Abstract
Transforming growth factor-β1 (TGF-β1) plays a central role in promoting extracellular matrix protein deposition by promoting the transformation of fibroblasts to myofibroblasts. To gain new insights into the transcriptional programs involved, we profiled human fetal lung fibroblast global gene expression in response to TGF-β1 up to 24 hours using oligonucleotide microarrays. In this report, we present data for 146 genes that were up-regulated at least twofold at two time points. These genes group into several major functional categories, including genes involved in cytoskeletal reorganization (n = 30), matrix formation (n = 25), metabolism and protein biosynthesis (n = 27), cell signaling (n = 21), proliferation and survival (n = 13), gene transcription (n = 9), and of uncertain function (n = 21). For 80 of these genes, this is the first report that they are TGF-β1-responsive. The early induction of two members of the inhibitor of differentiation (ID) family of transcriptional regulators, ID1 and ID3, was followed by the up-regulation of a number of genes that are usually expressed by highly differentiated smooth muscle cells, including smooth muscle myosin heavy chain, basic calponin, and smoothelin. These findings were confirmed at the protein level for primary adult lung fibroblasts. ID1 further behaved like a typical immediate-early gene and, unlike ID3, was expressed and induced at the protein level. Immunohistochemical analysis showed that ID1 was highly expressed by (myo)fibroblasts within fibrotic foci in experimentally induced pulmonary fibrosis. ID1 acts as a dominant-negative antagonist of basic helix-loop-helix transcription factors that drive cell lineage commitment and differentiation. These findings have important implications for our understanding of fibroblast transcriptional programming in response to TGF-β1 during development, oncogenesis, tissue repair, and fibrosis.
Transforming growth factor-β1 (TGF-β1) is the prototypic member of the TGF-β superfamily of pleotrophic cytokines that regulate a number of cellular processes, including proliferation, differentiation, apoptosis, and migration. 1,2 In addition to playing a central role in a number of developmental and immunological processes, TGF-β1 is one of the most potent promoters of connective tissue formation characterized to date. In the embryo, its fibrogenic effects are critical during both organogenesis and morphogenesis; whereas in the adult, they are central to wound healing and tissue repair. TGF-β1 production and activation is under tight regulatory control and dysregulation of this control is associated with a number of pathological conditions, including carcinogenesis, autoimmune diseases, and tissue fibrosis.
TGF-β elicits its biological effects by binding to high-affinity cell-surface receptors and initiating the assembly of a receptor complex consisting of two type I and two type II receptors. 3 These receptors contain cytoplasmic serine-threonine kinase domains; and once assembled into this complex, constitutively active type II receptors activate the type I receptor kinases through phosphorylation of juxtamembrane domains. Activated type I receptors in turn phosphorylate proteins of the Smad family of latent transcription factors that propagate the signal by binding to regulatory sequences within target genes and recruiting transcriptional co-activators or co-repressors. 4,5 There is accumulating in vitro evidence that TGF-β1 may also signal through MAPK and JAK/STAT pathways via both direct, indirect, and pathway cross-talk mechanisms. 1
In terms of its role in tissue fibrosis, TGF-β exerts its potent fibrogenic effects by up-regulating mesenchymal cell matrix protein synthesis and gene expression, decreasing intracellular degradation of procollagen; down-regulating matrix metalloproteinase production; stimulating the production of tissue inhibitors of matrix metalloproteinases; and promoting plasminogen activator inhibitor-1 expression. 6 In addition, TGF-β stimulates fibroblast proliferation at low concentration, 7 further enhancing the potential for increased matrix deposition at sites of tissue injury. The activation of fibroblasts by TGF-β is accompanied by their transformation into smooth muscle α-actin-expressing contractile myofibroblasts. 8 This is the most conspicuous fibroblast phenotype present in granulation tissue at sites of wound healing and is generally thought to be the major cell responsible for both extracellular matrix deposition and for the generation of contractile force associated with wound contraction.
Although TGF-β is essential for wound healing, overproduction of TGF-β plays a major role in promoting excess deposition of matrix proteins in a number of pathological conditions, including among others, pulmonary, liver, kidney, and cardiac fibrosis; scleroderma; keloid scars; and arterial intimal thickening. 2 The importance of TGF-β in tissue fibrosis is supported by studies in animals. Transient overexpression of active TGF-β1 in the lung induces a chronic fibrotic response; 9 whereas, tissue-specific overexpression of TGF-β1 causes both progressive glomerulosclerosis 10 and hepatic fibrosis. 11 Conversely, blocking TGF-β with either neutralizing antibodies, soluble receptors, or by adenovirally expressed dominant-negative type II TGF-β receptors or Smad 7 as antagonists of TGF-β signaling; inhibit experimentally induced fibrosis in the lung, skin, and liver. 12-14
Recent years have seen major advances in our understanding of the molecular mechanisms involved in the activation of fibroblasts by TGF-β but the global transcriptional profile of genes involved in this response has not yet been examined in detail, with the exception of a recent report focusing on the immediate-early transcriptional response of dermal fibroblasts. 15 The aim of our study was to further our understanding of the temporal expression, regulation, and function of genes involved in the fibroblast response to TGF-β1. To this end we profiled the global transcriptional response of human fetal lung fibroblasts (HFL1; American Type Culture Collection, Rockville, MD) at four time points up to 24 hours using oligonucleotide microarrays (Affymetrix GeneChip, Santa Clara, CA) providing gene expression data for >6000 full-length human sequences. In this report, we present data for genes selected on the basis of at least a twofold up-regulation at two time points. Data for seven genes encoding signaling molecules (ARHB, MAP2K1, RAC1) and transcription factors (C-MYC, FOSL2, HRY, ID3) with a fivefold up-regulation at a single time point are also included. In addition to identifying 80 new TGF-β-responsive genes, the main findings of this study are that TGF-β1 induces the rapid and transient expression of two members of the ID (inhibitor of DNA binding/inhibitor of differentiation) family of dominant-negative transcriptional repressors, followed by a number of genes that are usually expressed by highly differentiated smooth muscle cells. The induction of these genes was confirmed at the protein level for primary cultures of adult lung fibroblasts. The potential relevance of these observations in vivo was confirmed by demonstrating that ID1 was highly expressed by (myo)fibroblasts within fibrotic foci in a rat model of pulmonary fibrosis. These novel findings have important implications for our understanding of TGF-β1 as a fibroblast differentiation factor and of the phenotypic plasticity between fibroblasts, myofibroblasts, and smooth muscle cells. They further support the novel hypothesis that ID1 may play a role in regulating the fibroblast differentiation program induced in response to a fibrogenic stimulus.
Materials and Methods
Fibroblast Culture
Human fetal lung fibroblasts (HFL-1) were purchased from the American Type Culture Collection. Primary human adult lung fibroblasts (pHALF) grown from explant cultures of normal lung tissue were a kind gift from Dr. RJ McAnulty (University College London, London, UK). Cells were maintained in sterile Dulbecco’s modified Eagle’s medium (DMEM) supplemented with penicillin (100 U/ml), streptomycin (50 μg/ml), and 5% (v/v) newborn calf serum (NCS) (DMEM-5% NCS), in a humidified atmosphere of air containing 10% CO2. Cells were routinely passaged every 6 to 7 days and tested for mycoplasma infection. DMEM, tissue culture supplements, and tissue culture plates were all from Invitrogen Life Technologies (Paisley, UK).
Preparation of Samples for GeneChip Hybridization
HFL-1 cells were grown to confluence in T125 tissue culture flasks in DMEM-5% NCS for 5 days, quiesced by serum deprivation for 24 hours, and exposed to 1 ng/ml (40 pmol/L) of activated TGF-β1 (R&D Systems, Abingdon, UK) for 1.5, 6, 16, and 24 hours in serum-free DMEM. Samples for chip hybridization were prepared according to protocols supplied by Affymetrix. Briefly, at the end of the incubation period, total RNA was extracted using TRIzol reagent (Invitrogen Life Technologies) and mRNA was isolated using Oligotex according to the manufacturer’s instructions (Qiagen Inc., Valencia, CA). Double-stranded cDNA was synthesized by reverse transcription of twice-purified mRNA using T7-(T24) primers (Genset Corp., La Jolla, CA) and the Superscript II cDNA Synthesis System (Invitrogen Life Technologies). The cDNA was used as a template for the generation of biotin-labeled in vitro transcription products using the Ambion T7 Megascript System (Ambion, Austin, TX) and biotin-11-CTP and biotin-16-UTP (Enzo Diagnostics Inc., Farmingdale, NY). After fragmenting biotinylated in vitro transcription products (cRNA) in 40 mmol/L of Tris acetate, pH 8.1, 10 mmol/L of potassium acetate, and 30 mmol/L of magnesium acetate at 94°C for 35 minutes; targets for chip hybridization were prepared by combining 10 μg of fragmented cRNA with sonicated herring sperm DNA (0.1 mg/ml; Sigma, St. Louis, MO), plus a mixture of four control bacterial and phage cRNA (1.5 pmol/L BioB, 5 pmol/L BioC, 25 pmol/L BioD, and 100 pmol/L Cre) and 5 nmol/L of control oligonucleotide (3 nmol/L oligo B2) as internal controls for hybridization efficiency.
GeneChip Hybridization
For each sample, 10 μg of fragmented cRNA in 200 μl of hybridization buffer were hybridized to an Affymetrix HuGeneFL Array GeneChip for 16 hours at 45°C and 60 rpm. Stringency washing in 6× sodium chloride/sodium phosphate/ethylenediaminetetraacetic acid and streptavidin phycoerythrin (Molecular Probes Inc., Eugene, OR) staining with an anti-streptavidin phycoerythrin antibody amplification step was performed on an Affymetrix Fluidics station using an automated program and protocols supplied by Affymetrix. After extensive washing, GeneChips were scanned on a Hewlett Packard GeneArray scanner G2500A.
