Abstract
In situ hybridization is one of the most important techniques to visualize gene expression at the cellular level in various tissues. The in situ hybridization-AT tailing (ISH-AT) method uses a specially designed and synthesized oligonucleotide probe that has (AT)10 on the 3′ side. This (AT)10 of the probe is elongated by ΔTth DNA polymerase in the presence of dATP, dTTP, and labeled dUTP in the tissue after hybridization. Through this process the target is labeled with many hapten molecules. In this study, we detected human immunodeficiency virus type 1 RNA in formalin-fixed and paraffin-embedded tissues obtained from autopsied patients with acquired immunodeficiency syndrome by combining ISH-AT with the catalyzed signal amplification (CSA) system (ISH-AT-CSA), although we failed to detect signals from the same samples by conventional in situ hybridization using RNA probes (RISH) with CSA (RISH-CSA). We demonstrated that the ISH-AT-CSA method was superior to RISH-CSA in terms of both sensitivity and specificity, and that it was applicable to fluorescence in situ hybridization and double staining with immunohistochemistry for the characterization of cell phenotypes.
In the postgenomic era, it will be crucial to image both gene and protein expressions at the cellular level in a variety of tissues. In situ hybridization and immunohistochemistry are to be the important techniques to achieve this. Several methods of in situ hybridization have been developed, and its sensitivity and specificity of the signal-to-noise (S/N) ratio, preservation of tissue morphology, and preparation of probes have been improved. 1-7
RNA probes have generally been used as the most sensitive probe and are considered suitable for the detection of very low amounts of transcripts. Compared to oligonucleotide probes, however, their specificity, signal-to-noise (S/N) ratio, and penetration into cells and tissues are inferior and probe preparation is more complicated. The use of small-sized probes such as oligonucleotide probes reduces noise signals originating from nonspecifically bound probes. Moreover, it is noted that we are able to prepare the oligonucleotide probe without the cloning steps for construction of plasmid DNAs.
We developed a new type of probe, hybridization AT tailing (HybrAT) probe for filter hybridization, and a new in situ hybridization technique, ISH-AT, as described previously. 8,9 A HybrAT probe is an oligonucleotide probe consisting of a hybridization region and an AT tailing region, (AT)10, on the 3′ end. It should be noted that the probe was not labeled beforehand; however, after hybridization by incubation at a constant temperature (60°C) with a labeling solution consisting of dATP, dTTP, labeled dUTP (eg, biotin-16-dUTP, or digoxigenin-11-dUTP), and ΔTth DNA polymerase (Toyobo, Osaka, Japan), which mediated a rapid elongation of oligo-(AT)10 into high-molecular weight (AT)x copolymers with incorporation of labeled dUTP. 10 Furthermore, ISH-AT allowed detection of simian immunodeficiency virus (SIV) RNA in formalin-fixed and paraffin-embedded tissue sections of an experimentally infected monkey. 9 The sensitivity of this technique was at least comparable to that of in situ hybridization using RNA probes (RISH). 9
Recently, catalyzed signal amplification (CSA) or tyramide signal amplification systems have been introduced to amplify the signal detection by immunohistochemistry and in situ hybridization. 1,2,4,7,11-17 In this study, we attempted to combine ISH-AT with CSA and succeeded in detecting HIV-1 RNA with a high signal-to-noise (S/N) ratio in sections of formalin-fixed and paraffin-embedded autopsy tissues. We failed to detect any signals in the same samples by combining a conventional RISH with CSA (RISH-CSA). This ISH-AT-CSA allowed detection of the signals using both chromogen for conventional histopathology and fluorescence for confocal laser microscopy to characterize the phenotype of the cells.
Materials and Methods
Tissue Samples
The formalin-fixed and paraffin-embedded autopsy tissues selected in this study are summarized in Table 1 ▶ . M1, a rhesus monkey (Macaca mulatta) inoculated intravenously with 10 TCID50 of SHIV89.6PD (the gag, pol, nef regions of the gene are derived from SIV), showed CD4 + T cell depletion and high-plasma viral load, and was autopsied 8 months after infection. 18 M2 was inoculated with SIVmac32H 19 and was autopsied 1 year and 3 months after infection. They were serologically confirmed to be negative for SIV, simian type-D retrovirus, and simian T-cell leukemia virus type I before use and maintained in accordance with the institutional guidelines for laboratory animals. The lymph nodes were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) and brains were fixed in 10% buffered formalin.
Table 1.
Summary of Samples and Results of Immunohistochemistry, ISH-AT-CSA, and RISH-CSA
| Case | Tissue | Virus | Age* (sex) | SIVp27/HIVp24 | ISH-AT-CSA | RISH-CSA |
|---|---|---|---|---|---|---|
| Monkey | ||||||
| M1 | LN | SHIV89.6PD† | 8 months | + | +++ | + |
| Brain | + | +++ | ++ | |||
| M2 | Brain | SIVmac32H‡ | 1 year 3 months | + | +++ | ++ |
| Human | ||||||
| H1 | LN | HIV-1 | 6 months (M) | + | ++ | − |
| H2 | LN | HIV-1 | 6 months (M) | − | ++ | − |
| H3 | LN | HIV-1 | 7 months (M) | + | + | − |
| H4 | LN | HIV-1 | 9 months (M) | + | + | − |
| H5 | LN | HIV-1 | 1 year (M) | − | + | − |
| H6 | LN | HIV-1 | 1 year (F) | + | + | − |
| H7 | LN | HIV-1 | 1 year (M) | + | +++ | − |
| H8 | LN | HIV-1 | 1 year 1 month (F) | + | + | − |
| H9 | LN | HIV-1 | 1 year 2 months (M) | + | +++ | − |
| H10 | LN | HIV-1 | 1 year 4 months (M) | + | + | − |
| H11 | LN | HIV-1 | 1 year 9 months (M) | + | ++ | − |
| H12 | LN | HIV-1 | 2 years 1 month (F) | + | + | − |
| H13 | LN | HIV-1 | 2 years 4 months (F) | + | + | − |
| H14 | LN | HIV-1 | 7 years 4 months (M) | + | − | ND |
| H15 | LN | HIV-1 | 10 years (M) | − | − | ND |
| H16 | Brain | HIV-1 | 9 years (M) | + | ++ | − |
| H17 | Brain | HIV-1 | 33 years (M) | + | +++ | − |
| H18 | Brain | HIV-1 | 42 years (M) | + | + | − |
*Time after inoculation in cases of monkey.
†GenBank; U89134.
‡GenBank; D01065.
LN, lymph node; ND, not done.
The signal intensity is presented as +++, strong; ++, moderate; +, mild; −, no signal.
For detection of HIV-1 RNA, paraffin blocks of lymph node specimens (H1 to H15) were obtained from Romanian children who had died of pediatric acquired immunodeficiency syndrome (AIDS)-related illnesses. The brains (H16 to H18) were obtained from a Romanian child and Japanese adults who died of HIV-1 encephalitis.
