Abstract
We recently established an Epstein-Barr virus (EBV)-positive γδ T-cell line from a nasal T/natural killer (NK)-cell lymphoma (Nagata H, Konno A, Kimura N, Zhang Y, Kimura M, Demachi A, Sekine T, Yamamoto K, Shimizu N: Characterization of novel natural killer (NK)-cell and γδ T-cell lines established from primary lesions of nasal T/NK-cell lymphomas associated with the Epstein-Barr virus. Blood 2001, 97:708–713). Subsequently, we established two novel EBV-positive γδ T-cell lines from the peripheral blood of patients with chronic active EBV infection. Analysis of the terminal repeat of EBV showed that the three cell lines consisted of monoclonal populations, and flow cytometry showed that they had a common phenotype of γδ T cells: CD3+ CD4− CD8− CD16− CD19− CD56+ CD57− HLA-DR+ T-cell receptor (TCR) αβ− TCR γδ+. Analysis for the expression of TCR by flow cytometry showed that all three cell lines were Vγ9+/Vδ2+, but negative for VγI, Vδ1, or Vδ3 TCR. Southern blot analysis for TCR genes showed that the three cell lines had a common rearrangement of Vγ9-JγP and Jδ3 genes. Polymerase chain reaction and sequence analysis of the junction between Vδ and Jδ genes revealed that the Jδ3 genes were rearranged with the Vδ2 genes. In contrast, none of the EBV-negative γδ T-cell lines, Molt-14, Peer, or Loucy, which were analyzed for controls, had Vγ9 or Vδ2 TCR, or a rearrangement of Jδ3 genes. These results indicated that Vγ9-JγP/Vδ2-Jδ3+ γδ T cells were preferentially affected by EBV and expanded in patients with nasal γδ T-cell lymphoma and chronic active EBV infection. Jδ3+ γδ T cells are known to be a very minor population in γδ T cells of peripheral blood, whereas Vγ9-JγP/Vδ2-Jδ1+ cells are the major population. The close association of EBV with this particular γδ T-cell population may provide a key to the etiology of EBV-positive lymphoproliferative diseases.
γδ T cells comprise only a small proportion (1 to 5%) of the lymphocytes in the peripheral blood and organs, but are far more widespread within epithelial-rich tissues such as the skin, gut, and reproductive tract, where they comprise up to 50% of T cells. 1 Postthymic peripheral T-cell lymphomas of the γδ T-cell subset are rare, and most present as hepatosplenic γδ T-cell lymphomas. 2,3 Otherwise, they arise in the skin, nose, larynx, lung, or gastrointestinal tract. 4 They are believed to include some different disease entities based on their clinical and morphological features. Hepatosplenic γδ T-cell lymphomas show a specific sinusoidal localization of malignant cells in the spleen, liver, and bone marrow. Other nonhepatosplenic γδ T-cell lymphomas appear to be regarded as activated cytotoxic lymphomas arising in the mucosae and skin. Among these, nasal γδ T-cell lymphomas in particular show a strong association with Epstein-Barr virus (EBV), and together with natural killer (NK)-cell lymphomas, constitute the distinct disease category nasal T/NK-cell lymphoma. 4-7
The relationship between EBV and peripheral T/NK-cell neoplasms has been studied intensely since 1988, when Kikuta and associates 8 detected EBV in T lymphocytes in a boy with chronic active EBV infection (CAEBV), and Jones and colleagues 9 identified EBV in T-cell lymphomas arising in patients with CAEBV. As a result, it is now recognized that nasal T/NK-cell lymphoma has the strongest association with EBV among various kinds of peripheral T/NK-cell lymphomas. 2,3,10 However, studies on this disease have been limited because of the rarity of the disease and the tendency of the tumor to cause necrosis. To investigate this disease, we recently established two EBV-positive NK- and γδ T-cell lines, SNK-6 and SNT-8, from independent patients with nasal T/NK-cell lymphomas, showing that there are at least two phenotypes in this tumor, NK and γδ T cells. 7
Another important lymphoproliferative disease inevitably associated with EBV is CAEBV. In most CAEBV cases, the T and NK cells become positive for EBV, 8,9,11-25 and such EBV-positive T or NK cells showed monoclonal or oligoclonal expansion in many cases. 11,12,17,19,20,24,25 In addition, in some severe CAEBV cases, the establishment of cell lines of T- and NK-cell lineage has been reported, 16,21 suggesting that the clonal expansion of T/NK cells in CAEBV is in part because of an acquired great proliferative capacity brought by EBV. In an effort to understand the role of EBV in lymphoproliferative diseases of T/NK-cell lineage, we have been trying to establish cell lines from patients with CAEBV. Fortunately, we have to date successfully established six EBV-positive cell lines from the peripheral blood of patients with CAEBV. These include two NK–cell lines, two αβ T-cell lines, and two γδ T-cell lines.
In the present study, to clarify the role of EBV in the lymphoproliferative diseases of γδ T cells, we determined to analyze and compare the three EBV-positive γδ T-cell lines established. One was the previously reported SNT-8, and the other two were newly established lines from patients with CAEBV, SNT-13 and SNT-15. We aimed to analyze the phenotypes of the cell surface markers, the expression of T-cell receptors (TCRs) and the rearrangement of TCR genes in the cell lines. In addition, EBV-negative γδ T-cell lines, Molt-14, Peer, and Loucy were used for controls. We determined that Vγ9-JγP/Vδ2-Jδ3 TCR was commonly expressed in the three EBV-positive cell lines, but was not observed in the EBV-negative cell lines. The present study suggests that EBV probably affects γδ T cells expressing specific TCR to develop lymphoproliferative diseases.
Materials and Methods
Patients and Cell Lines
Three cell lines, designated as SNT-8, SNT-13, and SNT-15, established from patients with nasal T-cell lymphoma or CAEBV were used in this study. The clinical features of the patients are summarized in Table 1 ▶ . SNT-8 is a γδ T-cell line established from the primary lesion of a nasal T-cell lymphoma. Some of the characteristics of the cell line, as well as the case history, have been reported previously. 7 The other two cell lines were newly established from the peripheral blood of two unrelated patients with CAEBV. Both patients suffered from chronic mononucleosis-like symptoms, but showed no immunological abnormality or infection other than EBV infection; patient 2 had repeated fever and aphthae, and patient 3 had hepatosplenomegaly. They showed elevated titers for IgG to the Epstein-Barr viral capsid antigen (VCA-IgG) and diffuse and restricted components of early antigens (EA-DR IgG). They were thus diagnosed as having CAEBV in accordance with the criteria described by Straus. 26 They had increased γδ T-cell numbers among peripheral blood lymphocytes, and their peripheral blood was strongly positive for the EBV genome by polymerase chain reaction (PCR).
Table 1.
Clinical Profile of Patients
| Patient no. | Age/sex | Diagnosis | VCA IgG | Anti-EBV titers EA-DR IgG | EBNA | Peripheral blood findings | Cell line | Origin of cell line |
|---|---|---|---|---|---|---|---|---|
| 1* | 48/F | Nasal T/NK-cell lymphoma | 640 | 10 | 20 | WBC count 4400 (3% γδ T cells in total lymphocytes) | SNT-8 | Primary lesion in the nasal cavity |
| 2 | 13/F | CAEBV | 1280 | 320∼640 | 20∼40 | WBC count 3500 to 6000 γδ T-lymphocytosis (50 to 70% in total lymphocytes) | SNT-13 | Peripheral Blood |
| 3 | 15/F | CAEBV | 1280 | 160 | 10 | WBC count 1950 γδ T-lymphocytosis (23% in total lymphocytes) | SNT-15 | Peripheral Blood |
CAEBV, chronic active Epstein-Barr virus infection; VCA, viral capsid antigen; EA-DR, early antigens-diffuse and restricted; EBNA, Epstein-Barr nuclear antigen; WBC, white blood cell.
