of issues in pathology and oncology, such as tumor classification, 25,39-43 and as a useful gene discovery tool 44,45 to complement other similar technologies such as SAGE. 46,47 In this study, we used DNA microarrays to generate transcriptional profiles of benign and malignant adrenocortical tumors, as well as normal adrenal cortex and MNH. These profiles were used to discriminate normal and benign tissues from malignant tumors and to identify differentially expressed genes that may have diagnostic, pathogenetic, and therapeutic implications.
Select RNA and protein-based studies were performed to validate the array data; moreover, comparison of the array data to the published literature provides an additional and vital level of validation. For example, ample evidence exists that increased expression of IGF2 occurs in ACC 23 and this was robustly confirmed by this study. Furthermore, TOP2A has been reported to be up-regulated in ACC and used as a diagnostic marker 10,14 and was independently identified as one of the significantly up-regulated genes in this cohort of ACCs. Thus, it is reasonable to conclude our approach is informative and has identified numerous differentially expressed genes.
Although the histopathological diagnosis of ACC is usually not problematic, additional diagnostic markers would be useful in some challenging cases. Examination of the differentially expressed gene list yields several genes that may represent candidate diagnostic markers, including IGF2 and osteopontin, among others. Recent immunohistochemical studies of IGF2 protein as a marker are promising 48 and immunohistochemical validation studies using antibodies to osteopontin are currently underway.
Our results clearly identify increased IGF2 as a characteristic transcriptional event in the pathogenesis of ACC. This finding is consistent with a large body of evidence demonstrating that increased IGF2 expression is the result of dysregulation or rearrangement at 11p15.5. 21,49-52 The significance of the current findings is twofold: the magnitude of the increased IGF2 expression and the lack of other signal transduction-related changes identified in this albeit partial genome transcriptional survey. This suggests that interruption of IGF2-induced signal transduction may lead to a significant therapeutic advance, analogous to those related to STI-571 in certain leukemias 53 and gastrointestinal stromal tumors. 54 One of the ways to block IGF2 signaling might be to design a small molecule inhibitor against the tyrosine kinase domain of the IGF1 receptor. Interestingly, a recent study described the 2.1 A-resolution crystal structure of the activated form of the IGF1 receptor kinase. 55 Although the IGF1 receptor shares the nucleotide-binding cleft with the insulin receptor, the authors identified sequence differences in the nearby interlobe linker that might represent a target for anti-cancer drug design. Alternatively, proteins downstream in the IGF1 receptor signal transduction pathway may represent additional therapeutic targets.
Our study failed to identify significant differences in gene expression between the normal adrenal cortex and benign cortical processes such as ACA and MNH, a not entirely unexpected result given the histological similarity of these tissues. However, a few genes, such as serum glucocorticoid-regulated kinase (SGK) gene, may represent differentially expressed genes. Validation of some of these genes awaits additional protein-based studies.
The relative absence of growth factors and their receptors other than IGF2 is significant given the recent availability of targeted therapeutic agents, such as imatinib mesylate (Gleevec) and trastuzumab (Herceptin), and the expected availability of orally bioavailable EGFR inhibitors such as ZD1830 (Iressa). This suggests that the treatment of ACC with the available receptor tyrosine kinase inhibitors will likely not be effective and will have to await the development of additional compounds.