Analysis of GeneChip Data
Scanned output files were analyzed using Affymetrix Version 3.1 software as previously described. 16-18 Briefly, after visual inspection of files for hybridization or chip-related defects and correct alignment of the grid, baseline (hybridized with mRNA from media control cells at each time point) and experimental (hybridized with mRNA from TGF-β1-treated cells at each time point) chip files were analyzed by scaling to an average intensity of 150 per gene as recommended by Affymetrix. The expression value (average difference) for each gene was determined by calculating the average of differences of signal intensity between all probe pairs used (ie, perfect match signal intensity minus mismatch intensity). For comparison analysis and determination of fold changes in gene expression, each experimental chip file was analyzed using the corresponding baseline file at each time point examined.
Expression Data Selection Criteria and Data Visualization
After GeneChip 3.1 software analysis, HuGeneFL expression analysis files were migrated to Microsoft Access and linked to genome databases on the World Wide Web (National Heart, Lung and Blood Institute; Swiss Prot; and GeneCards). A value of 20 was assigned to all average difference values less than 20 and genes for further analysis and presentation in this report were chosen on the basis of at least one average intensity value more than 40 and a twofold up-regulation at two time points. For genes encoding transcription factors or signaling molecules, the data were also queried on the basis of a fivefold up-regulation at a single time point to capture genes that may only be up-regulated transiently in response to TGF-β1. The data are arranged according to groupings based on current known biological functions (Figure 1) ▶ , in which log-transformed values of the fold ratios are displayed according to the method developed by Eisen and colleagues 19 using Cluster and Treeview Programs, with permission (available at: http://rana.lbl.gov/). Genes are listed with accession numbers and symbols and named according to the nomenclature proposed by the Human Genome Organization (HUGO) Gene Nomenclature Committee using databases available on the internet, including GeneCards (http://bioinfo.weizmann.ac.il/cards) and The Source (http://genome-www4.stanford.edu/cgi-bin/SMD/source/) for batch analysis of multiple genes.
Figure 1.

Global profile of fibroblast genes up-regulated in response to TGF-β1. Figure ▶ shows fold changes in gene expression over time for media control and TGF-β1-exposed cells. Genes are grouped into functional categories based on our current understanding of their most likely function as follows: A, genes encoding transcription factors; B, genes encoding signaling molecules; C, genes influencing cell survival and proliferation; D, genes associated with matrix formation; E, genes associated with cytoskeletal reorganization; F, genes involved in cell metabolism and protein synthesis. For each panel, each row displays expression data for a single gene, whereas columns represent pairwise comparisons in gene expression for TGF-β1-treatments relative to matching media controls at each time point as indicated by the labeling at the top of each panel; where TGF-β1.5, TGF-β6, TGF-β16, and TGF-β24 represent the pairwise comparison of the TGF-β1-treatments at 1-5, 6, 16, and 24 hours relative to the matching media control at each time point. Also shown are expression data for media controls, where C1.5, C6, C16, and C24 represents the comparison value for each time point relative to baseline expression (media control at 1.5 hours). Changes in gene expression are based on log-transformed values of the fold ratios of the signal intensities (referred to as average difference by Affymetrix) using a visual analog in which progressively brighter shades of red or green correspond to progressively greater gene inductions or repressions, respectively, as described by Eisen and colleagues. 19 Genes are listed with their GenBank accession codes, followed by their symbols and full gene name according to the HUGO nomenclature. An asterisk denotes that the gene is a newly identified TGF-β-responsive gene. Genes were selected on the basis of at least a twofold increase in gene expression at two time points in response to TGF-β1 treatment compared with the corresponding media controls. Also included are three genes encoding signaling molecules (ARHB, MAP2K1, RAC1) and four genes encoding transcription factors (C-MYC, FOSL2, HRY, ID3), which were increased by at least fivefold at a single time point.
Northern Analysis of Fibroblast ID1 and ID3 mRNA Levels
HFL-1 and pHALF were seeded at 2 × 105 cells/ml in 6-cm-diameter dishes in DMEM-5% NCS. On reaching visual confluence, cells were quiesced in serum-free DMEM for 24 hours and incubated in fresh serum-free DMEM with and without TGF-β1 (1 ng/ml) for 45, 90, and 120 minutes. For cycloheximide experiments, cells were preincubated with cycloheximide (25 μg/ml, Sigma-Aldrich Company Ltd., Poole, UK) for 2 hours before exposure to serum-free control media or TGF-β1. At the end of the incubation, the media was removed and total RNA was isolated with TRIzol reagent (Invitrogen Life Technologies) according to the manufacturer’s instructions. Seven μg of total RNA were mixed with RNA loading buffer containing ethidium bromide (Sigma-Aldrich), heated to 65°C for 10 minutes, and electrophoresed on a formaldehyde 1% (w/v) agarose gel. RNA loading and integrity was visualized and quantitated by fluorescent scanning of the gel (FLA 3000, Fuji) before transfer to nylon membranes (Hybond N; Amersham International, High Wycombe, UK) by Northern transfer and fixation by UV crosslinking. Membranes were hybridized overnight at 65°C in a rotating hybridization oven in standard Denhardt’s-based hybridization solution in the presence of the [32P]-dCTP-labeled cDNA probes for either ID1 or ID3 (generous gifts from Professor F. Sablitzky, University of Nottingham, UK 20 ), generated by random priming using an oligolabeling kit (Pharmacia Biotech, Piscataway, NJ). At the end of the hybridization, filters were rinsed at low stringency (4× standard saline citrate, 0.1% sodium dodecyl sulfate for 30 minutes at 50°C) and once at medium stringency (2× standard saline citrate, 0.1% sodium dodecyl sulfate for 30 minutes at 50°C). Membranes were exposed to a phosphorimage storage screen (Fuji) for 16 hours and ID1 and ID3 mRNA levels were quantitated by phosphorimage analysis (FLA 3000, Fuji).
Western Analysis and Immunocytofluorescence of Fibroblast ID1 and ID3 and Smooth Muscle Cell Differentiation Proteins
Antibodies
All antibodies were used at a 1:1000 dilution and are listed in Table 1 ▶ .
Table 1.
Antibodies
| Antigen | Antibody | Clone |
|---|---|---|
| Vimentin | Mouse anti-human monoclonal | V9; Kappa Sigma, cat. no. V6630 |
| Desmin: | Mouse anti-desmin monoclonal | D33; DAKO, cat. no. M0724 |
| Smooth muscle α-actin | Mouse anti-human monoclonal | Clone 1A 4; Sigma, cat. no. A2547 |
| β-actin | Rabbit affinity-isolated | Sigma, cat. no. A2066 |
| Smooth muscle myosin heavy chain | Mouse anti-human monoclonal | Clone hSM-V; Sigma cat. no., M7786 |
| h-caldesmon | Mouse anti-human monoclonal | Clone hHCD; Sigma cat. no., C4562 |
| Smoothelin* | Mouse anti-human monoclonal | R4 |
| ID1 | Rabbit polyclonal IgG | C-20; Santa Cruz Biotech., cat. no. sc-488 |
| ID2 | Rabbit polyclonal IgG | C-20; Santa Cruz Biotech., cat. no. sc-489 |
| ID3 | Rabbit polyclonal IgG | C-20; Santa Cruz Biotech., cat. no. sc-490 |
| Negative control | Mouse IgG1 Kappa | MOPC21; Sigma, cat. no. I5381 |
*A kind gift from Dr. Guillaume van Eys, University of Masstricht, The Netherlands. 21
Western Analysis
HFL-1 and pHALF were seeded at 5 × 104 cells/ml in 2.4-cm-diameter dishes in DMEM-5% NCS. On reaching visual confluence, cells were quiesced in serum-free DMEM for 24 hours and exposed to control media (serum-free DMEM) or TGF-β1 (1 ng/ml for 2, 4, 6, or 36 hours). At the end of the incubation, the monolayer was washed twice with ice-cold PBS and cell lysis was performed by adding 100 μl of Laemmli sample buffer and scraping with a rubber policeman. Cell lysates were mixed several times to shear DNA and 25 μl aliquots were heated for 5 minutes at 95°C before electrophoresis (100 V for 3 hours) on a 12% sodium dodecyl sulfate-polyacrylamide gel with a 7% stacking gel for ID1, ID2, and ID3 and on Novex 4 to 20% Tris-glycine gradient gels (Invitrogen Life Technologies) for all cytoskeletal proteins. Separated proteins were transferred onto Hybond-ECL nylon membranes (Amersham International) for 1 hour at 25 V. The membrane was blocked with TBST (150 mmol/L NaCl, 50 mmol/L Tris-HCl, pH 7.4, 0.1% Tween 20) containing 5% dry milk for 1 hour and primary antibodies were added at a 1:1000 dilution overnight at 4°C. Horseradish peroxidase-conjugated anti-rabbit IgG (DAKO Ltd., Cambridge, UK) was added at a 1:2000 dilution for 1 hour followed by three washes in TBST for 15 minutes. Protein bands were visualized by enhanced chemiluminescence (ECL) according to Amersham International standard protocols. To confirm even protein loading, membranes were stripped and reprobed for β-actin levels.
Immunocytofluorescence
Cells were seeded at 4 × 103 cells per well in DMEM-10% NCS into chamberslides (Labtek, Nunc, Fisher Scientific UK, Ltd., Loughborough, UK), grown to subconfluence, quiesced in serum-free DMEM for 24 hours, and exposed to control media (DMEM) or TGF-β1 (1 ng/ml) for 36 hours. At the end of the incubation, the cell monolayer was gently washed three times with PBS, fixed for 45 seconds with methanol (100%; cooled to −20°C), and primary antibodies were added for 1 hour at room temperature. After extensive washing of the monolayer with PBS, normal goat serum in PBS (1:20) was added for 5 minutes and the monolayer was washed once in PBS before incubation with an fluorescein isothiocyanate-conjugated goat anti-mouse IgG (diluted 1:100 in PBS) for 30 minutes. After extensive washing with PBS, a single drop of CitiFluor AF1 (Chem. Lab., Canterbury, UK) was added and cells were photographed by UV light microscopy at ×1000 with oil immersion using a Zeiss Axioscop 2 microscope.