HybrAT Probes
The HybrAT probe consists of two regions, hybridization and elongation regions (Figure 1) ▶ . The hybridization region, a 40- to 50-base-long complementary sequence, is located on the 5′ side, and the AT tailing region for elongation, consisting of 10 repeats of AT, on the 3′ side. 8,9 The predicted Tm value of the hybridization sequence should be higher than the temperature for the elongation reaction (60°C) so that the probe will firmly bind to the target sequence during the elongation process.
Figure 1.
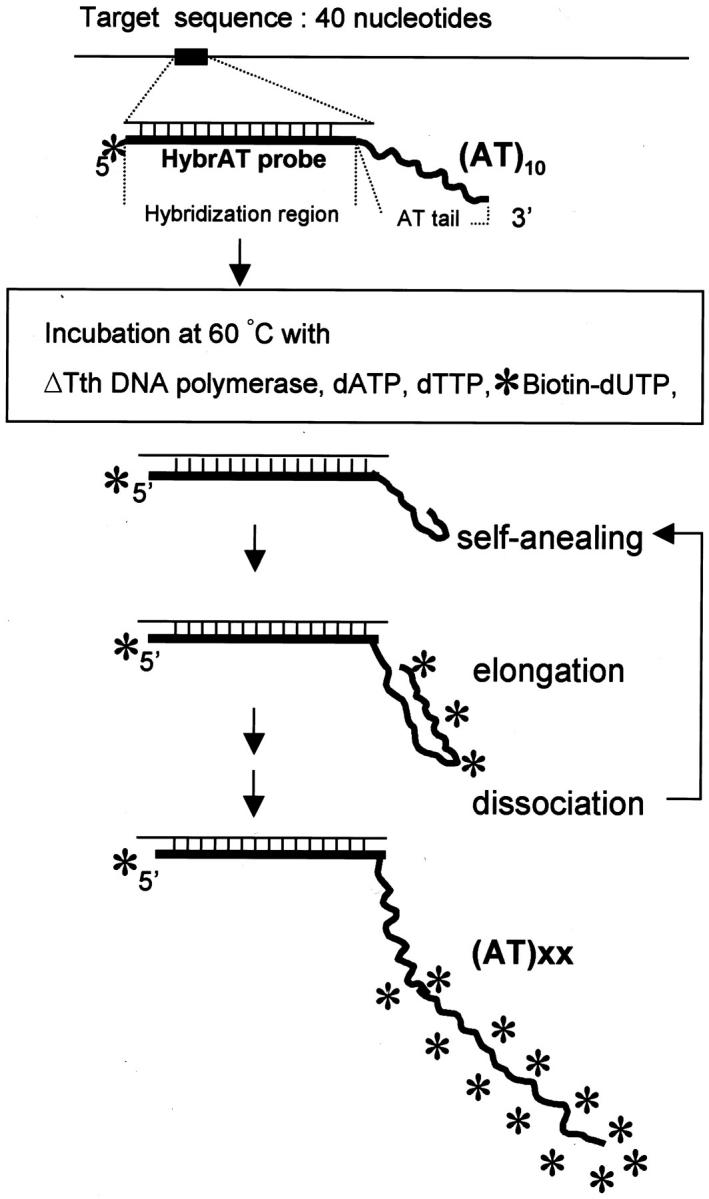
Schematic representation of ISH-AT. *, Biotin.
The sequences and positions of all oligonucleotides used in this study are presented in Table 2 ▶ . Each sequence of the hybridization region was checked for absence of cross homologies with human and simian sequences. One biotin molecule was conjugated on the 5′ side of all probes for control experiments. The GC% of the hybridization region was 50 to 60%. All of the HybrAT probes were synthesized at Nihon Gene Research Laboratories, Inc. (Sendai, Japan) on order.
Table 2.
Synthetic Oligonucleotides Used for Hybridization AT Tailing (HybrAT) Probes
| Probe | Position | Sequence |
|---|---|---|
| AS-5′ biotin-SIVnef(AT)10 | 9380–9419* | 5′ biotin-CCCATAAGTCTCCCCACGCGCCCGCAAGAGTCTCTGTCGCATATATATATATATATATAT-3′ |
| S-5′ biotin-SIVnef(AT)10 | 9419–9380 | 5′ biotin-GCGACAGAGACTCTTGCGGGCGCGTGGGGAGACTTATGGGATATATATATATATATATAT-3′ |
| AS-5′ biotin-HIVnef(AT)10 | 8910–8871† | 5′ biotin-GCTCCATGTTTTTCCAGGTCTCGGGATGCTGCTCCCACCCCATATATATATATATATATAT-3′ |
| S-5′ biotin-HIVnef(AT)10 | 8871–8910 | 5′ biotin-GGGGTGGGAGCAGCATCCCGAGACCTGGAAAAACATGGAGCATATATATATATATATATAT-3′ |
| AS-5′ biotin-HIVgag1(AT)10 | 1484–1447 | 5′ biotin-CTTGGTTCCTCATCTGGCCTGGTGCAATAGGCCCTGCATATATATATATATATATAT-3′ |
| S-5′ biotin-HIVgag1(AT)10 | 1447–1484 | 5′ biotin-GCAGGGCCTATTGCACCAGGCCAGATGAGAGAACCAAGATATATATATATATATATAT-3′ |
| AS-5′ biotin-HIVgag2(AT)10 | 1654–1617 | 5′ biotin-GTCCTTGTCTTATGTCCAGAATGCTGGTAGGGCTATACATATATATATATATATATAT-3′ |
| S-5′ biotin-HIVgag2(AT)10 | 1617–1654 | 5′ biotin-GTATAGCCCTACCAGCATTCTGGACATAAGACAAGGACATATATATATATATATATAT-3′ |
| AS-5′ biotin-HIVgag3(AT)10 | 1864–1827 | 5′ biotin-TATGGCCGGGTCCTCCCACTCCCTGACATGCTGTCATCATATATATATATATATATAT-3′ |
| S-5′ biotin-HIVgag3(AT)10 | 1827–1864 | 5′ biotin-GATGACAGCATGTCAGGGAGTGGGAGGACCCGGCCATAATATATATATATATATATAT-3′ |
RNA Probes
For preparation of SIVgag RNA probes and SIVnef RNA probes, the gag gene fragment (GenBank no. M33262, 1309 to 2841; 1533 bp) and the nef gene fragment (GenBank no. M33262, 9333 to 10,124; 792 bp) from plasmid pSIVmac239 20 were cloned into the transcription vector pGEM-T (Promega, Madison, WI). For HIV-1 RNA probes, the gag gene fragment (GenBank no. M19921, 790 to 2292; 1503 bp) from plasmid pNL43-2 21 was cloned into pBluescript KS (−) (Stratagene, La Jolla, CA) and the nef gene fragment (GenBank no. M19921, 8787 to 9407; 620 bp) from pNL43-2 was cloned into pGEM-T (Promega). Sense or anti-sense biotin-labeled RNA probes were generated using T3 or T7 or SP6 RNA polymerase by an RNA labeling kit (Roche, Indianapolis, IN) according to the manufacturer’s instructions as described before. 9 Each of them was confirmed to detect the SIV or HIV-1 DNA with equal efficiency in filter hybridization (data not shown). The sequence homology between SIVmac239 20 and SHIV89.6PD 18 was 100% in the gag region and 99.7% in the nef region. That between SIVmac239 20 and SIVmac32H 19 was 98.7% in the gag region and 97.1% in the nef region.