Examination of the list of genes differentially expressed between ACC and benign adrenocortical processes provides insight into the pathogenesis of ACC and possible therapeutic approaches. The serine-threonine kinase STK15 (also known as BTAK and aurora2) is associated with centrosomes, plays a role in the induction of centrosome abnormalities and thus aneuploidy and transformation in mammalian cells, and is up-regulated in some types of human tumors. 56 Our finding of almost fivefold increased STK15 transcripts in ACC suggests that this gene may play a role in the marked aneuploidy commonly observed in ACC. 57,58 Angiopoietin 2 (Ang-2), a member of the one of the two major classes of angiogenic factors, acts synergistically with vascular endothelial growth factor to induce angiogenesis. 59 Our results, which show a threefold to fourfold increase in Ang-2 transcripts in ACC, suggests that angiogenesis is an important aspect of adrenocortical tumorigenesis, as expected, and that anti-angiogenic therapies currently being developed for other more common cancers might be effective in ACC. The UbcH10 gene encodes a cyclin-selective ubiquitin carrier protein, E2-C, which catalyzes the destruction of mitotic cyclins (A and B) via ubiquitin-dependent proteolysis, permitting the completion of mitosis and entry into interphase of the next cell cycle. 60,61 Thus, it is possible to speculate that marked increased expression of UbcH10, as seen in our cohort of ACCs, may be a proproliferative factor and play a role in the pathogenesis of ACC. The ectodermal-neural cortex one (ENC-1) gene is up-regulated in colorectal carcinoma and was recently identified as a potential target of the β-catenin/T-cell factor complex. 62 Up-regulation of ENC-1 transcripts in ACC, as observed in our data, suggests that abnormal wnt signaling may play a role in ACC pathogenesis. The high mobility group A2 (HMG2) gene plays a role in the pathogenesis of common mesenchymal tumors 63 and is amplified and overexpressed in prolactinomas, likely related to chromosomal rearrangement of 12q14-15 resulting in genomic amplification. 64 Thus, our finding of a threefold increase in HMG2 transcripts in ACC is provocative and warrants further investigation. These examples represent just some of the interesting differentially expressed genes. Obviously, comprehensive gene expression profiles provide many interesting avenues for further investigation and will likely provide the foundation for significant therapeutic advances.
Finally, one of the uses of global gene expression data is correlation with existing genomic data such as conventional and microarray-based comparative genomic hybridization (CGH) data. For instance, a recent CGH study 65 of 35 adrenocortical tumors identified co-amplification of SAS/CDK4 and MDM2 in two advanced ACCs, suggesting that co-amplification of these genes may play a role in tumor progression. Our expression data does indicate increased expression of both SAS and CDK4 in one of the ACCs, but MDM2 expression is minimal or absent in all of the adrenocortical tissues based on data from four distinct probe sets. Thus, although increased expression of SAS and/or CDK4 via genomic amplification may play a role in ACC pathogenesis, a similar role for MDM2 is not likely.
Acknowledgments
We thank Doug Selby, Doug Johnson, John Weeks, and Enola Cushenberry, technicians of the University of Michigan Comprehensive Cancer Center Tissue Core for tissue procurement; Tina Fields for excellent technical immunohistochemical advice and assistance; and Dr. Kathleen Cho for useful discussions and critical reading of the manuscript.
Footnotes
Address reprint requests to Thomas J. Giordano, M.D., Ph.D., Department of Pathology, University Hospital, 2G332/0054, 1500 E. Medical Center Dr., Ann Arbor, MI 48109-0054 E-mail: giordano@umich.edu.
Supported by funds from the Millie Schembechler Adrenal Cancer Program of the University of Michigan Comprehensive Cancer Center, the Michigan National Institute of Diabetes and Digest and Kidney Diseases Biotechnology Center (National Institutes of Health no. DK58771), the University of Michigan Comprehensive Cancer Center Tissue Core (National Institutes of Health no. 46952), and indirectly by infrastructure provided by the National Cancer Institute Director’s Challenge project at the University of Michigan (National Institutes of Health no. 84952).