Immunohistochemical Localization of ID1 in Bleomycin-Induced Pulmonary Fibrosis
The model of bleomycin-induced pulmonary fibrosis has previously been described. 21,22 Briefly, male Lewis rats (6 weeks of age) were anesthetized and bleomycin sulfate (Lundbeck, Luton, UK) was administered by a single intratracheal injection (1.5 mg/kg in 0.3 ml of sterile saline). Control animals received saline alone. After 14 days, animals were killed and lungs were insufflated and fixed by intratracheal instillation of 4% paraformaldehyde. Tissues were further fixed by immersion in 4% paraformaldehyde, dehydrated, and embedded in paraffin wax. Four-μm-thick lung sections were mounted on slides, dewaxed in xylene, and rehydrated. Tissue endogenous peroxidase activity was blocked (1% H2O2 in distilled H2O) and slides were further blocked with normal goat serum. Sections were sequentially incubated with ID1 rabbit polyclonal IgG (1:800; 24 hours, 4°C), goat-anti-rabbit biotinylated secondary antibody (1:200, 1 hour at room temperature; DAKO, High Wycombe, UK) and the streptavidin/peroxidase conjugate (1:200, 30 minutes) followed by fresh 3′3-diaminobenzidine (10 minutes at room temperature) (DAB Vectorkit; Vector Laboratories Ltd., Peterborough, UK) and counterstained with Mayers’ hematoxylin according to standard immunohistochemical protocols. To confirm staining specificity, sections were also incubated with either nonimmune rabbit IgG isotype control (DAKO) or secondary antibody only or ID1 rabbit polyclonal IgG previously incubated with ID1 blocking peptide (catalog no. sc-488P; Santa Cruz Biotechnology Inc., Santa Cruz, CA) in fivefold excess for 2 hours at room temperature.
Statistical Analysis
All numerical data are presented as means ± SEM from three replicate cultures, unless otherwise indicated. Experiments were repeated at least three times and statistical evaluation was performed using an unpaired Student’s t-test. The mean values of various parameters were said to be significantly different when the probability of the differences of that magnitude, assuming the null hypothesis to be correct, fell below 5% (ie, P < 0.05).
Results
Global Expression Profiling in Response to TGF-β1
In this report, we present data for genes that were predominantly up-regulated in response to TGF-β1. The data are presented in Figure 1 ▶ and are arranged according to groupings based on current known biological functions as described in Materials and Methods. New TGF-β-responsive genes identified in this report are indicated by an asterisk placed next to their GenBank accession codes. A brief description of each of the functional gene categories is provided below.
Panel A: Genes Encoding Transcription Factors
Only 2 of the 9 genes in this functional category were previously known to be TGF-β-responsive (JUNB, C-MYC) at the time these experiments were performed. Most of these genes behaved like immediate-early response genes and were highly up-regulated (between 8- and 10-fold) in response to TGF-β1 at the earliest time point examined (eg, C-MYC, ID1, ID3, HRY). One notable exception was JUNB, which was highly up-regulated at 90 minutes and remained induced at subsequent time points. Of particular interest was the novel observation that TGF-β1 induced the expression of two members of the ID family of helix-loop-helix (HLH) transcriptional repressors, ID1 and ID3. This is also the first report to suggest that TGF-β1 up-regulates TCF8 and XBP1, associated with regulating the expression of genes involved in immune and inflammatory responses, as well as the gene encoding ubiquitous heterotrimeric protein CCAAT-binding factor (CBF/NF-Y).
Panel B: Genes Encoding Signaling Molecules
Only 2 of the 21 genes in this group were previously known to be up-regulated in response to TGF-β. This includes MADH7 encoding the inhibitory Smad, Smad7, involved in terminating signaling by TGF-β 23 and which was also the most highly up-regulated gene in this category. The up-regulation by TGF-β1 of genes encoding members of the MAPK pathway (eg, MAP2K1, MAPK6), the Rho/Ras family of small GTPases (eg, RSU1, LOC55969), and genes encoding regulators of G-protein signaling (CAV2) is consistent with previous studies, which report that in addition to Smad proteins, MAPK, Rho, and G-protein signaling pathways are activated in response to TGF-β. 24-26 A detailed discussion of all these genes is beyond the scope of this report but the finding that TGF-β1 induces the rapid expression of FZD2 (frizzled Drosophila homolog 2) encoding a G protein-coupled receptor that functions as a Wnt receptor, may be of particular interest in light of recent reports providing experimental evidence for the cooperation between TGF-β and Wnt/wingless signaling pathways. 27
Panel C: Genes Influencing Cell Survival and Proliferation
Most of the genes in this functional category were already known to be TGF-β-responsive. However, this study shows that the transcriptional effects on the insulin growth factor (IGF) system were particularly far-reaching and included IGF-1, its major binding protein, IGF-BP3; genes encoding low-affinity IGF binding proteins (Cyr61, IGFBP7/Mac25); and CTGF, which has been proposed to mediate a number of the profibrotic effects of TGF-β. 28 This is also the first evidence that TGF-β1 up-regulates the expression of PRSS11, encoding a protease named protease serine 11, which has been proposed to regulate the availability of IGFs by cleaving IGF-binding proteins. 29 Its induction by TGF-β1 may represent a novel mechanism by which TGF-β influences IGF/IGF-1 bioavailability. Other novel TGF-β-responsive genes include DAD1, encoding the anti-apoptotic protein, defender against cell death-1. Its induction along with that of two signaling genes (represented in Figure 1B ▶ ) involved in regulating cell growth and apoptosis, IER3 and PIM-1, 30,31 is consistent with recent reports that TGF-β1 promotes myofibroblast survival. 32
Panel D: Genes Associated with Matrix Formation
As expected, most of the genes in this functional category are known TGF-β-responsive genes but insights were gained in terms of the temporal nature of this response. For example, a number of procollagen genes (eg, COL1A1, COL1A2, COL4A2, COL5A1, and COL6A2) were already up-regulated within 90 minutes of exposure to TGF-β1 suggesting that they may be direct TGF-β/Smad target genes as has been recently reported. 15,33 As well as influencing matrix protein gene expression, TGF-β1 also promoted the expression of genes encoding proteins involved in extracellular matrix biosynthesis (eg, P4HA1; SERPINH2also known as HSP47; PLOD1 and PLOD2; LOX), including the novel TGF-β-responsive gene, PYCS. This gene encodes the enzyme, delta 1-pyrroline-5-carboxylate synthetase (P5CS), responsible for catalyzing the conversion of l-glutamate to glutamic γ-semialdehyde. Fibroblasts derive most of their proline for collagen biosynthesis from this pathway of proline biosynthesis, 34 so that up-regulation of this gene may represent an important mechanism by which proline is made available to meet the demands of increased procollagen synthesis in response to TGF-β1. The up-regulation of the novel TGF-β1-responsive gene, NDST1, encoding N-deacetylase/N-sulfotransferase involved in heparan sulfate proteoglycan biosynthesis, may similarly serve to ensure optimal cellular conditions for increased proteoglycan biosynthesis. Finally, in addition to up-regulating SERPINE encoding plasminogen activator inhibitor-1, 35 TGF-β1 also up-regulated PI7, encoding nexin 1, one of the major tissue inhibitors of thrombin and urokinase, raising the possibility that TGF-β may play a role in controlling coagulation events, in addition to promoting fibrin persistence at sites of tissue injury.
Panel E: Genes Associated with Cytoskeletal Reorganization
This is the largest functional group of genes up-regulated in response to TGF-β1 with the highest number of novel TGF-β-responsive genes (19 of 30 genes). Analysis of baseline gene expression revealed that the cells used in this study expressed some cytoskeletal genes typically associated with the myofibroblast phenotype (eg, ACTA2, TAGLN, CALD1, and so forth). These myofibroblast marker genes, as well as genes encoding proteins involved in reorganization of the actin cytoskeleton (eg, TPM1, FLNA, PLS3, ACTN1, ARPC2, GAL, and so forth), were highly up-regulated in response to TGF-β1, indicating the further phenotypic transformation of these fibroblast cultures toward myofibroblasts expressing a range of proteins involved in cell contraction. More surprising, TGF-β1 also induced the expression of a number of cytoskeletal genes, which are usually expressed by highly differentiated smooth muscle cells (eg, MYH11/SMMHC, SMTN, CNN1), including keratin 18, which is commonly used as a phenotypic marker of the functional switch from the contractile to the synthetic phenotype for vascular smooth muscle cells. 36 Of interest, maximal expression of most of these smooth-muscle cell phenotypic marker genes was delayed until at least 16 hours after exposure to TGF-β1 and coincides with that of CSRP2, encoding the smooth muscle cell-restricted signaling molecule, cysteine and glycine-rich protein 2 (also known as SmLIM). CSRP2 has been shown to be drastically down-regulated during periods of maximal smooth muscle cell proliferation in vitro and vessel wall injury in vivo and is therefore thought to play a role in smooth muscle cell differentiation. 37
Panel F: Genes Involved in Cell Metabolism and Protein Synthesis
Genes up-regulated in response to TGF-β1 within this functional category included genes encoding proteins involved in polyamine biosynthesis (eg, SRM), nucleic acid biosynthesis (eg, CTPS), protein biosynthesis (eg, EPRS, GARS, and IARS), and multimeric protein assembly (eg, HSPA5). Twenty-one of 27 of these genes were not previously known to be TGF-β-responsive and more than half were down-regulated in serum-free control conditions throughout time. As all expression comparisons are made between TGF-β1 and the corresponding serum-free control mRNA population at each time point, TGF-β1 may prevent these genes from being repressed. In addition, it is tempting to speculate that mRNA levels for these genes are maintained/up-regulated to provide the synthetic machinery to process the dramatic reprogramming of TGF-β1-treated cells both in terms of matrix protein production and cytoskeletal reorganization. Finally, TGF-β1 also up-regulated 21 genes of uncertain function, all of which were not previously known to be TGF-β-responsive. These genes will not be discussed further in this report.