Tissue Preparation and Pretreatment for in Situ Hybridization
Formalin-fixed and paraffin-embedded tissue blocks were cut in 4-μm-thick sections, and the sections were mounted on silane-coated clean glass slides. After deparaffinization in xylene and graded ethanol, the slides were immersed in diethylpyrocarbonate-treated double-distilled water. The sections were treated with a target retrieval solution (DAKO, Carpinteria, CA) at 95°C for 40 minutes, cooled to room temperature, and washed with diethylpyrocarbonate-treated double-distilled water. Then, they were treated with 0.1 to 1 μg/ml of proteinase K (DAKO) for 15 minutes at 37°C. The proteinase K was washed out and inactivated by immersing the sections twice in 0.2% glycine-0.1 mol/L Tris-HCl (pH 7.6) for 3 minutes and twice in diethylpyrocarbonate-treated double-distilled water for 3 minutes. Finally, the sections were immersed in 100% ethanol for 2 minutes and air-dried.
RNA-in Situ Hybridization Combined with CSA (RISH-CSA)
Deparaffinized sections were prehybridized in hybridization buffer at 37°C for 30 minutes and hybridized with anti-sense or sense RNA probes (a mixture of gag probe and nef probe) overnight at 50°C in 40 μl of hybridization buffer consisting of 50% formamide (WAKO, Tokyo, Japan); 3× standard saline citrate (SSC); 1× Denhardt’s solution (WAKO); 50 mmol/L Hepes, pH 7.0; 1 mmol/L ethylenediaminetetraacetic acid; and 500 μg/ml tRNA (Roche). After hybridization the sections were washed once in 0.1× SSC (1× SSC; 150 mmol/L NaCl, 15 mmol/L sodium citrate), followed by stringent washing in 0.01× SSC for 15 minutes at 55°C twice. Then the sections were treated with 0.3% H2O2/methanol for 30 minutes to block endogenous peroxidase activities and they were immersed in 25% Blockace (Snow Brand, Tokyo, Japan)/Tris-buffered saline [TBS; 100 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl] for 60 minutes at room temperature or overnight at 4°C.
For signal detection by the CSA method, the GenPoint system (DAKO) was used as follows. A 1000-fold diluted primary streptavidin-horseradish peroxidase (SA-HRP) solution was applied on each tissue section for 15 minutes at room temperature. The section was then washed three times with TBST [0.1 mol/L Tris-HCl (pH 7.5), 0.15 mol/L NaCl, 0.05% Tween 20] for 5 minutes. A solution of biotinyl tyramide was applied on the section, which was incubated for 15 minutes and then washed three times with TBST for 5 minutes. The secondary SA-HRP was applied on the section that was incubated for 15 minutes at room temperature and washed three times with TBST for 5 minutes. Signals were developed with 3,3′-diaminobenzidine tetrahydrochloride (DAB) (Dohjin, Kumamoto, Japan) chromogen solution, followed by hematoxylin counterstaining, and observation under a light microscope.
In Situ Hybridization AT Tailing Combined with CSA (ISH-AT-CSA)
ISH-AT procedures were performed as described previously. 9 Tissue sections were covered with 300 μl of hybridization buffer consisting of 10% formamide (WAKO), 2× SSC, 1× Denhardt’s solution (WAKO), 50 mmol/L NaH2PO4/Na2HPO4, 1 mmol/L ethylenediaminetetraacetic acid, and 500 μg/ml tRNA (Roche). Briefly, 2 vol of the oligonucleotide HybrAT probe (0.1 to 0.4 pmol/μl) and 1 vol of salmon sperm DNA (10 mg/ml, Sigma, St. Louis, MO) were mixed with 17 volumes of hybridization buffer and the mixture was heated at 95°C for 5 minutes. The solution in a volume of 40 μl was applied to the sections and covered with a coverglass. After hybridization at 50°C overnight in a moist chamber, the tissue was washed once in 0.1× SSC, followed by stringent washing in 0.01× SSC for 15 minutes at 55°C twice and rinsing in TBS. An appropriate size of Hybaid EasiSeal (Hybaid, Middlesex, UK) was attached to each slide so as to cover the section entirely, and the sections were saturated with ΔTth reaction buffer (10 mmol/L Tris-HCl, pH 8.9, 1.5 mmol/L MgCl2, 80 mmol/L KCl, 0.1% sodium deoxycholate, 0.1% Triton X-100, 0.5 mg/ml bovine serum albumin; Toyobo) at 60°C for 10 minutes. Then a HybrAT reaction mixture consisting of 200 μmol/L each of dATP and dTTP, 10 μmol/L of Bioin-16-dUTP (Roche), and 50 U of ΔTth DNA polymerase (Toyobo) in ΔTth reaction buffer was applied to the sections. The slides were then placed on a hot plate, OmniGene FlatBlock (Hybaid), and heated to 60°C for 30 minutes. The sections were washed with TBS and treated with 0.3% H2O2/methanol for 30 minutes. Then they were immersed in 25% Blockace/TBS for 60 minutes at room temperature or overnight at 4°C.
The procedure of signal detection by the CSA method was the same as described in the RISH-CSA method. For fluorescence detection, in place of the secondary SA-HRP, streptavidin-conjugated Alexa Fluor 488 (Molecular Probes, Eugene, OR) was applied on the tissue section for 30 minutes at 37°C. Alexa Fluor 488 dye was excited with a 488-nm line of the argon-krypton laser to exhibit green fluorescence. The sections were then washed with PBS, and covered with cover glasses after application of mounting medium for fluorescence with anti-fading reagents (Vectashild; Vector Laboratories, Burlingame, CA). Imaging was performed using a confocal microscope equipped with an argon-krypton laser (LSM-MicroSystem; Zeiss, Germany).
Negative controls included: 1) hybridization of lymph nodes and brain tissues from uninfected monkeys or humans, 2) hybridization with sense and irrelevant probes, 3) mock (no probe) hybridization, and 4) omission of ΔTth DNA polymerase. When there is too much target RNA or DNA in the section, omission of ΔTth DNA polymerase is unsuitable for negative control, because some signals might be obtained by only in situ hybridization using 5′ end-labeled HybrAT probe without a following elongation reaction by ΔTth DNA polymerase.