References
- 1.Brennan MF: Adrenocortical carcinoma. CA Cancer J Clin 1987, 37:348-365 [DOI] [PubMed] [Google Scholar]
- 2.Weiss LM: Comparative histologic study of 43 metastasizing and nonmetastasizing adrenocortical tumors. Am J Surg Pathol 1984, 8:163-169 [DOI] [PubMed] [Google Scholar]
- 3.Weiss LM, Medeiros LJ, Vickery AL, Jr: Pathologic features of prognostic significance in adrenocortical carcinoma. Am J Surg Pathol 1989, 13:202-206 [DOI] [PubMed] [Google Scholar]
- 4.Munro LM, Kennedy A, McNicol AM: The expression of inhibin/activin subunits in the human adrenal cortex and its tumours. J Endocrinol 1999, 161:341-347 [DOI] [PubMed] [Google Scholar]
- 5.McCluggage WG, Burton J, Maxwell P, Sloan JM: Immunohistochemical staining of normal, hyperplastic, and neoplastic adrenal cortex with a monoclonal antibody against alpha inhibin. J Clin Pathol 1998, 51:114-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arola J, Liu J, Heikkila P, Voutilainen R, Kahri A: Expression of inhibin alpha in the human adrenal gland and adrenocortical tumors. Endocr Res 1998, 24:865-867 [DOI] [PubMed] [Google Scholar]
- 7.Pelkey TJ, Frierson HF, Jr, Mills SE, Stoler MH: The alpha subunit of inhibin in adrenal cortical neoplasia. Mod Pathol 1998, 11:516-524 [PubMed] [Google Scholar]
- 8.Cho EY, Ahn GH: Immunoexpression of inhibin alpha-subunit in adrenal neoplasms. Appl Immunohistochem Mol Morphol 2001, 9:222-228 [DOI] [PubMed] [Google Scholar]
- 9.Renshaw AA, Granter SR: A comparison of A103 and inhibin reactivity in adrenal cortical tumors: distinction from hepatocellular carcinoma and renal tumors. Mod Pathol 1998, 11:1160-1164 [PubMed] [Google Scholar]
- 10.Goldblum JR, Shannon R, Kaldjian EP, Thiny M, Davenport R, Thompson N, Lloyd RV: Immunohistochemical assessment of proliferative activity in adrenocortical neoplasms. Mod Pathol 1993, 6:663-668 [PubMed] [Google Scholar]
- 11.Iino K, Sasano H, Yabuki N, Oki Y, Kikuchi A, Yoshimi T, Nagura H: DNA topoisomerase II alpha and Ki-67 in human adrenocortical neoplasms: a possible marker of differentiation between adenomas and carcinomas. Mod Pathol 1997, 10:901-907 [PubMed] [Google Scholar]
- 12.Nakazumi H, Sasano H, Iino K, Ohashi Y, Orikasa S: Expression of cell cycle inhibitor p27 and Ki-67 in human adrenocortical neoplasms. Mod Pathol 1998, 11:1165-1170 [PubMed] [Google Scholar]
- 13.Terzolo M, Boccuzzi A, Bovio S, Cappia S, De Giuli P, Ali A, Paccotti P, Porpiglia F, Fontana D, Angeli A: Immunohistochemical assessment of Ki-67 in the differential diagnosis of adrenocortical tumors. Urology 2001, 57:176-182 [DOI] [PubMed] [Google Scholar]
- 14.Wachenfeld C, Beuschlein F, Zwermann O, Mora P, Fassnacht M, Allolio B, Reincke M: Discerning malignancy in adrenocortical tumors: are molecular markers useful? Eur J Endocrinol 2001, 145:335-341 [DOI] [PubMed] [Google Scholar]
- 15.Gupta D, Shidham V, Holden J, Layfield L: Value of topoisomerase II alpha, MIB-1, p53, E-cadherin, retinoblastoma gene protein product, and HER-2/neu immunohistochemical expression for the prediction of biologic behavior in adrenocortical neoplasms Appl Immunohistochem Mol Morphol 2001, 9:215-221 [DOI] [PubMed] [Google Scholar]
- 16.Arola J, Salmenkivi K, Liu J, Kahri AI, Heikkila P: p53 and Ki67 in adrenocortical tumors. Endocr Res 2000, 26:861-865 [DOI] [PubMed] [Google Scholar]
- 17.Li FP, Fraumeni JF, Jr: Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann Intern Med 1969, 71:747-752 [DOI] [PubMed] [Google Scholar]
- 18.Ohgaki H, Kleihues P, Heitz PU: p53 mutations in sporadic adrenocortical tumors. Int J Cancer 1993, 54:408-410 [DOI] [PubMed] [Google Scholar]