ID1 and ID3 mRNA and Protein Levels
The novel observations that TGF-β1 induced the expression of two members of the ID family of dominant-negative HLH transcriptional repressors involved in regulating cell differentiation, as well as several genes that are usually expressed by differentiated smooth muscle cells, has implications for our current understanding of the role of TGF-β1 as a fibroblast differentiation factor. A series of microarray data confirmation experiments were therefore performed to investigate the expression of these genes in greater detail.
Figure 2 ▶ shows the induction of ID1 and ID3 in response to TGF-β1 assessed by Northern analysis. The top panels in Figure 2 ▶ show ID1 and ID3 mRNA levels throughout time and confirm the rapid and dramatic induction of both genes in response to TGF-β1 within 45 minutes of exposure. The induction was transient and ID1 and ID3 mRNA levels returned to baseline levels within 2 hours. Experiments performed in the presence of the protein synthesis inhibitor, cycloheximide (Figure 2 ▶ , middle panels), showed that up to two thirds of the stimulatory effects obtained for ID1 occur independently of de novo protein synthesis. In contrast, ID3 was induced in the presence of cycloheximide alone, but the effects of cycloheximide and TGF-β1 were not additive. This suggests that ID3 mRNA levels are highly regulated by a labile repressor at baseline, which may be overcome by the addition of TGF-β1. Similar experiments performed in the presence of an inhibitor of transcription, actinomycin D (Figure 2 ▶ , bottom panels), showed that the induction of ID1 and ID3 in response to TGF-β1 was completely abolished and therefore dependent on increased gene transcription.
Figure 2.
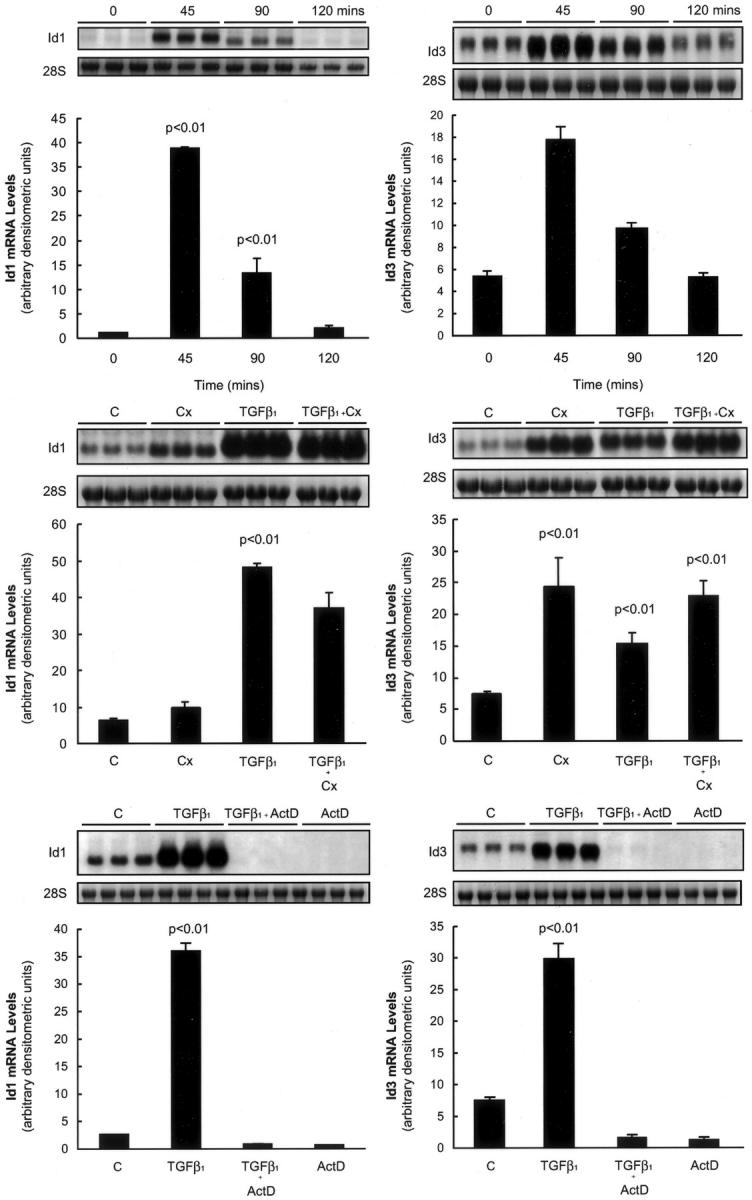
ID1 and ID3 behave like direct TGF-β1 target genes. Top: The effect of TGF-β1 on ID1 and ID3 mRNA levels over time assessed by Northern analysis. Middle: The effect of cycloheximide on TGF-β1-induced ID1 and ID3 mRNA levels. Bottom: The effect of TGF-β1 on ID1 and ID3 mRNA levels in cells pretreated with actinomycin D. Data for HFL-1 fibroblasts fetal is shown on the left; whereas the right shows data for primary adult lung fibroblasts (pHALF). Bar graphs represent the mean of three replicate cultures. Also shown are representative phosphorimages of the ID1 and ID3 transcripts and corresponding images of the 28S rRNA bands. P values represent comparisons made to serum-free media control-treated cells.
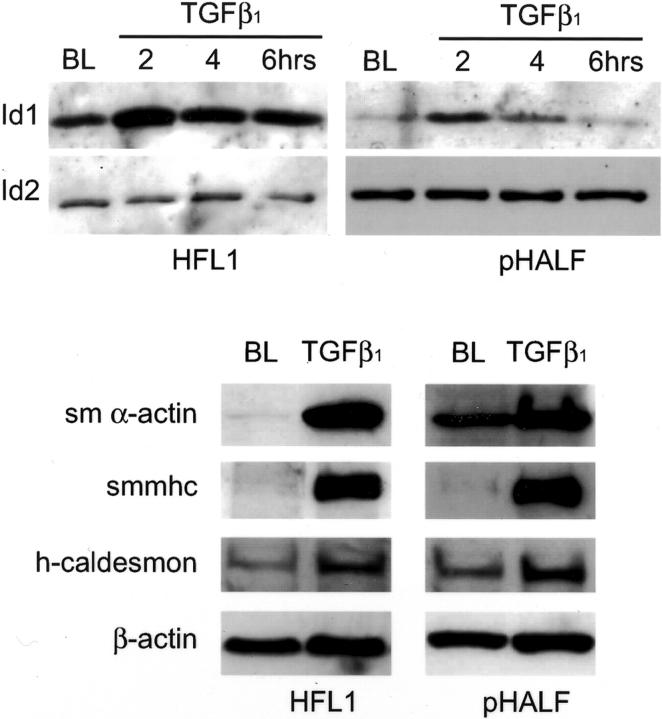
Protein confirmation experiments by Western analysis were similarly performed with both fetal lung fibroblasts and primary human adult lung fibroblasts (pHALF) exposed to TGF-β1 in serum-free conditions over time (Figure 3) ▶ . The intensity of the immunoreactive band obtained with an ID1-specific antibody was increased threefold and fivefold within 2 hours of exposure to TGF-β1 for fetal and adult lung fibroblasts, respectively. Also shown are the bands obtained with an ID2-specific antibody. Consistent with the lack of an effect in response to TGF-β1 in microarray experiments, ID2 protein levels were totally unaffected. In contrast, although ID3 was easily detectable in whole cell lysates from mixed blood lymphocyte cultures (positive control), we were unable to detect ID3 at the protein level for both fibroblast cultures (data not shown).
Figure 3.
TGF-β induces the expression of ID1 and smooth muscle cell differentiation marker proteins. Top: A representative Western blot for the effect of TGF-β1 in serum-free conditions on ID1 protein levels up to 6 hours for human fetal lung fibroblasts (HFL1) and primary human adult lung fibroblasts (pHALF) compared with baseline (BL). Also shown are ID2 protein levels that did not change on exposure to TGF-β1. In contrast, ID3 was undetectable at the protein level. Bottom: Representative Western blots for the effect of TGF-β1 for 36 hours on the indicated proteins for cultures of HFL-1 and pHALF compared with cultures exposed to serum-free control media (BL).
Myofibroblast and Smooth Muscle Cell Marker Protein Levels and Organization
The induction of typical myofibroblast and smooth muscle cell marker proteins 38 was also assessed by Western analysis (Figure 3) ▶ and immunocytofluorescent microscopy (Figure 4) ▶ . These experiments were performed after 36 hours of exposure to TGF-β1 to allow sufficient time for cytoskeletal proteins to be produced and organized into functional filamentous networks. Figure 3 ▶ shows that in accord with microarray data, fetal lung fibroblasts express low levels of smooth muscle α-actin and h-caldesmon in serum-free conditions. They are also vimentin-positive and desmin-negative (data not shown). On exposure to TGF-β1 for 36 hours, the intensity of the immunoreactive bands was markedly increased for all marker proteins examined with densitometric values normalized to β-actin increased 32-fold relative to media controls for smooth muscle α-actin and 1.5-fold for h-caldesmon. Again in accord with our microarray data, Western analysis showed that smooth muscle myosin heavy chain was virtually undetectable in media control-treated cells. In contrast, for cells exposed to TGF-β1 for 36 hours, this protein was detectable as a single high-intensity band with densitometric values increased ninefold relative to media control levels. Similar results were obtained with primary human adult lung fibroblasts (pHALFs, Figure 3 ▶ ) exposed to TGF-β1 with levels increased 2-, 1.8-, and 26-fold relative to media controls for smooth muscle α-actin, h-caldesmon, and smooth muscle myosin heavy chain, respectively.
Figure 4.