*Case 1 was previously reported. 7
For controls, three EBV-negative γδ T-cell lines, Molt-14, Peer, and Loucy, were analyzed in parallel. Molt-14 and Peer were obtained from Fujisaki Cell Center (Hayashibara Biochemical Laboratories, Inc., Okayama, Japan), and Loucy was kindly provided by Dr. Hannah Ben-Bassat (Hadassah University Hospital, Jerusalem, Israel). The origin of these EBV-negative cell lines has been described elsewhere. 27-29
Establishment of EBV-Positive γδ T-Cell Lines
The method used for establishing SNT-8 was described in our previous report. 7 Essentially the same method was used for the establishment of the SNT-13 and SNT-15 cell lines. Briefly, peripheral blood mononuclear cells were isolated from 10 ml of peripheral blood of the patients with CAEBV by the Ficoll-Hypaque method, and suspended in a 10-ml RPMI 1640 medium supplemented with 10% heat-inactivated human serum and 700 U/ml of interleukin (IL)-2. Then, CD4+ and CD8+ lymphocytes were removed using anti-CD4 and anti-CD8 monoclonal antibody-conjugated magnetic beads (Dynal, Oslo, Norway). The remaining CD4− CD8− cell fraction was seeded in a 96-well plate at 1 × 104 cells/well and cultured in a humidified atmosphere at 37°C with 5% CO2. Two days later, rapid cell growth was observed in all wells of the plate. After 1 week of culture, the growing cells were collected together. These cultured cells have been maintained in the presence of IL-2 for more than 40 months and 34 months, respectively, and were designated SNT-13 and SNT-15.
Flow Cytometric Analysis
The cell lines were analyzed by two-color immunofluorescence with a flow cytometer (EPICS XL; Beckman Coulter, Hialeah, FL) for the expression of surface markers. The following antibodies conjugated with fluorescein isothiocyanate or phycoerythrin (Becton Dickinson, San Jose, CA) were used: anti-CD3, -CD4, -CD8, -CD16, -CD19, -CD21, -CD56, -CD57, -HLA-DR, -TCR α/β, and -TCR γ/δ.
Flow cytometry was also performed to analyze variable regions of γδ TCR expressed on the cells. Antibodies used were anti-VγI and anti-Vδ1 monoclonal antibodies (Beckman Coulter), which required secondary antibodies to be detected: F(ab′)2 goat anti-mouse IgG conjugated with fluorescein isothiocyanate was used. Other antibodies used were fluorescein isothiocyanate-labeled anti-Vγ9, anti-Vδ2, and anti-Vδ3 monoclonal antibodies (Beckman Coulter).
Assessment of the Number of EBV Terminal Repeats
To determine the clonality of the EBV-positive cell lines, the number of EBV terminal repeats (EBV-TR) was assessed by Southern blot analysis. DNA extracted from the cell lines was digested with BamHI, separated on a 0.7% agarose gel, and transferred to a nylon membrane. A 1.9-kb XhoI subfragment of BamHI-Dhet derived from EBV termini was used as a probe for EBV-TR DNA. 30 Hybridization was visualized using a Fluorescein Gene Images System (Amersham, Buckinghamshire, UK) according to the manufacturer’s instructions. Raji and B95-8 cells were used as controls for monoclonal cell expansion with a single EBV-TR and polyclonal cell proliferation with EBV replication, respectively.
Southern Blot Analysis for TCR and Immunoglobulin Genes
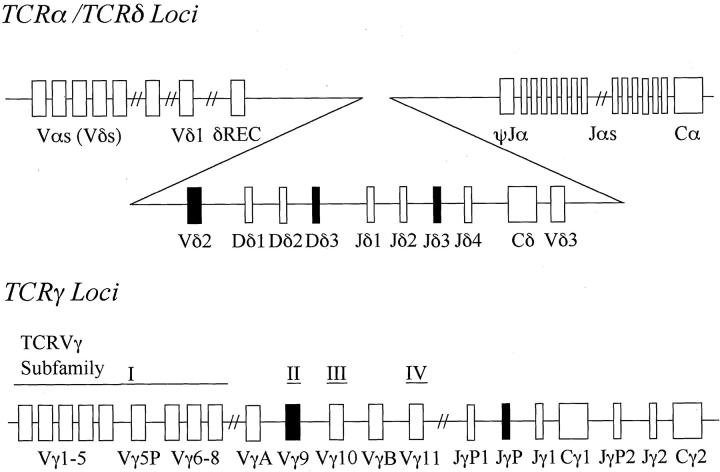
The cell lines were analyzed for rearrangements of the TCR β-, γ-, and δ-chain and the immunoglobulin heavy-chain genes by Southern blotting. Three μg of DNA extracted from each of the cell lines was digested with a restriction endonuclease BamHI, EcoRI, HindIII, or KpnI (Takara, Kyoto, Japan), electrophoresed through a 0.7% agarose gel, and transferred onto a nylon membrane. Human placental DNA was used as a germline control. The membrane was exposed to a fluorescein-labeled Cβ1, Jγ1, Jδ1, or Jδ3 probe for the TCR genes, or a JH probe for the immunoglobulin heavy-chain gene. 31-35 The Jγ1 probe, a 700-bp HindIII-EcoRI fragment, includes the Jγ1 segment and cross-hybridizes with Jγ2 but not with the additional Jγ segments. The Jδ1 probe consists of a 1-kb PstI-EcoRI segment. The Jδ3 probe was a 1.5-kb XbaI genomic fragment of the Jδ3 region. The organization of the human TCRγ and TCRδ loci is shown in Figure 1 ▶ . Hybridization was visualized by the same methods used for the Southern blot analysis of the EBV-TR.
Figure 1.
Schematic organization of the human TCRδ and TCRγ loci. The TCRδ locus is located entirely within the TCRα locus between the TCRVα and the TCRJα gene segments. TCRVγ genes are divided into four subfamilies (I to IV) based on sequence similarity. Each Vγ gene is assigned to its subgroup as indicated. The solid boxes are gene segments used in SNT cell lines.
PCR Analysis for Combinations of Vδ and Jδ Genes
To define Vδ-Jδ gene rearrangements in the cell lines, PCR analysis of the Vδ1, Vδ2, or Vδ3 gene and the Jδ1 or Jδ3 gene was performed. The sequences of primers used are listed in Table 2 ▶ .
Table 2.
Primers for PCR Analysis of TCR Rearrangement
| TCR δ gene | Primer | Nucleotide sequence (5′ to 3′) | |
|---|---|---|---|
| Sense | V δ 1 | L-V δ 1 | GTGTGTATTTGTGGCCTTCA |
| V δ 2 | L-V δ 2 | TCATCCATCTCTCTCTCTTC | |
| V δ 2 | GCACCATCAGAGAGAGATGA | ||
| V δ 3 | L-V δ 3 | TCTACAGGGGCACGCTGTGT | |
| Anti-sense | J δ 1 | 3′-J δ 1 | AAATGCTAGCTATTTCACCCA |
| J δ 3 | J δ 3 | GAGTTTGATGCCAGTTCCGAA | |
| Cδ | Cδ | TAACTTGGCAGTCAAGAGAAA |
Fifty ng of DNA extracted from each cell line was amplified with a pair of each combination of L-Vδ1, L-Vδ2, or L-Vδ3 and 3′-Jδ1 or Jδ3 primers. The PCR was performed with reagents composed of 500 nmol/L of primers, 200 μmol/L of dNTPs, 20 mmol/L of Tris-HCl, 1.2 mmol/L of MgCl2, and 2.5 U of TaqDNA polymerase (Takara) in a 100-μl reaction solution. The program was 94°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute for 40 cycles. The products were analyzed by electrophoresis on a 2% agarose gel after staining with ethidium bromide.