- 19.Lin SR, Lee YJ, Tsai JH: Mutations of the p53 gene in human functional adrenal neoplasms. J Clin Endocrinol Metab 1994, 78:483-491 [DOI] [PubMed] [Google Scholar]
- 20.Reincke M, Wachenfeld C, Mora P, Thumser A, Jaursch-Hancke C, Abdelhamid S, Chrousos GP, Allolio B: p53 mutations in adrenal tumors: Caucasian patients do not show the exon 4 “hot spot” found in Taiwan. J Clin Endocrinol Metab 1996, 81:3636-3638 [DOI] [PubMed] [Google Scholar]
- 21.Li M, Squire JA, Weksberg R: Molecular genetics of Wiedemann-Beckwith syndrome. Am J Med Genet 1998, 79:253-259 [PubMed] [Google Scholar]
- 22.Ilvesmaki V, Kahri AI, Miettinen PJ, Voutilainen R: Insulin-like growth factors (IGFs) and their receptors in adrenal tumors: high IGF-II expression in functional adrenocortical carcinomas. J Clin Endocrinol Metab 1993, 77:852-858 [DOI] [PubMed] [Google Scholar]
- 23.Gicquel C, Bertagna X, Schneid H, Francillard-Leblond M, Luton JP, Girard F, Le Bouc Y: Rearrangements at the 11p15 locus and overexpression of insulin-like growth factor-II gene in sporadic adrenocortical tumors. J Clin Endocrinol Metab 1994, 78:1444-1453 [DOI] [PubMed] [Google Scholar]
- 24.Logie A, Boulle N, Gaston V, Perin L, Boudou P, Le Bouc Y, Gicquel C: Autocrine role of IGF-II in proliferation of human adrenocortical carcinoma NCI H295R cell line. J Mol Endocrinol 1999, 23:23-32 [DOI] [PubMed] [Google Scholar]
- 25.Giordano TJ, Shedden KA, Schwartz DR, Kuick R, Taylor JM, Lee N, Misek DE, Greenson JK, Kardia SL, Beer DG, Rennert G, Cho KR, Gruber SB, Fearon ER, Hanash S: Organ-specific molecular classification of primary lung, colon, and ovarian adenocarcinomas using gene expression profiles. Am J Pathol 2001, 159:1231-1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beer DG, Kardia SL, Huang C-C, Giordano TJ, Levin AL, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JMG, Iannettoni MD, Orringer MB, Hanash S: Gene expression profiles predict survival of patients lung adenocarcinoma. Nat Med 2002, 62:4722-4729 [DOI] [PubMed] [Google Scholar]
- 27.Schwatrz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JMG, Misek DE, Wu R, Zhai Y, Darrah DM, Reed H, Ellenson LH, Giordano TJ, Hanash SM, Cho KR: Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res 2002, 8:816-824 [PubMed] [Google Scholar]
- 28.Young AN, Amin MB, Moreno CS, Lim SD, Cohen C, Petros JA, Marshall FF, Neish AS: Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol 2001, 158:1639-1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kihara C, Tsunoda T, Tanaka T, Yamana H, Furukawa Y, Ono K, Kitahara O, Zembutsu H, Yanagawa R, Hirata K, Takagi T, Nakamura Y: Prediction of sensitivity of esophageal tumors to adjuvant chemotherapy by cDNA microarray analysis of gene-expression profiles. Cancer Res 2001, 61:6474-6479 [PubMed] [Google Scholar]
- 30.Huang Y, Prasad M, Lemon WJ, Hampel H, Wright FA, Kornacker K, LiVolsi V, Frankel W, Kloos RT, Eng C, Pellegata NS, de la Chapelle A: Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci USA 2001, 98:15044-15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu XR, Huang J, Xu ZG, Qian BZ, Zhu ZD, Yan Q, Cai T, Zhang X, Xiao HS, Qu J, Liu F, Huang QH, Cheng ZH, Li NG, Du JJ, Hu W, Shen KT, Lu G, Fu G, Zhong M, Xu SH, Gu WY, Huang W, Zhao XT, Hu GX, Gu JR, Chen Z, Han ZG: Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci USA 2001, 98:15089-15094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertucci F, Houlgatte R, Nguyen C, Viens P, Jordan BR, Birnbaum D: Gene expression profiling of cancer by use of DNA arrays: how far from the clinic? Lancet Oncol 2001, 2:674-682 [DOI] [PubMed] [Google Scholar]