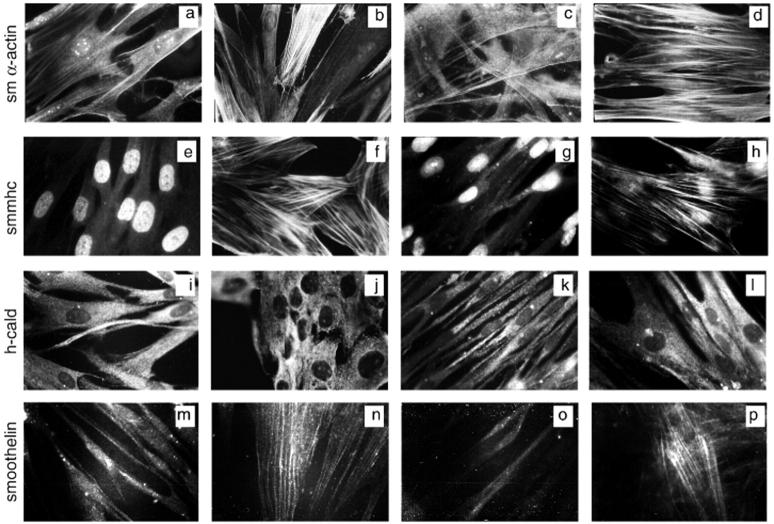
Effect of TGF-β1 on smooth muscle differentiation marker protein expression in fetal and primary adult lung fibroblasts by immunocytofluorescence. Images show human fetal lung fibroblasts (HFL-1) (first two columns) and primary human adult lung fibroblasts (pHALF) (last two columns) exposed to serum-free control media or TGF-β1 for 36 hours and stained for the indicated proteins by immunocytofluorescence. Images are representative of a minimum of 20 fields viewed at high magnification (×1000 under oil immersion) for three separate experiments performed. a–d: Smooth muscle α-actin. a, HFL1 in control media; b, HFL1 and TGF-β1; c, pHALF in control media; d, pHALF and TGF-β1. e–h: Smooth muscle myosin heavy chain. e, HFL1 in control media; f, HFL1 and TGF-β1; g, pHALF in control media; h, pHALF and TGF-β1. i–l: h-caldesmon. i, HFL1 in control media; j, HFL1 and TGF-β1; k, pHALF in control media; l, pHALF and TGF-β1.m–p: Smoothelin. m, HFL1 in control media; n, HFL1 and TGF-β1; o, pHALF in control media; p, pHALF and TGF-β1.
Figure 4 ▶ further confirmed that the cells used in this study express low levels of smooth muscle α-actin in serum-free conditions in that ∼8% fetal and ∼12% adult lung fibroblasts stained positively by immunocytofluorescent microscopy (Figure 4, a and c) ▶ . In cells exposed to TGF-β1, smooth muscle α-actin was visible as an extensive network of brightly stained fibers in most cells examined (Figure 4, b and d) ▶ . In contrast, smooth muscle myosin heavy chain was undetectable within the cytoplasm of media control cells (Figure 4, e and g) ▶ , but more than 90% fetal and adult lung fibroblasts expressed bright smooth muscle myosin heavy chain fibers after exposure to TGF-β1 (Figure 4, f and h) ▶ . h-Caldesmon, was visible as a faint diffuse protein expressed throughout the cytoplasm in media control cells (Figure 4, i and k) ▶ . On exposure to TGF-β1, all cells displayed tightly packed intensely stained granular h-caldesmon filaments (Figure 4, j and l) ▶ .
Finally, we also examined the appearance of the more recently characterized smooth muscle cell phenotypic marker, smoothelin, 39 because this was another highly induced gene in our microarray data set. As expected, smoothelin did not form filaments in media control-treated cells (Figure 4, m and o) ▶ but typical tenuous-looking filaments were clearly visible in greater than 80% fetal and primary adult lung fibroblasts exposed to TGF-β1 (Figure 4, n and p) ▶ .
ID1 Expression in Bleomycin-Induced Pulmonary Fibrosis
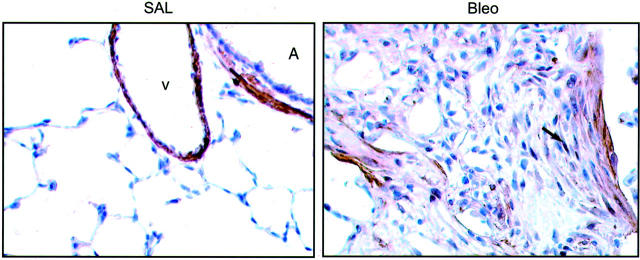
To establish the potential importance of these observations in an in vivo setting, we assessed the expression of ID1 in an established model of pulmonary fibrosis induced after a single intratracheal instillation of bleomycin in rats. 21,22 Figure 5 ▶ shows representative images for ID1 expression in sections from paraffin-embedded insufflated lungs of saline-treated control rats (SAL) compared with a typical fibrotic foci within the lung parenchyma observed 14 days after bleomycin injury (Bleo). In the normal lung, ID1 immunoreactivity is confined to smooth muscle bundles surrounding major airways (A) and blood vessels (V); whereas the lung parenchyma is completely negative. In contrast, in bleomycin-treated rat lungs, a high proportion of spindle-shaped cells with a typical (myo)fibroblast appearance stained positively within all fibrotic foci examined. Close examination further revealed that for a number of cells, ID1 immunoreactivity was predominantly localized to the nucleus.
Figure 5.
Immunohistochemical analysis of ID1 expression in experimental pulmonary fibrosis. Figure ▶ shows brown immunoperoxidase staining for ID1 in insufflated normal rat lung tissue 14 days after intratracheal instillation of saline (SAL) compared with rats given bleomycin (Bleo). In the normal lung, ID1 immunoreactivity is confined to smooth muscle bundles surrounding major airways (A) and blood vessels (V) but in bleomycin-induced pulmonary fibrosis, ID1 was also immunolocalized to (myo)fibroblasts within fibrotic foci, with some cells showing nuclear staining (arrow). Original magnifications, ×400; counterstained with Mayers hematoxylin.
Discussion
This global expression profiling study has allowed us to gain a number of insights into the transcriptional response of fibroblasts to one of their most potent activators. First, it has led to the identification of 80 novel TGF-β-responsive genes, including two genes encoding members of the ID family of transcription factors. Second, this study revealed that in addition to up-regulating a large number of genes associated with matrix formation, TGF-β1 promoted the expression of a much larger number of genes involved in cytoskeletal reorganization than previously realized. This includes a number of genes associated with myofibroblast transformation, but more surprisingly also genes associated with smooth muscle cell differentiation.
ID1 Is a Direct TGF-β1 Target Gene
Although several potential novel TGF-β1-responsive genes encoding transcription factors were identified by microarray analysis, we focused our attention on two members of the ID family of transcription factors, ID1 and ID3 (also known as HLH 1R21) (Figure 2A) ▶ . The ID family of proteins are HLH proteins that lack a basic region, and therefore do not bind DNA directly but act as dominant-negative inhibitors of DNA-binding basic HLH proteins that drive cell lineage commitment and differentiation. 40 Four mammalian ID proteins (ID1 to ID4) have been identified to date. Their expression patterns are partially overlapping and there seems to be a certain level of functional redundancy between ID1, ID2, and ID3; whereas ID4 expression is limited to specific tissues. 41 ID1 gene expression in a number of cell types has previously been reported to be strongly induced in response to serum, platelet-derived growth factor, 42,43 insulin-like growth factor-I (IGF-I), 44 and the bone morphogenic proteins, BMP-2 and BMP-4. 45,46 Until recently, TGF-β was not thought to induce ID1. 47 However, during the preparation of this manuscript, Goumans and colleagues, 48 reported that TGF-β was capable of inducing ID1 in endothelial cells via activation of the TGF-β type I receptor, ALK1 but not ALK5; whereas Lopez-Rovira and colleagues 49 reported that TGF-β was only a very modest inducer of ID1 promoter activity in C2C12 myoblasts. The induction of ID1 by TGF-β may therefore be exquisitely cell-specific and may further depend on the repertoire of TGF-β receptors expressed.
Time-course studies and experiments performed in the presence of actinomycin D and cycloheximide, showed that ID1 behaved like a typical immediate-early gene in response to TGF-β1. In contrast, data obtained for ID3 suggest that baseline expression is under regulatory control by a labile repressor protein, which may be overcome by the addition of TGF-β1. At the protein level, fetal and adult fibroblasts express both ID1 and ID2 at baseline but protein for ID3 was undetectable and only ID1 was up-regulated in response to TGF-β1. ID1 protein levels further primarily followed both the induction and decay in ID1 mRNA levels. This is consistent with previous reports that ID1 protein turnover is very rapid, with a half-life of 20 to 30 minutes. 50 Taken together these data indicate that ID1 is likely to be direct TGF-β1 target gene in fibroblasts, as has recently been reported for endothelial cells. 48
TGF-β1 Induces a Mixed Myofibroblast and Smooth Muscle Cell Differentiation Program
Global expression profiling of fetal fibroblast cultures further provided evidence that TGF-β1 induces several smooth muscle cell phenotypic marker genes associated with both the differentiated contractile (eg, SMMHC/MYH11, CNN1, SMTN, CSRP2) and the synthetic phenotype (eg, KRT 18, COL4A1, COL4A2), while maintaining the typical matrix synthetic program associated with myofibroblasts (eg, COL1A1, COL1A2, ELN, and so forth). Protein confirmation experiments by Western analysis and immunocytofluorescence were in full agreement with these data for myofibroblast/smooth muscle cell differentiation proteins (smooth muscle α-actin, smooth muscle myosin heavy chain, h-caldesmon), including the more recently described cytoskeletal protein smoothelin that is usually expressed by vascular smooth muscle cells in blood vessels that are capable of pulsatile contraction. 51 Smoothelin generally co-localizes with smooth muscle α-actin and is known to be induced by TGF-β in smooth muscle cells 52 but has never been reported to be expressed by non-smooth muscle cells. Finally, the observation that a number of these proteins adopted a typical cytoskeletal organization on exposure to TGF-β1 is in accord with our microarray data that TGF-β1 also up-regulated a number of genes encoding cytoskeletal-sorting, -targeting, and -processing proteins (eg, TPM1, FLNA, PLS3, ACTN1, ARPC2, GAL, and so forth).