Immunohistochemical Detection of Viral Antigens in Tissues
After deparaffinization with xylene and ethanol, the tissue sections were rehydrated in distilled water and treated with 1 mmol/L ethylenediaminetetraacetic acid (pH 8.0) at 121°C for 10 minutes. After washing with PBS, they were immersed in 0.1 mol/L glycine-HCl buffer (pH 2.2) at room temperature for 90 minutes, washed again with PBS, and treated with 0.3% H2O2/methanol at room temperature for 30 minutes. After washing in PBS, the tissue sections were incubated in 5% normal goat serum at room temperature for 20 minutes. Excess solutions were discarded and anti-SIVgag p27 polyclonal antibody (1:3000 diluted in PBS) or anti-HIV-1gag p24 mouse monoclonal antibody (NU24, 1:4000 diluted in PBS) were applied on the sections at 4°C overnight. After washing three times with PBS, biotin-conjugated goat anti-mouse IgG (LSAB kit, DAKO) and subsequently SA-HRP (LSAB kit, DAKO) were applied on the sections. The reaction was visualized using DAB or Vector VIP (Vector Laboratories, Burlingame, CA) chromogen solution.
Double Staining by ISH-AT-CSA and Immunohistochemistry
Immunohistochemistry was applied in combination with ISH-AT-CSA to identify cells expressing viral RNA. Endogenous biotin and biotinyl tyramide after ISH-AT-CSA staining with DAB as a chromogen were blocked with avidin solution and biotin solution (Biotin Blocking System, DAKO) according to the manufacturer’s instructions. Then, the tissue sections were incubated with 5% normal goat serum, followed by incubation with each mouse monoclonal anti-human cell marker antibody at 4°C overnight. The mouse monoclonal antibodies used were CD45RO (UCHL-1, DAKO), CD3 (PC3/188A, DAKO), and CD8 (NCL-CD8-295; Novocastra, Newcastle, UK) for T cells, CD68 (HAM56 for monkey, PGM1 for human; DAKO) for microglial/macrophage cells, CD20 (L26, DAKO) for B cells and CD21 (IF8, DAKO) for follicular dendritic cells (FDCs). After washing three times with PBS, biotin-conjugated goat anti-mouse IgG (DAKO) and subsequently SA-HRP (DAKO) were applied on the sections. The reaction was visualized using Vector VIP as a chromogen, followed by 2% methyl green counterstaining. For fluorescence detection, streptavidin-conjugated Alexa Fluor 568 (Molecular Probes) was used in place of SA-HRP. Alexa Fluor 568 dye was excited with a 543-nm line of the argon-krypton laser and exhibited red fluorescence. The ISH-AT-CSA signals exhibited green fluorescence. The emission patterns of the two fluorescent labels were collected separately and the data were overlaid on a computer to generate two-color images.
Results
Detection of SHIV or SIV RNA in Formalin-Fixed and Paraffin-Embedded Tissues of SHIV- or SIV-Infected Monkey
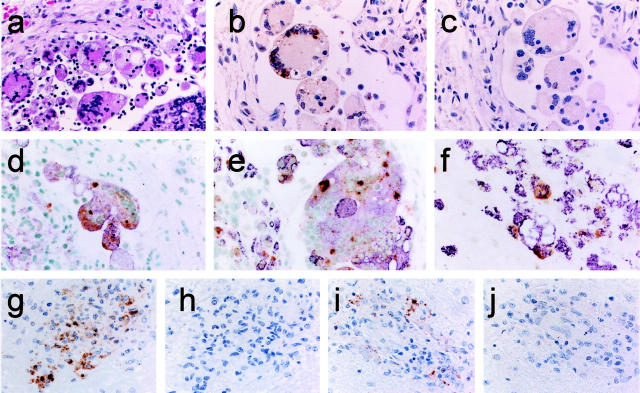
First, we attempted to detect viral RNA using the formalin-fixed and paraffin-embedded autopsy tissues from rhesus monkeys experimentally infected with SHIV89.6PD 18 or SIVmac32H 19 (Table 1 ▶ , M1 and M2). These specimens were fixed soon after death and a high copy number of viral RNAs were expected to remain in them. Histopathologically, the lymph node tissue of M1 was diagnosed as granulomatous lymphadenitis. 22,23 Epithelioid macrophages and multinucleated giant cells (MGCs) were found throughout the parenchyma, both within the subcapsular sinus and the cortex (Figure 2a) ▶ . Immunohistochemistry showed that the MGCs were CD68-positive macrophages, and SIVgag p27 antigen was detected in these MGCs. The ISH-AT-CSA signals were mainly detected over one or more nuclei and in the cytoplasm of MGCs, particularly in the subcapsular sinus using AS-5′ biotin-SIV nef (AT)10 (Table 2) ▶ (Figure 2b) ▶ . As the ISH-AT-CSA signals were not detected using sense HybrAT probes, S-5′ biotin-SIV nef (AT)10 (Figure 2c) ▶ , they specifically showed the plus strand SHIV genomic RNA or SHIV mRNA. They were not detected using unrelated AS-5′ biotin-HIV nef (AT)10 (data not shown). No signals were detected in the lymph nodes and brain tissues of monkeys without SHIV infection. When ΔTth DNA polymerase was omitted from the HybrAT mixture or the AT-tailing reaction step was omitted, the signals were also negative. The AT-tailing reaction was performed for only 5 minutes in the case of Figure 2, b and c ▶ . Double staining by ISH-AT-CSA and immunostaining of SIVgag p27 or CD68 antigen showed that almost all SHIV RNA-positive cells were identified as SIVgag p27-positive cells (Figure 2d) ▶ and CD68-positive cells (Figure 2e) ▶ .
Figure 2.
Localization of SHIV RNA by ISH-AT-CSA of lymph node sections (a–e) and a brain section (f) from SHIV-infected monkey (M1), and localization of SIV RNA by ISH-AT-CSA (g, h) or conventional RISH-CSA (i, j) of brain sections from SIV-infected monkey (M2). a: Granulomatous lymphadenitis (H&E). b: SHIV RNA detected by ISH-AT-CSA using anti-sense 5′ biotin-SIVnef(AT)10. c: No signals detected using sense 5′ biotin-SIVnef(AT)10. d: Double staining for SHIV RNA (brown signals) and SIVgag p27 antigen (purple). e and f: Double staining for SHIV RNA (ISH-AT-CSA, brown signals) and CD68 (immunostaining with HAM56, purple signals). g: SIV RNA detected by ISH-AT-CSA using anti-sense 5′ biotin-SIVnef(AT)10. h: No signals detected using sense 5′ biotin-SIVnef(AT)10. i: SIV RNA detected by RISH-CSA using a mixture of anti-sense SIVgag RNA probe and SIVnef RNA probe. j: No signals detected using a mixture of sense SIVgag RNA probe and SIVnef RNA probe. Original magnifications: ×200 (a–c, g–j), ×400 (d–f).