Reverse Transcriptase (RT)-PCR and Direct Sequencing for Junction of Vδ-Jδ Genes
Total RNA was extracted from each of the cell lines using TRIzol reagent (Life Technologies, Inc., Gaithersburg, MD), and 0.5 μg was reverse-transcribed into cDNA using a Thermoscript RT-PCR System (Life Technologies, Inc.). An oligo(dT)20 primer was used for the generation of cDNAs. After incubation at 60°C for 60 minutes, the samples were heated for 5 minutes at 85°C to terminate the reactions. PCR amplification of TCR δ-chain cDNA was accomplished by use of a Cδ anti-sense primer in combination with a Vδ2 sense primer. A human β-actin primer pair was obtained from Takara. The PCR cycling conditions were 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute for 40 cycles. Amplified products were identified by 2% agarose gel electrophoresis followed by staining with ethidium bromide. Amplification products were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany), and direct sequencing was performed using a BigDye Terminator Reaction kit version 2.0 and an ABI PRISM 377 genetic analyzer (Applied Biosystems, Foster City, CA), according to the manufacturer’s protocols.
Results
Phenotypes and Clonality of the Cell Lines
Flow cytometry showed that the SNT-8, SNT-13, and SNT-15 cells had an identical phenotype: CD3+ CD4− CD8− CD16− CD19− CD21− CD56+ CD57− HLA-DR+ TCR α/β− TCR γ/δ+ (Table 3) ▶ . Although they were positive for the NK-cell marker CD56, the expression of CD3 and γδ TCR indicated that they were derived from γδ T cells and distinct from NK-cell lines. We previously reported phenotypes of two NK-cell lines SNK-1 and SNK-6 as CD3− CD4− CD8− CD19− CD25+ CD56+ CD57− TCR α/β− TCR γ/δ−. 7,25
Table 3.
Immunophenotype of SNT Cells
| Cell line | CD3 | CD4 | CD8 | CD16 | CD19 | CD21 | CD56 | CD57 | HLA-DR | TCRαβ | TCRγδ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNT-8 | + | − | − | − | − | − | + | − | + | − | + |
| SNT-13 | + | − | − | − | − | − | + | − | + | − | + |
| SNT-15 | + | − | − | − | − | − | + | − | + | − | + |
| Molt-14 | + | − | − | − | − | + | − | − | − | − | + |
| Peer | + | + | − | − | − | − | − | − | − | − | + |
| Loucy | + | + | − | − | − | − | − | − | − | − | + |
Antibody reactivity: +, ≥75% of the cells were positive; −, <15% of the cells were positive.
Because the established EBV-positive cell lines proved to be of the γδ T-cell subset, we analyzed the expression of variable regions of γδ TCR by flow cytometry. We found the three EBV-positive cell lines to be positive for the anti-Vγ9 and anti-Vδ2 antibodies and negative for the anti-VγI, anti-Vδ1, and anti-Vδ3 antibodies (Table 4) ▶ . By contrast, the EBV-negative cell lines Molt-14, Peer, and Loucy were negative for the anti-Vδ2 antibodies, and the former two were negative for the anti-Vγ9 antibody as well; the last cell line weakly reacted with the anti-Vγ9 antibody (Table 4) ▶ .
Table 4.
Expression of TCR-Vγ and -Vδ Proteins by γδ T-Cell Lines
| Cell line | Antibodies | ||||
|---|---|---|---|---|---|
| VγI | Vγ9 | Vδ1 | Vδ2 | Vδ3 | |
| SNT-8 | − | +++ | − | +++ | − |
| SNT-13 | − | +++ | − | +++ | − |
| SNT-15 | − | +++ | − | +++ | − |
| Molt-14 | + | − | +++ | − | − |
| Peer | − | − | ++ | − | − |
| Loucy | + | + | − | − | − |
Antibody reactivity: +++, >90% of the cells were positive; ++, 50 to 90% of the cells were positive; +, 15 to 50% of the cells were positive; −, <15% of the cells were positive.
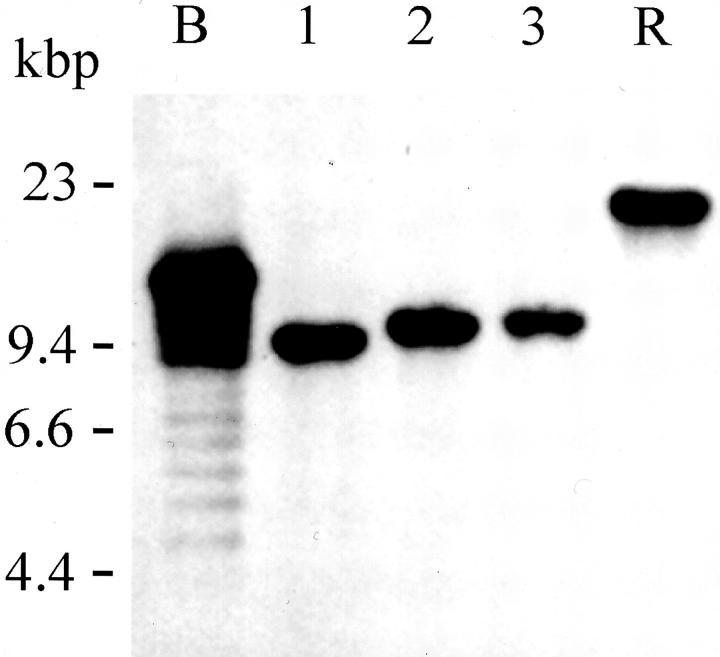
Regarding the clonality of the EBV-positive cell lines, the Southern blot analysis for the number of EBV-TR showed a single band in each of the SNT-8, SNT-13, and SNT-15 cells (Figure 2) ▶ . The results indicated that these cell lines consisted of respective monoclonal cell populations and were Vγ9/Vδ2+ γδ T cells. In addition, we previously reported that the clonality of SNT-8 was identical to that of the original tumor. 7 As to SNT-13, we could confirm that the clonality was the same as that detected in the peripheral blood mononuclear cells of the patient (data not shown).
Figure 2.
Clonality of cells assessed using EBV terminal repeats. Southern blot analysis of the number of EBV-TRs as evidence for monoclonal expansion of EBV-positive cells in SNT cell lines. Lane 1, SNT-8; lane 2, SNT-13, and lane 3, SNT-15 cells. B95-8 cells (B) and Raji cells (R) were used as controls for cells with productive EBV expansion and monoclonal EBV replication, respectively.