The observation that mesenchymal cells are capable of transforming into smooth muscle α-actin-positive contractile myofibroblasts during tissue repair and under certain experimental conditions has been extensively documented. 53 These cells are highly activated in terms of extracellular matrix protein synthesis and the production of fibrogenic cytokines 54 and generate the contractile force required for wound closure. Myofibroblasts are also the predominant fibroblast phenotype present within active fibrotic lesions in a number of internal organs, including the lung, 54,55 liver, 56,57 kidney, 58 and heart, 59 so that acquisition of the myofibroblast phenotype is thought to play a major role in excessive deposition of matrix proteins during the development of tissue fibrosis. This phenotype has further been associated with pathological contractures (eg, Dupuytren’s disease), hypertrophic scars, and stromal reactions to tumor growth and invasion. 60,61 Although, myofibroblasts have been shown to express a number of contractile proteins, in addition to smooth muscle α-actin, they are normally thought to arrest their cellular transition at the myofibroblast stage, 60 so that complete maturation to fully differentiated smooth muscle cells is rarely achieved. 62,63
The cellular conversion of mesenchymal cells to myofibroblasts is thought to be mediated by cytokines present during development, injury, and disease, with TGF-β1 being one of several key mediators involved. 64 There is also good in vitro and in vivo evidence that TGF-β1 promotes smooth muscle cell maturation, 65 and is capable of promoting the differentiation of multipotential 10T1/2 cells 66 and neural crest cells. 67 However, current evidence suggests that TGF-β1 is not capable of promoting the expression of late-stage smooth muscle differentiation marker genes (SMMHC/MYH11, CNN1) in fibroblasts and other non-smooth muscle cells. 38 Our study challenges this view in that TGF-β1 was capable of inducing the expression of a number of smooth muscle differentiation marker proteins, including smooth muscle myosin heavy chain and smoothelin. This finding was replicated in primary adult lung fibroblasts so that this differentiation potential was furthermore not restricted to fetal lung fibroblasts with a potentially less terminally differentiated phenotype. In terms of the lack of evidence for a similar effect observed in other studies, there may be a number of possible explanations, including critical differences in fibroblast isolation and culture conditions, the experimental approach adopted, and importantly the tissue from which the fibroblasts were derived. There is compelling evidence that fibroblasts exist as a number of subpopulations with unique phenotypes and functions. 68 The differentiation potential of fibroblasts derived from different organs, or even the same tissue, may therefore differ considerably and this may provide a very plausible explanation for the lack of a similar effect obtained with TGF-β1 in previous studies.
Potential Role of ID1 in Fibroblast Differentiation in Response to TGF-β1
The molecular mechanisms by which TGF-β1 induces the expression of smooth muscle genes remains partially understood, but much of the evidence points to a critical role for a novel TGF-β control element (TCE) present within the promoters of a number of these genes, which acts either independently (eg, SM22) 69 or in concert with CArG elements (eg, ACTA2 encoding smooth muscle α-actin) depending on the cell type examined. 70,71 The transcription factors involved in regulating the expression of these genes are also beginning to be identified and include the serum response element that binds the CArG elements 70 and both positive- and negative-acting Kruppel-like transcription factors that interact with the TCE in the SM22 promoter. 69
The dramatic induction of ID1 expression by TGF-β1 in our study raises the possibility that ID1 may play a novel role in regulating the transcriptional responses of fibroblasts to one of their most potent activators. One of the most well-characterized functions of ID1 is its ability to act as an inhibitor of myogenesis of skeletal tissue by associating with bHLH class A E proteins (E12, E47, E2-2, and HEB) and preventing them from forming active hetero-oligomeric complexes with the myogenic regulatory factors (myoD, myogenin, Myf-5, and MRF4/Myf-6). 72,73 Because ID1 is generally thought to function as an inhibitor of differentiation, the transient induction of ID1 may initially delay (rather than promote) the myofibroblast/smooth muscle cell differentiation program initiated in response to TGF-β1. Although, a previous study did not demonstrate a role for ID proteins in smooth muscle cell differentiation and maturation, 74 the regulation of smooth muscle genes has been shown to be highly cell type- and smooth muscle subtype-specific. 71,75 The idea that ID1 may play a role in regulating smooth muscle gene expression is further supported by reports that a number of myofibroblast/smooth muscle cell phenotypic marker genes that were up-regulated in response to TGF-β1 in this study, including the genes encoding smooth muscle α-actin, smooth muscle γ-actin, transgelin (SM22), and smooth muscle myosin heavy chain contain E box motifs within transcriptionally active regions. 76-79 The importance of E-box/bHLH transcriptional regulation of smooth muscle-specific genes is further strengthened by in vitro evidence that a bHLH protein, upstream stimulatory factor (USF), has been shown to be involved in E-box-dependent activation of the smooth muscle α-actin gene. 80 Moreover, E-boxes have recently also been shown to be involved in the control of smooth muscle myosin heavy chain gene expression in vivo. 75 In our experiments, the majority of myofibroblast/smooth muscle cell phenotypic marker genes were not induced until ID1 gene and protein expression had returned to baseline values. It is therefore tempting to speculate that the induction of ID1 may serve to coordinate and regulate the myofibroblast/smooth muscle cell differentiation program initiated by TGF-β1 by controlling the activities of bHLH transcriptional activators, both temporally and qualitatively. However, our microarray data suggests that the fetal fibroblasts used in this study do not express myogenic factors or other bHLH proteins known to be involved in E-box-dependent gene activation, at baseline or on addition of TGF-β. This does not however exclude the possibility that ID1 may control myofibroblast differentiation directly or indirectly by associating with an as yet undefined bHLH transcription factor (which was absent on the HuGeneFL array used). Of interest, our microarray data also showed that TGF-β1 induced the early and transient expression of HAIRY (HRY, L19314). HAIRY-related proteins are a distinct subfamily of bHLH proteins that also generally function as DNA-binding transcriptional repressors but act in opposition to bHLH transcriptional activators rather than interfering with activator proteins. 81 HAIRY has also been shown to inhibit skeletal tissue myogenesis and may therefore similarly play a role in regulating fibroblast differentiation in response to TGF-β1.
In addition to its role in regulating mesenchymal cell differentiation, there is good evidence that ID1 can promote G1 cell-cycle progression 43 and oppose cellular senescence. 82 Although, TGF-β1 is not mitogenic at the dose and conditions of cellular confluence used in this study, our microarray data revealed that TGF-β1 up-regulated a number of genes associated with cell survival (DAD-1, PIM-1, and IER3). This is in accord with recent reports that TGF-β1 is anti-apoptotic for myofibroblasts, 32 raising the possibility that ID1 may also have a role in controlling cell survival in response to TGF-β1.
ID1 Is Expressed by Myofibroblasts in Vivo
In this study we also examined the immunolocalization of ID1 in a rat model of bleomycin-induced lung fibrosis, in which there is good evidence that both TGF-β1 and myofibroblasts play a major role in promoting excessive deposition of matrix proteins within the lung parenchyma. 12 Sections were examined on day 14 after intratracheal instillation of bleomycin, when the number of myofibroblasts expressing procollagen α1(I) mRNA within fibrotic foci is maximal 83 and total lung collagen content is close to double that of control lungs. 22 In the normal rat lung, ID1 expression was confined to airway and vascular smooth muscle cells. In contrast, in the lungs of bleomycin-instilled rats, ID1 was also highly expressed by (myo)fibroblasts within multiple fibrotic foci. This is to our knowledge the first evidence that (myo)fibroblasts express this transcriptional regulator within active fibrotic lesions and further supports the idea that ID1 may play a role in regulating fibroblast transcriptional responses during the progression to fibrosis in vivo.
Summary and Conclusions
Myofibroblasts play an essential role in organogenesis and normal tissue repair. However, inappropriate control of myofibroblast function contributes to oncogenesis, inflammation, and tissue fibrosis, so that the identification of genes involved in regulating fibroblast differentiation is of primary importance. The ability of TGF-β1 to promote fibroblast to myofibroblast transformation is well recognized. In this report, we provide evidence that TGF-β1 may be capable of promoting fibroblast differentiation beyond the myofibroblast stage toward cells expressing a number of late smooth muscle cell differentiation marker proteins. Our data may be of particular interest in light of the recent finding that global expression profiling of human pulmonary fibrosis [or more precisely usual interstitial pneumonia (UIP)] revealed that the genes encoding smooth muscle myosin heavy chain and basic calponin were among some of the most highly up-regulated in the lungs of these patients. 84 The finding that TGF-β1 induces the transient expression of ID1 and that this transcriptional regulator is also expressed by (myo)fibroblasts within active fibrotic lesions further raises the interesting possibility that ID1 may regulate fibroblast transcriptional reprogramming during fibrogenesis in vivo. The precise role of ID1 in coordinating fibroblast responses to TGF-β may be revealed by performing similar global expression profiling studies using ID1 blocking strategies in conjunction with ID1 overexpression studies. The results of these studies should provide valuable information on the target genes under ID1 regulatory control and on the role of ID proteins in regulating fibroblast transcriptional responses to one of their most potent activators.
Acknowledgments
We thank Dr. David Howell and Mr. Steve Bottoms for excellent assistance with bleomycin studies, and Dr. Robin McAnulty (University College London) for helpful discussions. We are also grateful to Dr. Hal Van Wart and Dr. Robert Booth (Roche Bioscience, Palo Alto, CA) for their enthusiastic support of this project, our collaborators for providing essential research material, and to the staff at Media Resources for assistance with the preparation of figures.
Footnotes
Address reprint requests to Rachel C. Chambers, Centre for Respiratory Research, University College London, Rayne Institute, 5 University St., London WC1E 6JJ, UK. E-mail: r.chambers@ucl.ac.uk.
Supported by the Wellcome Trust (postdoctoral fellowship ref 040921 to R. C. C. and program grant no. 051154 to G. J. L.).