The monkeys, M1 and M2, showed giant cell encephalitis characterized by multifocal perivascular aggregates of macrophages and MGCs 24 in the brain tissues. The cells expressing SHIV RNA in the brain were found mainly in perivascular macrophages and MGCs identified as CD68-positive cells (Figure 2f) ▶ . The distribution of SIVgag p27-positive cells was nearly the same as that of ISH-AT-CSA-positive signals (data not shown).
These SIV RNA signals detected by the ISH-AT-CSA method (Figure 2g) ▶ were strong and widely distributed, but had a similar pattern to those detected by RISH-CSA using a mixture of anti-sense SIVgag RNA probe and SIVnef RNA probe (Figure 2i) ▶ . These results are summarized in Table 1 ▶ .
Detection of HIV-1 RNA in Formalin-Fixed and Paraffin-Embedded AIDS Autopsy Tissues
Next, we attempted to detect HIV-1 RNA or/and DNA in formalin-fixed and paraffin-embedded tissue sections obtained from autopsied AIDS patients using conventional RISH-CSA and ISH-AT-CSA. The paraffin block specimens of the lymph nodes were obtained from Romanian autopsied children who had died of pediatric AIDS between 1989 to 1990 (Table 1 ▶ , H1 to H15).
Histological findings of the lymph nodes of H9 in Table 1 ▶ showed the follicular involution stage (Figure 3, a and h) ▶ . Germinal centers (GCs) were not involuted completely but lymphoid cells were mostly depleted from the GC area.
Figure 3.
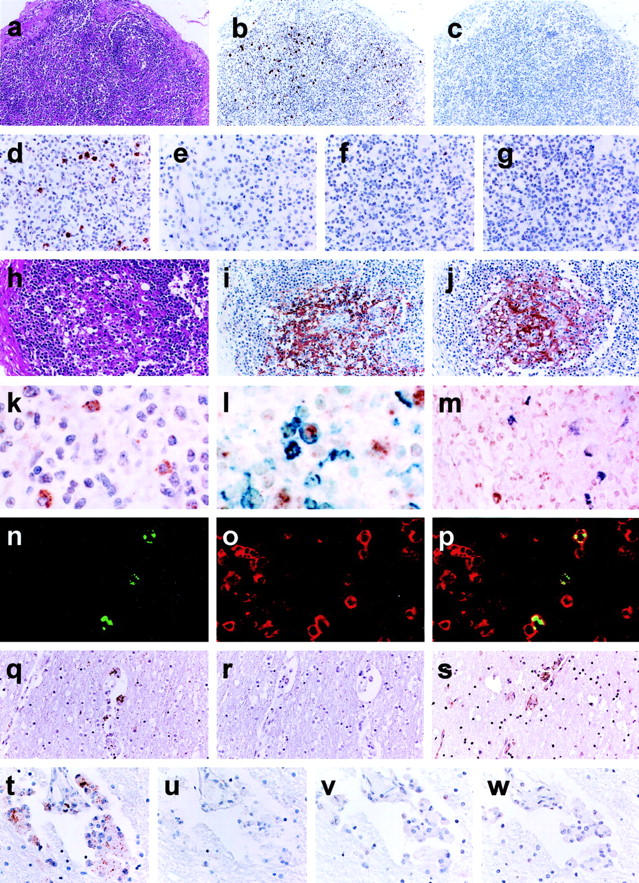
a–p: Lymph node sections from a pediatric AIDS case (H9). HIV-1 RNA detected by ISH-AT-CSA (b-e, k) or conventional RISH-CSA (f, g). a and h: The lymph node showed the follicular involution stage of AIDS (H&E). b, d, and k: The ISH-AT-CSA signals using the mixture of HIV-1 anti-sense HybrAT probes. c and e: No signals detected using the mixture of HIV-1 sense HybrAT probes. No signals were detected by RISH-CSA using either anti-sense (f) or sense (g) HIV-1 gag and HIV-1 nef RNA probes. i: Immunostaining of HIV-1 gag p24 antigen. j: Immunostaining of CD21 for FDCs. l: Double staining for HIV-1 RNA (brown signals) and CD45RO (blue signals). m: Double staining for HIV-1 RNA (brown signals) and CD68 (purple signals). n: Fluorescence ISH-AT-CSA signals (green). o: CD3 for T lymphocytes (red). p: Merge. q–w: Brain sections from an adult HIV-1 encephalitis case (H17). q and t: The ISH-AT-CSA signals using the mixture of HIV-1 anti-sense HybrAT probes. r and u: No signals were detected using the mixture of HIV-1 sense HybrAT probes. s: Immunostaining of HIV-1gag p24. v and w: No signals were detected by RISH-CSA using either an anti-sense (v) or sense (w) mixture of HIV-1gag RNA probe and HIV-1nef RNA probe. Original magnifications: ×100 (a–c, q–s); ×200 (d–j, t–w); ×400 (k–m); ×1000) (n–p).
Four HIV-1 anti-sense HybrAT probes [AS-5′ biotin-HIVnef(AT)10, AS-5′ biotin-HIVgag1(AT)10, AS-5′ biotin-HIVgag2(AT)10, and AS-5′ biotin-HIVgag3(AT)10] were synthesized (Table 2) ▶ . Each of them was confirmed to detect the HIV-1 DNA with equal efficiency in filter hybridization (data not shown). All probes showed similar distributions of ISH-AT-CSA signals and the signals obtained using AS-5′ biotin-HIVnef(AT)10 showed the strongest intensity (data not shown). The mixture of these four HIV-1 anti-sense HybrAT probes detected the ISH-AT-CSA signals more clearly over individual cells located mainly in the interfollicular area (Figure 3; b, d, and k) ▶ , but not in the GC area. No signals were detected when the mixture of HIV-1 sense HybrAT probes (Figure 3, c and e) ▶ or unrelated AS-5′ biotin-SIVnef(AT)10 was used. In addition, no signals were detected in other control tissues without HIV-1 infection. Furthermore, when ΔTth DNA polymerase or the HybrAT probe was omitted from the HybrAT mixture, the signals turned negative (data not shown). As the signals were not detected by senseHybrAT probes, they did not show HIV-1 DNA but did show the plus strand HIV-1 genomic RNA or HIV-1 mRNA, suggesting that the signals may correspond to replicating HIV-1. For comparison of the sensitivity, we also applied RISH-CSA method but were unable to detect any signals in the consecutive sections (Figure 3, f and g) ▶ .