Southern Blot Analysis for TCR and Immunoglobulin Genes
TCR γ-Chain Genes
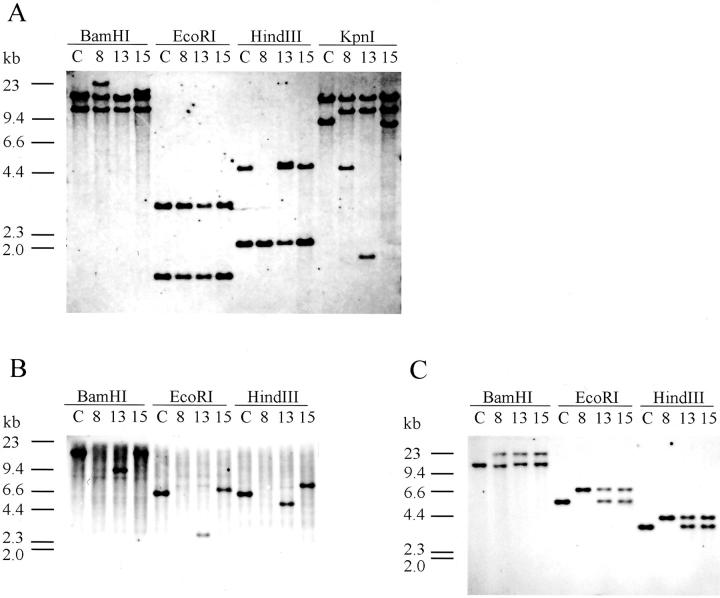
Southern blot hybridization of the KpnI digests of DNA with the Jγ1 probe detected rearrangements of TCR γ-chain genes in the three EBV-positive cell lines (Figure 3A) ▶ . The EBV-positive cell lines showed three bands in the KpnI digests, whereas the germline control showed two bands of 16 kb and 9 kb. The sizes of bands detected in the cell lines were as follows; 16 kb, 12 kb, and 4.7 kb in SNT-8; 16 kb, 12 kb, and 1.8 kb in SNT-13; 16 kb, 12 kb, and 8.5 kb in SNT-15. Thus, the three cell lines commonly showed 12-kb bands, which are known to be generated by rearrangement of the Vγ9 gene to the JγP gene. Rearranged genes were deduced from the sizes of KpnI digests, because it was reported that the Vγ9-JγP rearrangement frequently observed in rearranging γ-loci can be detected with the KpnI digest, but the BamHI-rearranged bands are virtually the same size as the germline bands and the EcoRI and HindIII bands are in a germline configuration in this rearrangement for their restriction sites. 36,37 The results, thus, showed that the three cell lines had the Vγ9-JγP rearrangement in one allele. Additionally, the other bands were supposed to be generated by the following rearrangement of the γ-chain genes in the other alleles: the 4.7-kb band in SNT-8 was generated by rearrangement of the VγI or VγIII gene to the JγP2 gene, the 1.8-kb band in SNT-13 was made by rearrangement of the VγI or VγIII gene to the Jγ2 gene, and the 8.5-kb band in SNT-15 was derived from rearrangement of the VγI or VγIII gene to the JγP1 gene. Although the three cell lines showed the rearrangement of γ-chain genes in both alleles, it was suggested that the Vγ9-JγP chain was the expressed allele because flow cytometry showed that the three cell lines expressed Vγ9-encoded receptors.
Figure 3.
Southern blot analysis for TCR genes. Shown are the hybridizations with the Jγ1 probe (A), the Jδ1 probe (B), and the Jδ3 probe (C). Southern blot analysis shows rearrangement of the Jγ1, Jδ1, and Jδ3 genes in all SNT cell lines. Human placental DNA was used as a germline configuration (c); 8, SNT-8; 13, SNT-13; 15, SNT-15 cells.
The EBV-negative cell lines Molt-14, Peer, and Loucy were analyzed in the same manner, but none showed the 12-kb band in KpnI digests, a hallmark of the Vγ9-JγP rearrangement. Molt-14 showed rearrangements in both alleles: VγI or VγIII to JγP1, and VγI or VγIII to Jγ2. Peer and Loucy had the same rearrangement, Vγ9 to Jγ2 in one allele and VγI or VγIII to Jγ2 in the other (data not shown). The results of the Southern blot analyses are summarized in Table 6 ▶ .
Table 6.
Summary of Analysis for γδ TCR
| Cell line | EBV | Southern blot analysis | PCR, sequencing, and Southern blot analysis | Flow cytometry | |||
|---|---|---|---|---|---|---|---|
| TCR Jγ | TCR Jδ1 | TCR Jδ3 | TCR γ-chain gene rearrangement | TCR δ-chain gene rearrangement | γδ TCR-protein expression | ||
| SNT-8 | + | R | D | R | Vγ9-JγP | Vδ2-Jδ3 | Vγ9/Vδ2 |
| VγI/III-JγP2 | |||||||
| SNT-13 | + | R | R | R | Vγ9-JγP | Vδ2-Jδ3 | Vγ9/Vδ2 |
| VγI/III-Jγ2 | Dδ2-Jδ1 | ||||||
| SNT-15 | + | R | R | R | Vγ9-JγP | Vδ2-Jδ3 | Vγ9/Vδ2 |
| VγI/III-JγP1 | Vδ2-Dδ3 | ||||||
| Molt-14 | − | R | R | G | VγI/III-JγP1 | Vδ1-Jδ1 | VγI/Vδ1 |
| VγI/III-Jγ2 | Unknown R | ||||||
| Peer | − | R | R | G | Vγ9-Jγ2 | Vδ1-Jδ1 | Unknown/Vδ1* |
| VγI/III-Jγ2 | |||||||
| Loucy | − | R | R | G | Vγ9-Jγ2 | Dδ2-Jδ1 | ND† |
| VγI/III-Jγ2 | Unknown R | ||||||
| Unknown R | |||||||
G, germline band; R, rearrangement band; D, deletion of germline band (s); ND, not determined.
*Vγ protein was not determined. Peer was negative for anti-Vγ9 and -VγI antibodies, although it was positive for anti-TCR γ/δ antibody.
†Not determined, but Vγ9/Vδ2 TCR was not detected.
TCR δ-Chain Genes
Using the Jδ1 probe, we detected no band in SNT-8 and a rearranged band in each digest of DNA of SNT-13 and SNT-15 (Figure 3B) ▶ . The sizes of the bands were 9.4 kb in the BamHI digest, 2.8 kb in the EcoRI digest, and 4.8 kb in the HindIII digest in SNT-13, and 19.5 kb in the BamHI digest, 6.9 kb in the EcoRI digest, and 7.1 kb in the HindIII digest in SNT-15. The deduced rearrangement was an incomplete rearrangement of Dδ2-Jδ1 in SNT-13 and Vδ2-Dδ3 in SNT-15. 38,39 Therefore, a complete Jδ1 rearrangement to be expressed was not detected in the three cell lines.
Hybridization with the Jδ3 probe revealed that the three cell lines showed rearranged bands of an identical size in each of the BamHI, EcoRI, and HindIII digests (Figure 3C) ▶ . The results suggest that the three cell lines have a common rearrangement of Jδ3 genes. Because of the lack of a precise map for restriction enzymes in this region, the Southern blot analysis could not determine how Jδ3 genes were rearranged with Vδ genes.
SNT-8 showed only rearranged bands in the BamHI, EcoRI, and HindIII digests, and these results suggested that SNT-8 had the rearrangement of Jδ3 genes in both alleles, or the rearrangement of a Jδ3 gene in one allele with a deletion of Jδ genes in the other. In addition, the SNT-8 cells were found to express γδTCR by flow cytometry, indicating that deletion of the Jδ1 gene was a consequence of the usage of the Jδ3 gene at the same δ locus.
Both SNT-13 and SNT-15 showed the rearranged band and a germline band in each digest obtained with the Jδ3 probe. These results suggested that SNT-13 and SNT-15 had a rearrangement of Jδ3 genes, which was common to that in SNT-8, in one allele, and incomplete rearrangements in the other allele upstream of the Jδ3 segment as described above.
The three EBV-negative cell lines Molt-14, Peer, and Loucy showed rearranged bands with the Jδ1 probe, but not with the Jδ3 probe (data not shown). The deduced rearrangement in these cell lines is summarized in Table 6 ▶ .
TCR β-Chain and Immunoglobulin Genes
All of the EBV-positive and -negative cell lines showed rearrangements of the Cβ genes, whereas none showed a rearrangement of the immunoglobulin heavy-chain genes (data not shown). It is known that rearrangements of the β-chain gene are commonly found in peripheral γδ T-cell lymphomas. 40,41
PCR for Combinations of Vδ and Jδ Genes
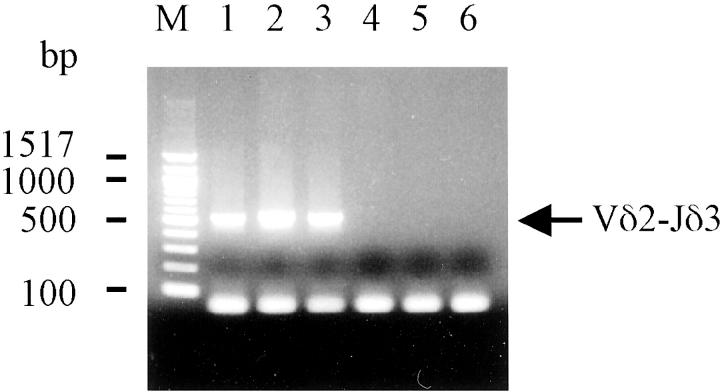
To elucidate the rearrangement of Vδ and Jδ genes, PCR analysis was performed for each combination of the Vδ1, Vδ2, or Vδ3 gene and the Jδ1 or Jδ3 genes. It was found that, only PCR using the L-Vδ2 and Jδ3 primers amplified fragments with an expected size of ∼500 bp in the three EBV-positive cell lines, indicating that the cell lines had the Vδ2-Jδ3 rearrangement, but not the Vδ1-Jδ1, Vδ1-Jδ3, Vδ2-Jδ1, Vδ3-Jδ1, and Vδ3-Jδ3 rearrangements (Figure 4) ▶ . In terms of the EBV-negative cell lines, the DNA of Molt-14 and Peer was amplified with the L-Vδ1 and 3′-Jδ1 primers. This result indicated that these two cell lines had the Vδ1-Jδ1 rearrangement (data not shown).