- 33.Rickman DS, Bobek MP, Misek DE, Kuick R, Blaivas M, Kurnit DM, Taylor J, Hanash SM: Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res 2001, 61:6885-6891 [PubMed] [Google Scholar]
- 34.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP: Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998, 4:844-847 [DOI] [PubMed] [Google Scholar]
- 35.Sheibani K, Tubbs RR: Enzyme immunohistochemistry: technical aspects. Semin Diagn Pathol 1984, 1:235-250 [PubMed] [Google Scholar]
- 36.Heid CA, Stevens J, Livak KJ, Williams PM: Real time quantitative PCR. Genome Res 1996, 6:986-994 [DOI] [PubMed] [Google Scholar]
- 37.Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang SI, Polikoff D, Nonet G, Cochran J, Myambo K, Jay KE, Froula J, Cloutier T, Kuo WL, Yaswen P, Dairkee S, Giovanola J, Hutchinson JB, Isola J, Kallioniemi OP, Palazzolo M, Martin C, Ericsson C, Pinkel D, Albertson DL, Li WB, Gray JW: Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc Natl Acad Sci USA 1998, 95:8703-8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown FM, Gaffey TA, Wold LE, Lloyd RV: Myxoid neoplasms of the adrenal cortex: a rare histologic variant. Am J Surg Pathol 2000, 24:396-401 [DOI] [PubMed] [Google Scholar]
- 39.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES: Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999, 286:531-537 [DOI] [PubMed] [Google Scholar]
- 40.Lakhani SR, Ashworth A: Microarray and histopathological analysis of tumours: the future and the past? Nature Rev Cancer 2001, 1:151-157 [DOI] [PubMed] [Google Scholar]
- 41.Liotta L, Petricoin E: Molecular profiling of human cancer. Nat Rev Genet 2000, 1:48-56 [DOI] [PubMed] [Google Scholar]
- 42.Yeang CH, Ramaswamy S, Tamayo P, Mukherjee S, Rifkin RM, Angelo M, Reich M, Lander E, Mesirov J, Golub T: Molecular classification of multiple tumor types. Bioinformatics 2001, 17:S316-S322 [DOI] [PubMed] [Google Scholar]
- 43.Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF, Jr, Hampton GM: Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res 2001, 61:7388-7393 [PubMed] [Google Scholar]
- 44.Ricci MS, el-Deiry WS: Novel strategies for therapeutic design in molecular oncology using gene expression profiles. Curr Opin Mol Ther 2000, 2:682-690 [PubMed] [Google Scholar]
- 45.Meltzer PS: Spotting the target: microarrays for disease gene discovery. Curr Opin Genet Dev 2001, 11:258-263 [DOI] [PubMed] [Google Scholar]
- 46.Green CD, Simons JF, Taillon BE, Lewin DA: Open systems: panoramic views of gene expression. J Immunol Methods 2001, 250:67-79 [DOI] [PubMed] [Google Scholar]
- 47.Madden SL, Wang CJ, Landes G: Serial analysis of gene expression: from gene discovery to target identification. Drug Discov Today 2000, 5:415-425 [DOI] [PubMed] [Google Scholar]
- 48.Erickson LA, Jin L, Sebo TJ, Lohse C, Pankratz VS, Kendrick ML, van Heerden JA, Thompson GB, Grant CS, Lloyd RV: Pathologic features and expression of insulin-like growth factor-2 in adrenocortical neoplasms. Endocr Pathol 2001, 12:429-435 [DOI] [PubMed] [Google Scholar]
- 49.Gicquel C, Le Bouc Y: Insulin-like growth factor II (IGF II) and adrenocortical tumorigenesis. Annu Endocrinol 1995, 56:617-618 [PubMed] [Google Scholar]
- 50.Gicquel C, Raffin-Sanson ML, Gaston V, Bertagna X, Plouin PF, Schlumberger M, Louvel A, Luton JP, Le Bouc Y: Structural and functional abnormalities at 11p15 are associated with the malignant phenotype in sporadic adrenocortical tumors: study on a series of 82 tumors. J Clin Endocrinol Metab 1997, 82:2559-2565 [DOI] [PubMed] [Google Scholar]