In contrast, the HIV-1gag p24 antigen was detected exclusively within the GCs of the lymph nodes with a reticular staining pattern (Figure 3i) ▶ . Immunohistochemistry showed that the cells in the GCs were mainly CD21-positive FDCs (Figure 3j) ▶ . IgG and IgM antigens were also detected mainly in the same region with the same pattern as that of CD21 (data not shown), suggesting that the immune complex, which consisted of HIV-1gag p24 protein and immunoglobulin, precipitated at the surface of FDCs, but without replicating HIV-1. CD45RO- and CD68-positive cells were distributed in the interfollicular area of the lymph nodes, and CD20-positive cells were rather abundant in the cortex (data not shown).
The paraffin blocks of brain were obtained from AIDS autopsied patients who presented symptoms and signs of HIV-1 encephalitis (Table 1 ▶ , H16 to H18). Case H16 was a 9-year-old Romanian boy who died in 2001 and cases H17 and H18 were Japanese homosexual males who died in 1988 and 1992, respectively. Each brain section showed lesions characteristic of HIV-associated giant cell encephalitis in the white matter. The ISH-AT-CSA method was applied on consecutive tissue sections. The mixture of HIV-1 anti-sense HybrAT probes detected clearly the HIV-1 RNA signals in MGCs located in the perivascular area (Figure 3, q and t) ▶ but not the mixture of sense HybrAT probes (Figure 3, r and u) ▶ . Control studies similar to those described above were performed, and it was found that the signals were specific for HIV-1 RNA. The MGCs were also positive for HIV-1gag p24 antigen (Figure 3s) ▶ . In contrast, the RISH-CSA signals were not detected in the tissues of H16 to H18 (Figure 3, v and w) ▶ .
As summarized in Table 1 ▶ , these analyses were done with a total of 18 cases. At least in 16 AIDS autopsy cases, the ISH-AT-CSA method was superior in detection sensitivity to the conventional RISH-CSA using a mixture of HIV-1gag RNA probe and HIV-1nef RNA probes. In two cases (H2 and H5), ISH-AT-CSA signals were positive but HIV-1gag p24 antigen was not detected.
Identification of Infected Cells in Lymph Nodes by Double Staining
To analyze the phenotype of HIV-1-infected cells, immunohistochemistry was used in combination with the ISH-AT-CSA method. After HIV-1 RNA-positive cells were stained as brown signals by ISH-AT-CSA, they were further double stained as blue or purple signals using either anti-CD45RO (UCHL1) or anti-CD68 (PGM1) antibodies, respectively. There were several HIV-1 RNA-positive cells detected by ISH-AT-CSA, which were also stained by immunohistochemistry using the anti-CD45RO antibody (Figure 3l) ▶ , but not with the anti-CD68 antibody (Figure 3m) ▶ . To confirm these findings, ISH-AT-CSA signals were stained further with Alexa 488 (Figure 3n) ▶ and the cell surface antigens were stained with Alexa 568 (Figure 3o) ▶ for analysis by confocal laser microscopy. As shown in Figure 3p ▶ , the HIV-1 RNA-positive cells were identified as CD3-positive cells, but not as CD8- or CD68-positive cells (data not shown).
Discussion
For visualization of the localization of gene expression within a specific cell and tissue, in situ hybridization is an indispensable molecular technique. We previously developed the ISH-AT method and showed that the sensitivity of ISH-AT with alkaline phosphatase-based visualization was at least comparable to that of conventional in situ hybridization using an RNA probe (RISH). 9
The potential problem of nonspecific incorporation of a labeled nucleotide into fragmented endogenous DNA should be considered also in the ISH-AT method, although this problem is much less serious when compared with the in situ polymerase chain reaction method. 25-29 The reason for this is as follows. First, the activity of polymerization by ΔTth DNA polymerase is primer/template-dependent 10,30 and ΔTth DNA polymerase is exonuclease-free DNA polymerase. Second, the ΔTth DNA polymerase recognized the AT tailing region as a primer/template and elongated (AT)10 with dATPs, dTTPs, and labeled dUTPs (Figure 1) ▶ . So, only dATPs and dTTPs are used in the ISH-AT method except for labeled dUTPs. Third, the elongation reaction is performed at a constant temperature (60°C) without a denaturing step at 94°C. So the tissues are less damaged than those are in the in situ polymerase chain reaction method. 28 Finally, the labeled probe will firmly bind to the target sequence. Infact, the primer/template-independent polymerization by TaqDNA polymerase 30 and the signal diffusion observed in the in situ polymerase chain reaction method 28 were completely absent in the ISH-AT method. 9 To exclude false-positive signals, control experiments should be done at least with irrelevant probes and with omission of probes.
In this study, to improve the signal resolution and the detection sensitivity of the ISH-AT method, we combined it with the CSA system using DAB-based visualization (ISH-AT-CSA). We attempted to detect viral RNA in formalin-fixed and paraffin-embedded tissues from a rhesus monkey model system and autopsied patients with AIDS by both the ISH-AT-CSA method and conventional in situ hybridization using RNA probes with CSA (RISH-CSA).
The condition of the experimentally infected animal tissue specimens is generally much better than that of clinical autopsy tissue specimens and there are usually many residual transcripts, thus it is not so difficult to detect mRNA in situ. So, first, we used formalin-fixed and paraffin-embedded autopsy tissue specimens of SHIV- or SIV-infected monkeys to confirm that ISH-AT-CSA worked well. The SHIV RNA localizations detected by ISH-AT-CSA were in agreement with those in previous reports 22-24 and were similar to those detected by conventional RISH-CSA (Figure 2; g to j) ▶ . In addition, the signals were apparently improved to be stronger and to have a wider distribution.
Next, we applied ISH-AT-CSA for the detection of HIV-1 RNA or DNA in formalin-fixed and paraffin-embedded tissue specimens of autopsied patients with AIDS. No signals were detected in any of the specimens, comprised of 15 lymph nodes and 3 brains, by conventional RISH-CSA using a mixture of HIV-1gag RNA probe and HIV-1nef RNA probe. In contrast, the specific signals were detected in 13 lymph nodes and 3 brains by the ISH-AT-CSA method (Table 1 ▶ and Figure 3; f, g, v, and w ▶ ). The HIV-1 RNA signals detected in three brain specimens by ISH-AT-CSA were located in MGCs, which were also positive for HIV-1gag p24 antigen. So, ISH-AT-CSA was superior to RISH-CSA in detection sensitivity for formalin-fixed and paraffin-embedded HIV encephalitis autopsy specimens. The HIV-1 RNA signals in the lymphoid tissues were positive in 13 pediatric AIDS cases. They were from children 6 months to 2 years and 4 months of age, and their duration of HIV-1 infection had been supposed to be very short. The HIV-1 RNA signals detected by ISH-AT-CSA were scattered in the interfollicular area, and rare in the germinal centers (GCs) (Figure 3b) ▶ . This result was consistent with previous reports on in situ hybridization using especially radiolabeled RNA probes showing that HIV-1 RNA signals in autopsy specimens of AIDS patients with late-stage disease were rare in GCs but were dispersed over individual cells in the involuted follicles or paracortical areas of the lymph node. 31,32 In these reports, however, the HIV-1gag p24 antigens were not detected at the late stage of AIDS. In the case of pediatric AIDS shown in this study, HIV-1 gag p24 antigens were detected in the CD21-positive FDCs in GCs (Figure 3i) ▶ . There was a discrepancy that gag antigens were retained in FDCs without comparable in situ hybridization signals. We speculated that HIV-1 genomic RNA in virions was less stable than viral antigens, and degenerated to levels below the detection limit of ISH-AT-CSA. 33 As for HIV-1 mRNA, there were conflicting reports that support and refute FDC’s role in HIV-1 replication. 34-37 Absence of ISH-AT-CSA signals of HIV-1 mRNA in GCs agrees with the latter view.