Figure 4.
PCR analysis for the junction of Vδ and Jδ genes. PCR analysis shows the presence of the Vδ2-Jδ3 TCR gene rearrangement in the three SNT cell lines. Only the combination of a L-Vδ2 sense primer and a Jδ3 anti-sense primer amplified ∼500-bp PCR products identified as Vδ2-Jδ3. Lane 1, SNT-8; lane 2, SNT-13; lane 3, SNT-15 cells. No signal was detected in EBV-negative γδ T-cell lines; lane 4, Molt-14; lane 5, Peer; lane 6, Loucy.
Analysis of TCR δ-Chain Transcripts
RT-PCR and direct sequencing were performed to confirm the results of flow cytometry, Southern blotting and PCR analysis showing that the three cell lines had the Vδ2-Jδ3 rearrangement. RT-PCR with the Vδ2 and Cδ primers commonly amplified ∼550-bp single bands in the three cell lines (data not shown), and β-actin transcripts were amplified in all of the samples and served as a control for amplifiable mRNA. Direct sequencing of the amplified RT-PCR products clearly showed that the three cell lines had sequences for the Vδ2-Dδ3-Jδ3 rearrangement (Table 5) ▶ . The results demonstrate that the Vδ2-Jδ3 rearrangement took place, and mRNA for Vδ2-Jδ3-Cδ receptor was then transcribed in the SNT cell lines. In addition, the same RT-PCR and sequence analysis was performed on peripheral blood of the patient from which SNT-13 was derived at primary diagnosis. The patient’s blood had completely the same sequence as that of SNT-13 (data not shown).
Table 5.
Nucleotide Sequence of Vδ2-Jδ3 Transcripts Expressed by γδ T-Cell Lines
| Vδ2 | N/P | Dδ3 | N/P | Jδ3 | |
|---|---|---|---|---|---|
| Germline | GCCTGTGACACC | ACTGGGGGATACG | CTCCTGGGACACCC | ||
| SNT-8 | GCCTGTGA | TCT | ACTGG | CGGG | CTCCTGGGACACCC |
| SNT-13 | GCCTGTGACACC | GTGA | ACTGGGGGATACG | CGG | CTCCTGGGACACCC |
| SNT-15 | GCCTGTGACAC | GT | ACTGGGGGA | G | CTCCTGGGACACCC |
The germline sequences of the 3′ end of Vδ2, the Dδ3, and the 5′ end of Jδ3 gene segments are at the top. The sequences of TCR δ-chain transcripts of the γδ T-cell lines are aligned with the germline sequences. N/P; template-independent “N” nucleotide insertions or palindromic “P” synthesis.
The results of the flow cytometric, Southern blot, PCR, RT-PCR, and direct sequencing analyses are summarized in Table 6 ▶ . The table shows that the results are consistent. Overall, they demonstrate that the three EBV-positive cell lines always express Vγ9-JγP/Vδ2-Jδ3 TCR, a phenomenon not observed in the EBV-negative γδ T-cell lines.
Discussion
In this study, we reported characteristics of three EBV-positive cell lines of γδ T cells. One of these was the previously-reported γδ T-cell line SNT-8, which was established from the primary lesion of a nasal T/NK-cell lymphoma, 7 whereas the other two, SNT-13 and SNT-15, were newly established lines from the peripheral blood of patients with CAEBV. In the previous study, SNT-8 was demonstrated to have developed from the lymphoma cells in the patient, because SNT-8 had the same immunophenotype and monoclonal EBV clones as the original lesion. 7 In patient 2, SNT-13 had the same EBV clone and TCR δ-chain as the peripheral blood mononuclear cells, indicating that SNT-13 had developed from cells predominantly expanding in the patient and responsible for her disease. These data, thus, suggest that it is the cells proliferating in patients with nasal T/NK-cell lymphoma and CAEBV that exclusively grow in our culture system.
The three cell lines showed a phenotype typical of γδ T cells, and were associated with a consistent rearrangement of the TCR genes Vγ9-JγP/Vδ2-Jδ3. To the best of our knowledge, these are the only γδ T-cell lines that have been found to be positive for EBV. Moreover, this study presented the first evidence of the existence of cases of CAEBV of the γδ T-cell subset.
Not only SNT-8 cells but also SNT-13 and SNT-15 cells proliferated in vitro, and an analysis of the number of EBV-TR showed that every cell line consisted of monoclonal cells. The monoclonal expansion of the cells suggested that not only SNT-8 cells but also SNT-13 and SNT-15 cells were transformed to a certain degree and acquired a greater proliferative capacity compared with normal T cells. Many studies have documented monoclonal and oligoclonal proliferation of EBV-positive T cells and NK cells in patients with CAEBV. 11,12,17,19,20,24,25 In addition, the development of malignant lymphomas of T/NK-cell lineage from patients with CAEBV has been reported. 9,14,18,19,23,25,42 Furthermore, the establishment of cell lines of αβ T-cell 16 and NK-cell lineage 21 has been reported in some severe CAEBV cases, and the present study described the establishment of γδ T-cell lines. Therefore, the present study, together with those previous reports, suggests that most cases of CAEBV are considered as lymphoproliferative diseases of NK cells, αβ T cells, or γδ T cells, rather than as infectious diseases.
The most interesting finding in the present study was that the EBV-positive γδ T-cell lines SNT-8, SNT-13, and SNT-15 expressed exactly the same TCR, Vγ9-JγP/Vδ2-Jδ3. This was, on the other hand, not observed in the EBV-negative γδ T-cell lines, which were established from T-lymphoblastic leukemia patients and expressed Vδ1 protein with the Vδ1-Jδ1 gene rearrangement. Consistent with our observation, Arnulf and colleagues 4 have reported that five EBV-positive cases (three nasal, one laryngeal, and one intestinal lymphomas) in a series of nonhepatosplenic γδ T-cell lymphomas were invariably Vδ2+, whereas five EBV-negative or -weakly positive cases expressed one of the Vδ1 (one case in the lung), Vδ2 (two cases in the skin), or Vδ3 (two cases in the gastrointestinal tract) chain. Thus, it was suggested that EBV has a close affinity to Vδ2+ cells in the lymphoproliferative diseases, especially to Vγ9-JγP/Vδ2-Jδ3+ cells.