- 51.Wilkin F, Gagne N, Paquette J, Oligny LL, Deal C: Pediatric adrenocortical tumors: molecular events leading to insulin-like growth factor II gene overexpression. J Clin Endocrinol Metab 2000, 85:2048-2056 [DOI] [PubMed] [Google Scholar]
- 52.Gicquel C, Bertagna X, Gaston V, Coste J, Louvel A, Baudin E, Bertherat J, Chapuis Y, Duclos JM, Schlumberger M, Plouin PF, Luton JP, Le Bouc Y: Molecular markers and long-term recurrences in a large cohort of patients with sporadic adrenocortical tumors. Cancer Res 2001, 61:6762-6767 [PubMed] [Google Scholar]
- 53.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL: Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 2001, 344:1031-1037 [DOI] [PubMed] [Google Scholar]
- 54.Joensuu H, Roberts PJ, Sarlomo-Rikala M, Andersson LC, Tervahartiala P, Tuveson D, Silberman S, Capdeville R, Dimitrijevic S, Druker B, Demetri GD: Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med 2001, 344:1052-1056 [DOI] [PubMed] [Google Scholar]
- 55.Favelyukis S, Till JH, Hubbard SR, Miller WT: Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Biol 2001, 8:1058-1063 [DOI] [PubMed] [Google Scholar]
- 56.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S: Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet 1998, 20:189-193 [DOI] [PubMed] [Google Scholar]
- 57.Mertens F, Kullendorff CM, Moell C, Alumets J, Mandahl N: Complex karyotype in a childhood adrenocortical carcinoma. Cancer Genet Cytogenet 1998, 105:190-192 [DOI] [PubMed] [Google Scholar]
- 58.Limon J, Dal Cin P, Kakati S, Huben RP, Sandberg AA: Cytogenetic findings in a primary adrenocortical carcinoma. Cancer Genet Cytogenet 1987, 26:271-277 [DOI] [PubMed] [Google Scholar]
- 59.Visconti RP, Richardson CD, Sato TN: Orchestration of angiogenesis and arteriovenous contribution by angiopoietins and vascular endothelial growth factor (VEGF). Proc Natl Acad Sci USA 2002, 99:8219-8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aristarkhov A, Eytan E, Moghe A, Admon A, Hershko A, Ruderman JV: E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc Natl Acad Sci USA 1996, 93:4294-4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Townsley FM, Aristarkhov A, Beck S, Hershko A, Ruderman JV: Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc Natl Acad Sci USA 1997, 94:2362-2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujita M, Furukawa Y, Tsunoda T, Tanaka T, Ogawa M, Nakamura Y: Up-regulation of the ectodermal-neural cortex 1 (ENC1) gene, a downstream target of the beta-catenin/T-cell factor complex, in colorectal carcinomas. Cancer Res 2001, 61:7722-7726 [PubMed] [Google Scholar]
- 63.Fedele M, Battista S, Manfioletti G, Croce CM, Giancotti V, Fusco A: Role of the high mobility group A proteins in human lipomas. Carcinogenesis 2001, 22:1583-1591 [DOI] [PubMed] [Google Scholar]
- 64.Finelli P, Pierantoni GM, Giardino D, Losa M, Rodeschini O, Fedele M, Valtorta E, Mortini P, Croce CM, Larizza L, Fusco A: The high mobility group A2 gene is amplified and overexpressed in human prolactinomas. Cancer Res 2002, 62:2398-2405 [PubMed] [Google Scholar]
- 65.Zhao J, Roth J, Bode-Lesniewska B, Pfaltz M, Heitz PU, Komminoth P: Combined comparative genomic hybridization and genomic microarray for detection of gene amplifications in pulmonary artery intimal sarcomas and adrenocortical tumors. Genes Chromosom Cancer 2002, 34:48-57 [DOI] [PubMed] [Google Scholar]
- 66.Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998, 95:14863-14868 [DOI] [PMC free article] [PubMed] [Google Scholar]