We identified these HIV-1-infected cells by double-fluorescence staining with ISH-AT-CSA and immunohistochemistry, and showed that the HIV-1 RNA-positive cells in the lymphoid tissues at the late stage of AIDS were CD3-positive T cells (Figure 3p) ▶ . The double-fluorescence staining of specimens is essential in molecular pathological analysis. From this viewpoint, ISH-AT-CSA is a promising technique for the purpose. The ISH-AT-CSA method has the advantages of convenient probe preparation, better accessibility to target mRNA and lower background. Although the sensitivity of ISH-AT-CSA has not reached a level at which it can be used to detect integrated viral DNA (provirus), this issue might be resolved using a cocktail of several HybrAT probes or by developing a new HybrAT probe similar to the branched oligonucleotide probe reported previously. 5
In conclusion, the above data suggest that ISH-AT-CSA can be used as an alternative to the RISH-CSA. The ISH-AT-CSA method is significantly simplified and should be applicable broadly in the fields of cell biology and pathology.
Acknowledgments
We thank Drs. T. Matano and L. Bhoopat for tissue specimens and Drs. T. Iwasaki and H. Hasegawa for helpful discussion.
Footnotes
Address reprint requests to Tetsutaro Sata, M.D., Ph.D., Department of Pathology, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku, Tokyo 162-8640, Japan. E-mail: tsata@nih.go.jp.
Supported by a grant-in-aid from the Ministry of Health, Labor, and Welfare of Japan (to T. S.).
References
- 1.Corput MPC, Dirks RW, Gijlswijk PM, Binnendijk E, Hattinger CM, Paus RA, Landegent JE, Raap AK: Sensitive mRNA detection by fluorescence in situ hybridization using horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. J Histochem Cytochem 1998, 46:1249-1259 [DOI] [PubMed] [Google Scholar]
- 2.Speel EJM, Saremaslani P, Roth J, Hopman AHN, Komminoth P: Improved mRNA in situ hybridization on formaldehyde-fixed and paraffin-embedded tissue using signal amplification with different haptenized tyramides. Histochem Cell Biol 1998, 110:571-577 [DOI] [PubMed] [Google Scholar]
- 3.Egger D, Bolton R, Rahner C, Bienz K: Fluorochrome-labeled RNA as a sensitive, strand-specific probe for direct fluorescence in situ hybridization. Histochem Cell Biol 1999, 111:319-324 [DOI] [PubMed] [Google Scholar]
- 4.Speel EJM: Detection and amplification systems for sensitive, multiple-target DNA and RNA in situ hybridization: looking inside cells with a spectrum of colors. Histochem Cell Biol 1999, 112:89-113 [DOI] [PubMed] [Google Scholar]
- 5.Luehrsen KR, Davidson S, Lee YJ, Rouhani R, Soleimani A, Raich T, Cain CA, Collarini EJ, Yamanishi DT, Pearson J, Magee K, Madlansacay MR, Bodepudi V, Davoudzadeh D, Schueler PA, Mahoney W: High-density hapten labeling and HRP conjugation of oligonucleotides for use as in situ hybridization probes to detect mRNA targets in cells and tissues. J Histochem Cytochem 2000, 48:133-145 [DOI] [PubMed] [Google Scholar]
- 6.Murakami T, Hagiwara T, Yamamoto K, Hattori J, Kasami M, Utsumi M, Kaneda T: A novel method for detecting HIV-1 by non-radioactive in situ hybridization: application of a peptide nucleic acid probe and catalyzed signal amplification. J Pathol 2001, 194:130-135 [DOI] [PubMed] [Google Scholar]
- 7.Qian X, Bauer RA, Xu HS, Lloyd RV: In situ hybridization detection of calcitonin mRNA in routinely fixed, paraffin-embedded tissue sections: a comparison of different types of probes combined with tyramide signal amplification. Appl Immunohistochem Mol Morphol 2001, 9:61-69 [PubMed] [Google Scholar]
- 8.Nakajima N, Hanaki K, Shimizu YK, Ohnishi S, Gunji T, Nakajima A, Nozaki C, Mizuno K, Odawara T, Yoshikura H: Hybridization-AT-tailing (HybrAT) method for sensitive and strand-specific detection of DNA and RNA. Biochem Biophys Res Commun 1998, 248:613-620 [DOI] [PubMed] [Google Scholar]
- 9.Nakajima N, Sata T, Hanaki K, Kurata K, Yoshikura H: Application of hybridization AT-tailing method for detection of human immunodeficiency virus RNA in cells and simian immunodeficiency virus RNA in formalin-fixed and paraffin-embedded tissues. J Virol Methods 1999, 81:169-177 [DOI] [PubMed] [Google Scholar]
- 10.Hanaki K, Odawara T, Nakajima N, Shimizu YK, Nozaki C, Mizuno K, Muramatsu T, Kuchino Y, Yoshikura H: Two different reactions involved in the primer/template independent polymerization of dATP and dTTP by Taq DNA polymerase. Biochem Biophys Res Commun 1998, 244:210-219 [DOI] [PubMed] [Google Scholar]
- 11.Bobrow MN, Harris TD, Shaughnessy KJ, Litt GJ: Catalyzed reporter deposition, a novel method of signal amplification. J Immunol Methods 1989, 125:279-285 [DOI] [PubMed] [Google Scholar]
- 12.Van Gijlswijk RPM, Zijlmans HJMAA, Wiegant J, Bobrow MN, Erickson TJ, Adler KE, Tanke HJ, Raap AK: Fluorochrome-labeled tyramides: use in immunohistochemistry and fluorescence in situ hybridization. J Histochem Cytochem 1997, 45:375-382 [DOI] [PubMed] [Google Scholar]
- 13.Schmidt BF, Chao J, Zhu Z, DeBiasio RL, Fischer G: Signal amplification in the detection of single-copy DNA and RNA by enzyme-catalyzed deposition (CARD) of the novel fluorescent reporter substrate Cy3.29-tyramide. J Histochem Cytochem 1997, 45:365-373 [DOI] [PubMed] [Google Scholar]
- 14.Speel EJM, Hopman AHN, Komminoth P: Amplification methods to increase the sensitivity of in situ hybridization: play CARD(S). J Histochem Cytochem 1999, 47:281-288 [DOI] [PubMed] [Google Scholar]