It is of interest when and where EBV affected the Vγ9-JγP/Vδ2-Jδ3+ cells in our patients. It is known that the usage of TCRs in human γδ T cells has limited diversity, and the type of cells that constitute the majority of changes during fetal and postnatal life in individual organs. 1 For example, in the postnatal thymus and other epithelial-rich tissues such as the intestine and spleen, Vδ1-Jδ1+ cells are the major population. 1,43,44 It is thus reasonable that cases of γδ T-cell acute lymphoblastic leukemia, cells of which are supposed to be the malignant counterpart of thymocytes, expressed Vδ1-Jδ1 TCR. 45 Likewise, most hepatosplenic γδ T-cell lymphomas were reported to express Vδ1 proteins and/or rearranged Vδ1 genes. 40,41,46,47 In contrast, Vδ2+ cells are known to be the major population in the peripheral blood during adulthood. 1,43,44 In the peripheral blood, Vδ1+ cells constitute the majority at birth, and Vδ2+ cells increase from a mean of 20% in the cord blood to ∼60 to 70% before the age of 10. 44 In addition, Vγ9-JγP/Vδ2+ cells, which are capable of recognizing mycobacterium and Daudi Burkitt’s lymphoma cells, 48,49 are believed to be selected and expand in number in the periphery. 1 With regard to the nose, the major population among the nasal intraepithelial γδ T cells was reported to express Vγ1/Vδ1 genes. 50 Thus, it is likely that EBV affected the Vγ9-JγP/Vδ2-Jδ3+ cells in the peripheral blood in our patients with nasal T-cell lymphoma and CAEBV. Nonetheless, although Vδ2-Jδ3+ cells constitute the major population in cord blood, Vδ2-Jδ1+ cells comprise the largest population, ∼50 to 98% of γδ T cells, in the peripheral blood of healthy adults. 1,43 Therefore, Vγ9-JγP/Vδ2-Jδ3+ cells are a minor population in the peripheral blood in adulthood in the context of Jδ3 expression, although probably more common in infants and youth. Thus, it may be reasonable that EBV affected these specific γδ T cells in the peripheral blood of our two patients with CAEBV because they were young people. It is unknown, however, why EBV affected these specific γδ T cells in SNT-8 because the patient was 48-years-old. It may be that EBV has particular affinity to the Jδ3 TCR. Otherwise, EBV might affect Vγ9-JγP/Vδ2-Jδ3+ cells when the patient is young, and the cells transform later to form the nasal T-cell lymphoma.
It is thus an intriguing question how EBV affected the Vγ9-JγP/Vδ2-Jδ3+ cells in our patients. A possible explanation for the close affinity between EBV and this specific cell population is that there is a specific receptor for EBV other than CD21 on the Vγ9-JγP/Vδ2-Jδ3+ cells because the three EBV+ cell lines were negative for the CD21 antigen, a known receptor for EBV. 51,52 One approach to assessing such a possibility is to study whether isolated Vγ9-JγP/Vδ2-Jδ3+ cells can be infected with EBV.
Alternatively, the relationship between EBV and Vγ9-JγP/Vδ2-Jδ3+ cells may be explained as follows: the population of these cells expanded in response to a certain antigen and infiltrated a tissue to then be infected with resident EBV. Indeed, Vγ9-JγP-Cγ1/Vδ2+ cells were reported to have a great ability to respond to Daudi cells or mycobacterial antigens and comprise a large proportion of the γδ T cells in the peripheral blood. 49 This also suggests that they are highly responsive to antigens structurally identical or related to Daudi or mycobacterial antigens. Alternatively, Vγ9-JγP/Vδ2-Jδ3+ cells might expand in number during EBV infection because γδ T cells were reported to increase in acute and convalescent phases of infectious mononucleosis. 53,54 Therefore, it seems to be worth investigating how Vγ9-JγP/Vδ2-Jδ3+ cells react with Daudi antigens, mycobacterial antigens, or EBV.
Another possible mechanism for the expansion of the Vγ9-JγP/Vδ2-Jδ3+ cell population in our three patients is that EBV affected immature T-lymphocytes and drove them to differentiate into this population. Indeed, it has been reported that EBV infects immature T lymphocytes. Imai and co-workers 16 established four EBV-positive T-cell lines from patients with CAEBV, and one of the four had an immature T-cell phenotype without TCR gene rearrangement. It has also been reported that immature thymocytes and an immature T-cell line HPB-ALL expressed CD21 and could be infected with EBV. 55,56 Moreover, it has been shown that infections of EBV in HPB-ALL altered the expression of T-cell surface molecules involved in antigen recognition. It has thus been suggested that EBV infection of T cells at early stage of differentiation may lead to a failure in the development of the normal T-cell repertoire. 56 Therefore, we believe that the effect of EBV infection of immature thymocytes on the development of the T-cell repertoire is an important issue to be addressed.
The present study raised another important question as to why the infection by EBV of an identical γδ T-cell subset resulted in different diseases, ie, nasal T/NK-cell lymphoma and CAEBV; the former is a malignant lymphoma and the latter a nonmalignant lymphoproliferative disease. We recently reported a patient with CAEBV of NK-cell lineage who subsequently developed nasal NK-cell lymphoma. 25 There were three NK-cell clones in the patient, and it was suggested that one of the three was involved in the formation of the malignant lymphoma. We concluded that the type and/or degree of transformation of EBV-positive NK cells could be variable even in a single patient. Similar conditions showing variability in the transformation of EBV-positive cells have been reported in the posttransplantation lymphoproliferative disorders (PT-LPD) of B lymphocytes. 57-61 These reports hypothesized that the development of PT-LPD is a multistep process, initiates as polyclonal expansions of EBV-carrying B cells on the basis of immunosuppression, and may progress to lymphomas. The present study together with these previous reports, suggests that transformation of cells with EBV only is not sufficient for the development of a malignant lymphoma, and that an additional abnormality such as genetic mutation is likely to be involved in the malignant transformation of EBV-affected cells.
In conclusion, this study demonstrated that the three EBV-positive γδ T-cell lines expressed a common TCR, the Vγ9-JγP/Vδ2-Jδ3 TCR, and shed light on the etiology of nasal T-cell lymphoma and CAEBV of the γδ T-cell subset. The establishment of the cell lines allowed us to focus on these diseases. Thus, we believe that the results warrant the use of these cell lines in future studies, and necessitate further efforts to develop more cell lines from lymphoproliferative diseases caused by EBV.
Acknowledgments
We thank Dr. Junko H. Ohyashiki for valuable discussion and to Mrs. Aya Kasuya for technical assistance.
Footnotes
Address reprint requests to Dr. Norio Shimizu, Department of Virology, Division of Medical Science, Medical Research Institute, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo 113-8510, Japan. E-mail: nshivir@mri.tmd.ac.jp.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan (no. 11670207 and no. 09253103), the Sasakawa Scientific Research Grant from the Japan Science Society, and Grants-in-Aid for Education and Research Promotion Program 1999 from the Tokyo Medical and Dental University.