G. J. L. and R. A. H. contributed equally to this work.
References
- 1.Massague J, Blain SW, Lo RS: TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 2000, 103:295-309 [DOI] [PubMed] [Google Scholar]
- 2.Blobe GC, Schiemann WP, Lodish HF: Role of transforming growth factor beta in human disease. N Engl J Med 2000, 342:1350-1358 [DOI] [PubMed] [Google Scholar]
- 3.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J: Mechanism of activation of the TGF-beta receptor. Nature 1994, 370:341-347 [DOI] [PubMed] [Google Scholar]
- 4.Heldin CH, Miyazono K, ten Dijke P: TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 1997, 390:465-471 [DOI] [PubMed] [Google Scholar]
- 5.Massague J: TGF-beta signal transduction. Annu Rev Biochem 1998, 67:753-791 [DOI] [PubMed] [Google Scholar]
- 6.McAnulty RJ, Laurent GJ: Pathogenesis of lung fibrosis and potential new therapeutic strategies. Exp Nephrol 1995, 3:96-107 [PubMed] [Google Scholar]
- 7.McAnulty RJ, Hernandez-Rodriguez NA, Mutsaers SE, Coker RK, Laurent GJ: Indomethacin suppresses the anti-proliferative effects of transforming growth factor-beta isoforms on fibroblast cell cultures. Biochem J 1997, 321:639-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serini G, Gabbiani G: Mechanisms of myofibroblast activity and phenotypic modulation. Exp Cell Res 1999, 250:273-283 [DOI] [PubMed] [Google Scholar]
- 9.Sime PJ, Xing Z, Graham FL, Csaky KG, Gauldie J: Adenovector-mediated gene transfer of active transforming growth factor-beta1 induces prolonged severe fibrosis in rat lung. J Clin Invest 1997, 100:768-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopp JB, Factor VM, Mozes M, Nagy P, Sanderson N, Bottinger EP, Klotman PE, Thorgeirsson SS: Transgenic mice with increased plasma levels of TGF-beta 1 develop progressive renal disease. Lab Invest 1996, 74:991-1003 [PubMed] [Google Scholar]
- 11.Sanderson N, Factor V, Nagy P, Kopp J, Kondaiah P, Wakefield L, Roberts AB, Sporn MB, Thorgeirsson SS: Hepatic expression of mature transforming growth factor beta 1 in transgenic mice results in multiple tissue lesions. Proc Natl Acad Sci USA 1995, 92:2572-2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giri SN, Hyde DM, Hollinger MA: Effect of antibody to transforming growth factor beta on bleomycin induced accumulation of lung collagen in mice. Thorax 1993, 48:959-966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormick LL, Zhang Y, Tootell E, Gilliam AC: Anti-TGF-beta treatment prevents skin and lung fibrosis in murine sclerodermatous graft-versus-host disease: a model for human scleroderma. J Immunol 1999, 163:5693-5699 [PubMed] [Google Scholar]
- 14.Nakamura T, Sakata R, Ueno T, Sata M, Ueno H: Inhibition of transforming growth factor beta prevents progression of liver fibrosis and enhances hepatocyte regeneration in dimethylnitrosamine-treated rats. Hepatology 2000, 32:247-255 [DOI] [PubMed] [Google Scholar]
- 15.Verrecchia F, Chu ML, Mauviel A: Identification of novel TGF-beta/Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J Biol Chem 2001, 276:17058-17062 [DOI] [PubMed] [Google Scholar]
- 16.Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ: Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol 1997, 15:1359-1367 [DOI] [PubMed] [Google Scholar]
- 17.Der SD, Zhou A, Williams BR, Silverman RH: Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA 1998, 95:15623-15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminski N, Allard JD, Pittet JF, Zuo F, Griffiths MJ, Morris D, Huang X, Sheppard D, Heller RA: Global analysis of gene expression in pulmonary fibrosis reveals distinct programs regulating lung inflammation and fibrosis. Proc Natl Acad Sci USA 2000, 97:1778-1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998, 95:14863-14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sablitzky F, Moore A, Bromley M, Deed RW, Newton JS, Norton JD: Stage- and subcellular-specific expression of Id proteins in male germ and Sertoli cells implicates distinctive regulatory roles for Id proteins during meiosis, spermatogenesis, and Sertoli cell function. Cell Growth Differ 1998, 9:1015-1024 [PubMed] [Google Scholar]
- 21.Mutsaers SE, Foster ML, Chambers RC, Laurent GJ, McAnulty RJ: Increased endothelin-1 and its localization during the development of bleomycin-induced pulmonary fibrosis in rats. Am J Respir Cell Mol Biol 1998, 18:611-619 [DOI] [PubMed] [Google Scholar]
- 22.Howell DC, Goldsack NR, Marshall RP, McAnulty RJ, Starke R, Purdy G, Laurent GJ, Chambers RC: Direct thrombin inhibition reduces lung collagen, accumulation, and connective tissue growth factor mRNA levels in bleomycin-induced pulmonary fibrosis. Am J Pathol 2001, 159:1383-1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P: Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 1997, 389:631-635 [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Rovira T, Chalaux E, Rosa JL, Bartrons R, Ventura F: Interaction and functional cooperation of NF-kappa B with Smads. Transcriptional regulation of the junB promoter. J Biol Chem 2000, 275:28937-28946 [DOI] [PubMed] [Google Scholar]
- 25.McAnulty RJ, Chambers RC, Laurent GJ: Regulation of fibroblast procollagen production. Transforming growth factor-beta 1 induces prostaglandin E2 but not procollagen synthesis via a pertussis toxin-sensitive G-protein. Biochem J 1995, 307:63-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atfi A, Djelloul S, Chastre E, Davis R, Gespach C: Evidence for a role of Rho-like GTPases and stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) in transforming growth factor beta-mediated signaling. J Biol Chem 1997, 272:1429-1432 [DOI] [PubMed] [Google Scholar]
- 27.Nishita M, Hashimoto MK, Ogata S, Laurent MN, Ueno N, Shibuya H, Cho KW: Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann’s organizer. Nature 2000, 403:781-785 [DOI] [PubMed] [Google Scholar]
- 28.Kothapalli D, Frazier KS, Welply A, Segarini PR, Grotendorst GR: Transforming growth factor beta induces anchorage-independent growth of NRK fibroblasts via a connective tissue growth factor-dependent signaling pathway. Cell Growth Differ 1997, 8:61-68 [PubMed] [Google Scholar]
- 29.Zumbrunn J, Trueb B: Primary structure of a putative serine protease specific for IGF-binding proteins. FEBS Lett 1996, 398:187-192 [DOI] [PubMed] [Google Scholar]
- 30.Kumar R, Kobayashi T, Warner GM, Wu Y, Salisbury JL, Lingle W, Pittelkow MR: A novel immediate early response gene, IEX-1, is induced by ultraviolet radiation in human keratinocytes. Biochem Biophys Res Commun 1998, 253:336-341 [DOI] [PubMed] [Google Scholar]
- 31.Lilly M, Sandholm J, Cooper JJ, Koskinen PJ, Kraft A: The PIM-1 serine kinase prolongs survival and inhibits apoptosis-related mitochondrial dysfunction in part through a bcl-2-dependent pathway. Oncogene 1999, 18:4022-4031 [DOI] [PubMed] [Google Scholar]
- 32.Zhang HY, Phan SH: Inhibition of myofibroblast apoptosis by transforming growth factor beta(1). Am J Respir Cell Mol Biol 1999, 21:658-665 [DOI] [PubMed] [Google Scholar]
- 33.Chen SJ, Yuan W, Mori Y, Levenson A, Trojanowska M, Varga J: Stimulation of type I collagen transcription in human skin fibroblasts by TGF-beta: involvement of Smad 3. J Invest Dermatol 1999, 112:49-57 [DOI] [PubMed] [Google Scholar]
- 34.Bellon G, Monboisse JC, Randoux A, Borel JP: Effects of preformed proline and proline amino acid precursors (including glutamine) on collagen synthesis in human fibroblast cultures. Biochim Biophys Acta 1987, 930:39-47 [DOI] [PubMed] [Google Scholar]
- 35.Keski-Oja J, Raghow R, Sawdey M, Loskutoff DJ, Postlethwaite AE, Kang AH, Moses HL: Regulation of mRNAs for type-1 plasminogen activator inhibitor, fibronectin, and type I procollagen by transforming growth factor-beta. Divergent responses in lung fibroblasts and carcinoma cells. J Biol Chem 1988, 263:3111-3115 [PubMed] [Google Scholar]
- 36.Denger S, Jahn L, Wende P, Watson L, Gerber SH, Kubler W, Kreuzer J: Expression of monocyte chemoattractant protein-1 cDNA in vascular smooth muscle cells: induction of the synthetic phenotype: a possible clue to SMC differentiation in the process of atherogenesis. Atherosclerosis 1999, 144:15-23 [DOI] [PubMed] [Google Scholar]
- 37.Jain MK, Fujita KP, Hsieh CM, Endege WO, Sibinga NE, Yet SF, Kashiki S, Lee WS, Perrella MA, Haber E, Lee ME: Molecular cloning and characterization of SmLIM, a developmentally regulated LIM protein preferentially expressed in aortic smooth muscle cells. J Biol Chem 1996, 271:10194-10199 [DOI] [PubMed] [Google Scholar]
- 38.Hautmann MB, Adam PJ, Owens GK: Similarities and differences in smooth muscle alpha-actin induction by TGF-beta in smooth muscle versus non-smooth muscle cells. Arterioscler Thromb Vasc Biol 1999, 19:2049-2058 [DOI] [PubMed] [Google Scholar]
- 39.van der Loop FT, Schaart G, Timmer ED, Ramaekers FC, van Eys GJ: Smoothelin, a novel cytoskeletal protein specific for smooth muscle cells. J Cell Biol 1996, 134:401-411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H: The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 1990, 61:49-59 [DOI] [PubMed] [Google Scholar]
- 41.Norton JD: ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci 2000, 113:3897-3905 [DOI] [PubMed] [Google Scholar]