- 15.Strappe PM, Wang TH, McKenzie CA, Lowrie S, Simmonds P, Bell JE: Enhancement of immunohistochemical detection of HIV-1 p24 antigen in brain by tyramide signal amplification. J Virol Methods 1997, 67:103-112 [DOI] [PubMed] [Google Scholar]
- 16.Wang G, Achim CL, Hamilton RL, Wiley CA, Soontornniyomkij V: Tyramide signal amplification methods in multiple-label immunofluorescence confocal microscopy. Methods 1999, 18:459-464 [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Wanner IB, Roper SD, Chaudhari N: An optimized method for in situ hybridization with signal amplification that allows the detection of rare mRNAs. J Histochem Cytochem 1999, 47:431-445 [DOI] [PubMed] [Google Scholar]
- 18.Matano T, Kano M, Nakamura H, Takeda A, Nagai Y: Rapid appearance of secondary immune response and protection from acute CD4 depletion after highly pathogenic immunodeficiency virus challenge in macaques vaccinated with a DNA prime/Sendai virus vector boost regimen. J Virol 2001, 75:11891-11896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rud EW, Cranage M, Yon J, Quirk J, Ogilvie L, Cook N, Webster S, Dennis M, Clarke BE: Molecular and biological characterization of simian immunodeficiency virus macaque strain 32H proviral clones containing nef size variants. J Gen Virol 1994, 75:529-543 [DOI] [PubMed] [Google Scholar]
- 20.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N: Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science 1990, 248:1109-1112 [DOI] [PubMed] [Google Scholar]
- 21.Adachi A, Gendelman HE, Koenig S, Folks T, Willy R, Rabson A, Martin MA: Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 1986, 59:284-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ringler DJ, Wyand MS, Walsh DG, Mackey JJ, Chalifoux LV, Popovic M, Minassian AA, Sehgal PK, Daniel MD, Desrosiers RC, King NW: Cellular localization of simian immunodeficiency virus in lymphoid tissues. I. Immunochemistry and electron microscopy. Am J Pathol 1989, 134:373-383 [PMC free article] [PubMed] [Google Scholar]
- 23.Wyand MS, Ringler DJ, Naidu YM, Mattmuller M, Chalifoux LV, Sehgal PK, Daniel MD, Desrosiers RC, King NW: Cellular localization of simian immunodeficiency virus in lymphoid tissues. II. In situ hybridization. Am J Pathol 1989, 134:385-393 [PMC free article] [PubMed] [Google Scholar]
- 24.Lackner AA, Smith MO, Munn RJ, Martfeld DJ, Gardner MB, Marx PA, Dandekar S: Localization of simian immunodeficiency virus in the central nervous system of rhesus monkeys. Am J Pathol 1991, 139:609-621 [PMC free article] [PubMed] [Google Scholar]
- 25.Nuovo GJ: PCR in Situ Hybridization: Protocols and Applications. 1994. Raven Press, New York
- 26.Nuovo GJ, Becker J, Burk MW, Margiotta M, Fuhrer J, Steigbigel RT: In situ detection of PCR-amplified HIV-1 nucleic acids in lymph nodes and peripheral blood in patients with asymptomatic HIV-1 infection and advanced-stage AIDS. J AIDS 1994, 7:916-923 [PubMed] [Google Scholar]
- 27.Bagasra O, Lavi E, Bobroski L, Khalili K, Pestaner JP, Tawadros R, Pomerantz RJ: Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS 1996, 10:573-585 [DOI] [PubMed] [Google Scholar]
- 28.Long AA, Komminoth P: In situ PCR. An overview. Methods Mol Biol 1997, 71:141-161 [DOI] [PubMed] [Google Scholar]
- 29.Strappe PM, Wang TH, McKenzie CA, Lowrie S, Simmonds P, Bell JE: In situ polymerase chain reaction amplification of HIV-1 DNA in brain tissue. J Virol Methods 1998, 70:119-127 [DOI] [PubMed] [Google Scholar]
- 30.Hanaki K, Odawara T, Muranatsu T, Kuchino Y, Masuda M, Yamamoto K, Nozaki C, Mizuno K, Yoshikura H: Primer/template-independent synthesis of poly d(A-T) by Taq polymerase. Biochem Biophys Res Commun 1997, 238:113-118 [DOI] [PubMed] [Google Scholar]
- 31.Fox CH, Tenner-Racz K, Racz P, Firpo A, Pizzo PA, Fauci AS: Lymphoid germinal centers are reservoirs of human immunodeficiency virus type 1 RNA. J Infect Dis 1991, 164:1051-1057 [DOI] [PubMed] [Google Scholar]
- 32.Pantaleo G, Graziosi C, Demarest JF, Butini L, Montroni M, Fox CH, Orenstein JM, Kotler DP, Fauci AS: HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature 1993, 362:355-358 [DOI] [PubMed] [Google Scholar]
- 33.Takahashi H, Sawa H, Hasegawa H, Sata T, Hall WW, Nagashima K, Kurata T: Reconstitution of cleavage of human immunodeficiency virus type-1 (HIV-1) RNAs. Biochem Biophys Res Commun 2002, 293:1084-1091 [DOI] [PubMed] [Google Scholar]
- 34.Parmentier HK, Wichen D, Sie-Go DMDS, Goudsmit J, Borleffs JCC, Schuurman HJ: HIV-1 infection and virus production in follicular dendric cells in lymph nodes. Am J Pathol 1990, 137:247-251 [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz J, Lunzen J, Tenner-Racz K, Grobschupff G, Racz P, Scmitz H, Dietrich M, Hufert FT: Follicular dendric cells retain HIV-1 particles on their plasma membrane, but are not productively infected in asymptomatic patients with follicular hyperplasia. J Immunol 1994, 153:1352-1359 [PubMed] [Google Scholar]
- 36.Tacchetti C, Favre A, Moresco L, Meszaros P, Luzzi P, Truini M, Rizzo F, Grossi CE, Ciccone E: HIV is trapped and masked in the cytoplasm of lymph node follicular dendric cells. Am J Pathol 1997, 150:533-542 [PMC free article] [PubMed] [Google Scholar]
- 37.Spiegel H, Herbst H, Niedobitek G, Foss HD, Stein H: Follicular dendric cells are a major reservoir for human immunodeficiency virus type 1 in lymphoid tissues facilitating infection of CD4+ T-helper cells. Am J Pathol 1992, 140:15-22 [PMC free article] [PubMed] [Google Scholar]