References
- 1.McVay LD, Carding SR: Generation of human γδ T-cell repertoires. Crit Rev Immunol 1999, 19:431-460 [PubMed] [Google Scholar]
- 2.Chan JK: Peripheral T-cell and NK-cell neoplasms: an integrated approach to diagnosis. Mod Pathol 1999, 12:177-199 [PubMed] [Google Scholar]
- 3.Jaffe ES, Krenacs L, Kumar S, Kingma DW, Raffeld M: Extranodal peripheral T-cell and NK-cell neoplasms. Am J Clin Pathol 1999, 111:S46-S55 [PubMed] [Google Scholar]
- 4.Arnulf B, Copie-Bergman C, Delfau-Larue MH, Lavergne-Slove A, Bosq J, Wechsler J, Wassef M, Matuchansky C, Epardeau B, Stern M, Bagot M, Reyes F, Gaulard P: Nonhepatosplenic γδ T-cell lymphoma: a subset of cytotoxic lymphomas with mucosal or skin localization. Blood 1998, 91:1723-1731 [PubMed] [Google Scholar]
- 5.Kanavaros P, Lescs MC, Briere J, Divine M, Galateau F, Joab I, Bosq J, Farcet JP, Reyes F, Gaulard P: Nasal T-cell lymphoma: a clinicopathologic entity associated with peculiar phenotype and with Epstein-Barr virus. Blood 1993, 81:2688-2695 [PubMed] [Google Scholar]
- 6.Harabuchi Y, Imai S, Wakashima J, Hirao M, Kataura A, Osato T, Kon S: Nasal T-cell lymphoma causally associated with Epstein-Barr virus: clinicopathologic, phenotypic, and genotypic studies. Cancer 1996, 77:2137-2149 [DOI] [PubMed] [Google Scholar]
- 7.Nagata H, Konno A, Kimura N, Zhang Y, Kimura M, Demachi A, Sekine T, Yamamoto K, Shimizu N: Characterization of novel natural killer (NK)-cell and γδ T-cell lines established from primary lesions of nasal T/NK-cell lymphomas associated with the Epstein-Barr virus. Blood 2001, 97:708-713 [DOI] [PubMed] [Google Scholar]
- 8.Kikuta H, Taguchi Y, Tomizawa K, Kojima K, Kawamura N, Ishizaka A, Sakiyama Y, Matsumoto S, Imai S, Kinoshita T, Koizumi S, Osato T, Kobayashi I, Hamada I, Hirai K: Epstein-Barr virus genome-positive T lymphocytes in a boy with chronic active EBV infection associated with Kawasaki-like disease. Nature 1988, 333:455-457 [DOI] [PubMed] [Google Scholar]
- 9.Jones JF, Shurin S, Abramowsky C, Tubbs RR, Sciotto CG, Wahl R, Sands J, Gottman D, Katz BZ, Sklar J: T-cell lymphomas containing Epstein-Barr viral DNA in patients with chronic Epstein-Barr virus infections. N Engl J Med 1988, 318:733-741 [DOI] [PubMed] [Google Scholar]
- 10.Jaffe ES, Chan JK, Su IJ, Frizzera G, Mori S, Feller AC, Ho FC: Report of the Workshop on Nasal and Related Extranodal Angiocentric T/Natural Killer Cell Lymphomas. Definitions, differential diagnosis, and epidemiology. Am J Surg Pathol 1996, 20:103-111 [DOI] [PubMed] [Google Scholar]
- 11.Kawa-Ha K, Ishihara S, Ninomiya T, Yumura-Yagi K, Hara J, Murayama F, Tawa A, Hirai K: CD3-negative lymphoproliferative disease of granular lymphocytes containing Epstein-Barr viral DNA. J Clin Invest 1989, 84:51-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishihara S, Tawa A, Yumura-Yagi K, Murata M, Hara J, Yabuuchi H, Hirai K, Kawa-Ha K: Clonal T-cell lymphoproliferation containing Epstein-Barr (EB) virus DNA in a patient with chronic active EB virus infection. Jpn J Cancer Res 1989, 80:99-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukunaga Y, Asano T, Takehana J, Ambo K, Matsuoka K, Yamamoto M: CD3-negative lymphoproliferative disease of granular lymphocytes in a girl with an unusual pattern of anti-Epstein-Barr virus antibodies. Eur J Pediatr 1994, 153:894-897 [DOI] [PubMed] [Google Scholar]
- 14.Tanaka Y, Sasaki Y, Kurozumi H, Hyodo Y, Nishi T, Nakatani Y, Imai S, Osato T: Angiocentric immunoproliferative lesion associated with chronic active Epstein-Barr virus infection in an 11-year-old boy. Clonotopic proliferation of Epstein-Barr virus-bearing CD4+ T lymphocytes. Am J Surg Pathol 1994, 18:623-631 [DOI] [PubMed] [Google Scholar]
- 15.Kanegane H, Wado T, Nunogami K, Seki H, Taniguchi N, Tosato G: Chronic persistent Epstein-Barr virus infection of natural killer cells and B cells associated with granular lymphocytes expansion. Br J Haematol 1996, 95:116-122 [DOI] [PubMed] [Google Scholar]
- 16.Imai S, Sugiura M, Oikawa O, Koizumi S, Hirao M, Kimura H, Hayashibara H, Terai N, Tsutsumi H, Oda T, Chiba S, Osato T: Epstein-Barr virus (EBV)-carrying and -expressing T-cell lines established from severe chronic active EBV infection. Blood 1996, 87:1446-1457 [PubMed] [Google Scholar]
- 17.Ishihara S, Okada S, Wakiguchi H, Kurashige T, Hirai K, Kawa-Ha K: Clonal lymphoproliferation following chronic active Epstein-Barr virus infection and hypersensitivity to mosquito bites. Am J Hematol 1997, 54:276-281 [DOI] [PubMed] [Google Scholar]
- 18.Kanegane H, Bhatia K, Gutierrez M, Kaneda H, Wada T, Yachie A, Seki H, Arai T, Kagimoto S, Okazaki M, Oh-ishi T, Moghaddam A, Wang F, Tosato G: A syndrome of peripheral blood T-cell infection with Epstein-Barr virus (EBV) followed by EBV-positive T-cell lymphoma. Blood 1998, 91:2085-2091 [PubMed] [Google Scholar]
- 19.Ohshima K, Suzumiya J, Sugihara M, Nagafuchi S, Ohga S, Kikuchi M: Clinicopathological study of severe chronic active Epstein-Barr virus infection that developed in association with lymphoproliferative disorder and/or hemophagocytic syndrome. Pathol Int 1998, 48:934-943 [DOI] [PubMed] [Google Scholar]
- 20.Ohga S, Kimura N, Takada H, Nagano M, Ohshima K, Nomura A, Muraoka K, Take H, Yamamori S, Hara T: Restricted diversification of T-cells in chronic active Epstein-Barr virus infection: potential inclination to T-lymphoproliferative disease. Am J Hematol 1999, 61:26-33 [DOI] [PubMed] [Google Scholar]
- 21.Tsuge I, Morishima T, Morita M, Kimura H, Kuzushima K, Matsuoka H: Characterization of Epstein-Barr virus (EBV)-infected natural killer (NK) cell proliferation in patients with severe mosquito allergy; establishment of an IL-2-dependent NK-like cell line. Clin Exp Immunol 1999, 115:385-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamura T, Hatsukawa Y, Arai H, Inoue M, Kawa K: Blood stem-cell transplantation for chronic active Epstein-Barr virus with lymphoproliferation. Lancet 2000, 356:223-224 [DOI] [PubMed] [Google Scholar]
- 23.Quintanilla-Martinez L, Kumar S, Fend F, Reyes E, Teruya-Feldstein J, Kingma DW, Sorbara L, Raffeld M, Straus SE, Jaffe ES: Fulminant EBV(+) T-cell lymphoproliferative disorder following acute/chronic EBV infection: a distinct clinicopathologic syndrome. Blood 2000, 96:443-451 [PubMed] [Google Scholar]
- 24.Kimura H, Hoshino Y, Kanegane H, Tsuge I, Okamura T, Kawa K, Morishima T: Clinical and virologic characteristics of chronic active Epstein-Barr virus infection. Blood 2001, 98:280-286 [DOI] [PubMed] [Google Scholar]
- 25.Nagata H, Numata T, Konno A, Mikata I, Kurasawa K, Hara S, Nishimura M, Yamamoto K, Shimizu N: Presence of natural killer-cell clones with variable proliferative capacity in chronic active Epstein-Barr virus infection. Pathol Int 2001, 51:778-785 [DOI] [PubMed] [Google Scholar]
- 26.Straus SE: The chronic mononucleosis syndrome. J Infect Dis 1988, 157:405-412 [DOI] [PubMed] [Google Scholar]