- 42.Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D: An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci USA 1991, 88:1815-1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hara E, Uzman JA, Dimri GP, Nehlin JO, Testori A, Campisi J: The helix-loop-helix protein Id-1 and a retinoblastoma protein binding mutant of SV40 T antigen synergize to reactivate DNA synthesis in senescent human fibroblasts. Dev Genet 1996, 18:161-172 [DOI] [PubMed] [Google Scholar]
- 44.Belletti B, Drakas R, Morrione A, Tu X, Prisco M, Yuan T, Casaburi I, Baserga R: Regulation of Id1 protein expression in mouse embryo fibroblasts by the type 1 insulin-like growth factor receptor. Exp Cell Res 2002, 277:107-118 [DOI] [PubMed] [Google Scholar]
- 45.Ogata T, Wozney JM, Benezra R, Noda M: Bone morphogenetic protein 2 transiently enhances expression of a gene, Id (inhibitor of differentiation), encoding a helix-loop-helix molecule in osteoblast-like cells. Proc Natl Acad Sci USA 1993, 90:9219-9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A: Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem 1999, 274:19838-19845 [DOI] [PubMed] [Google Scholar]
- 47.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T: Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol 1994, 127:1755-1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P: Balancing the activation state of the endothelium via two distinct TGF-beta type I receptors. EMBO J 2002, 21:1743-1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez-Rovira T, Chalaux E, Massague J, Rosa JL, Ventura F: Direct binding of Smad1 and Smad4 to two distinct motifs mediates BMP-specific transcriptional activation of Id1 gene. J Biol Chem 2001, 277:3176-3185. [DOI] [PubMed] [Google Scholar]
- 50.Norton JD, Deed RW, Craggs G, Sablitzky F: Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol 1998, 8:58-65 [PubMed] [Google Scholar]
- 51.van der Loop FT, Gabbiani G, Kohnen G, Ramaekers FC, van Eys GJ: Differentiation of smooth muscle cells in human blood vessels as defined by smoothelin, a novel marker for the contractile phenotype. Arterioscler Thromb Vasc Biol 1997, 17:665-671 [DOI] [PubMed] [Google Scholar]
- 52.Christen T, Bochaton-Piallat ML, Neuville P, Rensen S, Redard M, van Eys G, Gabbiani G: Cultured porcine coronary artery smooth muscle cells. A new model with advanced differentiation. Circ Res 1999, 85:99-107 [DOI] [PubMed] [Google Scholar]
- 53.Gabbiani G: Some historical and philosophical reflections on the myofibroblast concept. Curr Top Pathol 1999, 93:1-5 [DOI] [PubMed] [Google Scholar]
- 54.Phan SH, Zhang K, Zhang HY, Gharaee-Kermani M: The myofibroblast as an inflammatory cell in pulmonary fibrosis. Curr Top Pathol 1999, 93:173-182 [DOI] [PubMed] [Google Scholar]
- 55.Nouchi T, Tanaka Y, Tsukada T, Sato C, Marumo F: Appearance of alpha-smooth-muscle-actin-positive cells in hepatic fibrosis. Liver 1991, 11:100-105 [DOI] [PubMed] [Google Scholar]
- 56.Zhang HY, Gharaee-Kermani M, Zhang K, Karmiol S, Phan SH: Lung fibroblast alpha-smooth muscle actin expression and contractile phenotype in bleomycin-induced pulmonary fibrosis. Am J Pathol 1996, 148:527-537 [PMC free article] [PubMed] [Google Scholar]
- 57.Cassiman D, Libbrecht L, Desmet V, Denef C, Roskams T: Hepatic stellate cell/myofibroblast subpopulations in fibrotic human and rat livers. J Hepatol 2002, 36:200-209 [DOI] [PubMed] [Google Scholar]
- 58.Yang J, Liu Y: Dissection of key events in tubular epithelial to myofibroblast transition and its implications in renal interstitial fibrosis. Am J Pathol 2001, 159:1465-1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Y, Weber KT: Angiotensin converting enzyme and myofibroblasts during tissue repair in the rat heart. J Mol Cell Cardiol 1996, 28:851-858 [DOI] [PubMed] [Google Scholar]
- 60.Chiavegato A, Bochaton-Piallat ML, D’Amore E, Sartore S, Gabbiani G: Expression of myosin heavy chain isoforms in mammary epithelial cells and in myofibroblasts from different fibrotic settings during neoplasia. Virchows Arch 1995, 426:77-86 [DOI] [PubMed] [Google Scholar]
- 61.Lazard D, Sastre X, Frid MG, Glukhova MA, Thiery JP, Koteliansky VE: Expression of smooth muscle-specific proteins in myoepithelium and stromal myofibroblasts of normal and malignant human breast tissue. Proc Natl Acad Sci USA 1993, 90:999-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roelofs M, Wein AJ, Monson FC, Passerini-Glazel G, Koteliansky VE, Sartore S, Levin RM: Contractility and phenotype transitions in serosal thickening of obstructed rabbit bladder. J Appl Physiol 1995, 78:1432-1441 [DOI] [PubMed] [Google Scholar]
- 63.Faggian L, Pampinella F, Roelofs M, Paulon T, Franch R, Chiavegato A, Sartore S: Phenotypic changes in the regenerating rabbit bladder muscle. Role of interstitial cells and innervation on smooth muscle cell differentiation. Histochem Cell Biol 1998, 109:25-39 [DOI] [PubMed] [Google Scholar]
- 64.Serini G, Gabbiani G: Mechanisms of myofibroblast activity and phenotypic modulation. Exp Cell Res 1999, 250:273-283 [DOI] [PubMed] [Google Scholar]
- 65.Grainger DJ, Metcalfe JC, Grace AA, Mosedale DE: Transforming growth factor-beta dynamically regulates vascular smooth muscle differentiation in vivo. J Cell Sci 1998, 111:2977-2988 [DOI] [PubMed] [Google Scholar]
- 66.Hirschi KK, Rohovsky SA, D’Amore PA: PDGF, TGF-beta, and heterotypic cell-cell interactions mediate endothelial cell-induced recruitment of 10T1/2 cells and their differentiation to a smooth muscle fate. J Cell Biol 1998, 141:805-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shah NM, Groves AK, Anderson DJ: Alternative neural crest cell fates are instructively promoted by TGFbeta superfamily members. Cell 1996, 85:331-343 [DOI] [PubMed] [Google Scholar]
- 68.Fries KM, Blieden T, Looney RJ, Sempowski GD, Silvera MR, Willis RA, Phipps RP: Evidence of fibroblast heterogeneity and the role of fibroblast subpopulations in fibrosis. Clin Immunol Immunopathol 1994, 72:283-292 [DOI] [PubMed] [Google Scholar]
- 69.Adam PJ, Regan CP, Hautmann MB, Owens GK: Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J Biol Chem 2000, 275:37798-37806 [DOI] [PubMed] [Google Scholar]
- 70.Hautmann MB, Madsen CS, Owens GK: A transforming growth factor beta (TGFbeta) control element drives TGFbeta-induced stimulation of smooth muscle alpha-actin gene expression in concert with two CArG elements. J Biol Chem 1997, 272:10948-10956 [DOI] [PubMed] [Google Scholar]
- 71.Roy SG, Nozaki Y, Phan SH: Regulation of alpha-smooth muscle actin gene expression in myofibroblast differentiation from rat lung fibroblasts. Int J Biochem Cell Biol 2001, 33:723-734 [DOI] [PubMed] [Google Scholar]
- 72.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H: The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 1990, 61:49-59 [DOI] [PubMed] [Google Scholar]
- 73.Sun XH, Copeland NG, Jenkins NA, Baltimore D: Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol 1991, 11:5603-5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kemp PR, Metcalfe JC, Grainger DJ: ID—a dominant negative regulator of skeletal muscle differentiation—is not involved in maturation or differentiation of vascular smooth muscle cells. FEBS Lett 1995, 368:81-86 [DOI] [PubMed] [Google Scholar]
- 75.Manabe I, Owens GK: The smooth muscle myosin heavy chain gene exhibits smooth muscle subtype-selective modular regulation in vivo. J Biol Chem 2001, 276:39076-39087 [DOI] [PubMed] [Google Scholar]
- 76.Jung F, Johnson AD, Kumar MS, Wei B, Hautmann M, Owens GK, McNamara C: Characterization of an E-box-dependent cis element in the smooth muscle alpha-actin promoter. Arterioscler Thromb Vasc Biol 1999, 19:2591-2599 [DOI] [PubMed] [Google Scholar]
- 77.Kovacs AM, Zimmer WE: Cell-specific transcription of the smooth muscle gamma-actin gene requires both positive- and negative-acting cis elements. Gene Expr 1998, 7:115-129 [PMC free article] [PubMed] [Google Scholar]
- 78.Osbourn JK, Weissberg PL, Shanahan CM: A regulatory element downstream of the rat SM22 alpha gene transcription start point enhances reporter gene expression in vascular smooth muscle cells. Gene 1995, 154:249-253 [DOI] [PubMed] [Google Scholar]
- 79.Katoh Y, Loukianov E, Kopras E, Zilberman A, Periasamy M: Identification of functional promoter elements in the rabbit smooth muscle myosin heavy chain gene. J Biol Chem 1994, 269:30538-30545 [PubMed] [Google Scholar]
- 80.Johnson AD, Owens GK: Differential activation of the SMalphaA promoter in smooth vs. skeletal muscle cells by bHLH factors. Am J Physiol 1999, 276:C1420-C1431 [DOI] [PubMed] [Google Scholar]
- 81.Fisher AL, Ohsako S, Caudy M: The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol 1996, 16:2670-2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alani RM, Young AZ, Shifflett CB: Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc Natl Acad Sci USA 2001, 98:7812-7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang K, Rekhter MD, Gordon D, Phan SH: Myofibroblasts and their role in lung collagen gene expression during pulmonary fibrosis. A combined immunohistochemical and in situ hybridization study. Am J Pathol 1994, 145:114-125 [PMC free article] [PubMed] [Google Scholar]
- 84.Zuo F, Kaminski N, Eugui E, Allard J, Yakhini Z, Ben Dor A, Lollini L, Morris D, Kim Y, DeLustro B, Sheppard D, Pardo A, Selman M, Heller RA: Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci USA 2002, 99:6292-6297 [DOI] [PMC free article] [PubMed] [Google Scholar]