- 27.Matsuo Y, Minowada J: Human leukemia cell lines—clinical and theoretical significances. Hum Cell 1988, 1:263-274 [PubMed] [Google Scholar]
- 28.Ravid Z, Goldblum N, Zaizov R, Schlesinger M, Kertes T, Minowada J, Verbi W, Greaves M: Establishment and characterization of a new leukaemic T-cell line (Peer) with an unusual phenotype. Int J Cancer 1980, 25:705-710 [DOI] [PubMed] [Google Scholar]
- 29.Ben-Bassat H, Shlomai Z, Kohn G, Prokocimer M: Establishment of a human T-acute lymphoblastic leukemia cell line with a (16;20) chromosome translocation. Cancer Genet Cytogenet 1990, 49:241-248 [DOI] [PubMed] [Google Scholar]
- 30.Raab-Traub N, Flynn K: The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell 1986, 47:883-889 [DOI] [PubMed] [Google Scholar]
- 31.Sims JE, Tunnacliffe A, Smith WJ, Rabbitts TH: Complexity of human T-cell antigen receptor β-chain constant- and variable-region genes. Nature 1984, 312:541-545 [DOI] [PubMed] [Google Scholar]
- 32.Quertermous T, Strauss W, Murre C, Dialynas DP, Strominger JL, Seidman JG: Human T-cell γ genes contain N segments and have marked junctional variability. Nature 1986, 322:184-187 [DOI] [PubMed] [Google Scholar]
- 33.Tkachuk DC, Griesser H, Takihara Y, Champagne E, Minden M, Feller AC, Lennert K, Mak TW: Rearrangement of T-cell δ locus in lymphoproliferative disorders. Blood 1988, 72:353-357 [PubMed] [Google Scholar]
- 34.Korsmeyer SJ, Arnold A, Bakhshi A, Ravetch JV, Siebenlist U, Hieter PA, Sharrow SO, LeBien TW, Kersey JH, Poplack DG, Leder P, Waldmann TA: Immunoglobulin gene rearrangement and cell surface antigen expression in acute lymphocytic leukemias of T cell and B cell precursor origins. J Clin Invest 1983, 71:301-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takihara Y, Tkachuk D, Michalopoulos E, Champagne E, Reimann J, Minden M, Mak TW: Sequence and organization of the diversity, joining, and constant region genes of the human T-cell δ-chain locus. Proc Natl Acad Sci USA 1988, 85:6097-6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huck S, Lefranc MP: Rearrangements to the JP1, JP and JP2 segments in the human T-cell rearranging gamma gene (TRGγ) locus. FEBS Lett 1987, 224:291-296 [DOI] [PubMed] [Google Scholar]
- 37.Triebel F, Faure F, Graziani M, Jitsukawa S, Lefranc MP, Hercend T: A unique V-J-C-rearranged gene encodes a gamma protein expressed on the majority of CD3+ T cell receptor-α/β− circulating lymphocytes. J Exp Med 1988, 167:694-699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breit TM, Wolvers-Tettero IL, Beishuizen A, Verhoeven MA, van Wering ER, van Dongen JJ: Southern blot patterns, frequencies, and junctional diversity of T-cell receptor-delta gene rearrangements in acute lymphoblastic leukemia. Blood 1993, 82:3063-3074 [PubMed] [Google Scholar]
- 39.Kimura N, Akiyoshi T, Uchida T, Okamura J, Kozuru M, Nishimura J, Ohyashiki JH, Kikuchi M, Niho Y, Okumura M: High prevalence of T cell receptor Dδ2(Dδ)Jδ rearrangement in CD7-positive early T cell acute lymphoblastic leukemia. Leukemia 1996, 10:650-657 [PubMed] [Google Scholar]
- 40.Gaulard P, Bourquelot P, Kanavaros P, Haioun C, Le Couedic JP, Divine M, Goossens M, Zafrani ES, Farcet JP, Reyes F: Expression of the α/β and γ/δ T-cell receptors in 57 cases of peripheral T-cell lymphomas. Identification of a subset of γ/δ T-cell lymphomas. Am J Pathol 1990, 137:617-628 [PMC free article] [PubMed] [Google Scholar]
- 41.Mastovich S, Ratech H, Ware RE, Moore JO, Borowitz MJ: Hepatosplenic T-cell lymphoma: an unusual case of a γδ T-cell lymphoma with a blast-like terminal transformation. Hum Pathol 1994, 25:102-108 [DOI] [PubMed] [Google Scholar]
- 42.Bonagura VR, Katz BZ, Edwards BL, Valacer DJ, Nisen P, Gloster E, Mir R, Lanzkowsky P: Severe chronic EBV infection associated with specific EBV immunodeficiency and an EBNA+ T-cell lymphoma containing linear, EBV DNA. Clin Immunol 1990, 57:32-44 [DOI] [PubMed] [Google Scholar]
- 43.Casorati G, De Libero G, Lanzavecchia A, Migone N: Molecular analysis of human gamma/delta+ clones from thymus and peripheral blood. J Exp Med 1989, 170:1521-1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker CM, Groh V, Band H, Porcelli SA, Morita C, Fabbi M, Glass D, Strominger JL, Brenner MB: Evidence for extrathymic changes in the T cell receptor gamma/delta repertoire. J Exp Med 1990, 171:1597-1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langerak AW, Wolvers-Tettero IL, van den Beemd MW, van Wering ER, Ludwig WD, Hahlen K, Necker A, van Dongen JJ: Immunophenotypic and immunogenotypic characteristics of TCRgammadelta+ T cell acute lymphoblastic leukemia. Leukemia 1999, 13:206-214 [DOI] [PubMed] [Google Scholar]
- 46.Ohno T, Komada F, Yamaguchi M, Oka K, Nishii K, Tsuda M, Katsuta K, Yamaguchi T, Kita K, Shirakawa S: Gamma/delta T-cell lymphoma with hepatosplenomegaly: report of a case. Int J Hematol 1993, 57:269-276 [PubMed] [Google Scholar]
- 47.Yamaguchi M, Ohno T, Nakamine H, Oka K, Matsuzuka F, Miwa H, Shiku H, Kimura N, Nanba K, Kita K: Gamma delta T-cell lymphoma: a clinicopathologic study of 6 cases including extrahepatosplenic type. Int J Hematol 1999, 69:186-195 [PubMed] [Google Scholar]
- 48.Ohmen JD, Barnes PF, Uyemura K, Lu SZ, Grisso CL, Modlin RL: The T cell receptors of human gamma delta T cells reactive to Mycobacterium tuberculosis are encoded by specific V genes but diverse V-J junctions. J Immunol 1991, 147:3353-3359 [PubMed] [Google Scholar]
- 49.Davodeau F, Peyrat MA, Hallet MM, Gaschet J, Houde I, Vivien R, Vie H, Bonneville M: Close correlation between Daudi and mycobacterial antigen recognition by human gamma delta T cells and expression of V9JPC1 gamma/V2DJC delta-encoded T cell receptors. J Immunol 1993, 151:1214-1223 [PubMed] [Google Scholar]
- 50.Pawankar RU, Okuda M, Suzuki K, Okumura K, Ra C: Phenotypic and molecular characteristics of nasal mucosal gamma delta T cells in allergic and infectious rhinitis. Am J Respir Crit Care Med 1996, 153:1655-1665 [DOI] [PubMed] [Google Scholar]
- 51.Moore MD, Cooper NR, Tack BF, Nemerow GR: Molecular cloning of the cDNA encoding the Epstein-Barr virus/C3d receptor (complement receptor type 2) of human B lymphocytes. Proc Natl Acad Sci USA 1987, 84:9194-9198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanner J, Weis J, Fearon D, Whang Y, Kieff E: Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 1987, 50:203-213 [DOI] [PubMed] [Google Scholar]
- 53.De Paoli P, Gennari D, Martelli P, Cavarzerani V, Comoretto R, Santini G: Gamma delta T cell receptor-bearing lymphocytes during Epstein-Barr virus infection. J Infect Dis 1990, 161:1013-1016 [DOI] [PubMed] [Google Scholar]
- 54.Hassan J, Feighery C, Bresnihan B, Whelan A: Elevated T cell receptor gamma delta + T cells in patients with infectious mononucleosis. Br J Haematol 1991, 77:255-256 [DOI] [PubMed] [Google Scholar]
- 55.Paterson RL, Kelleher C, Amankonah TD, Streib JE, Xu JW, Jones JF, Gelfand EW: Model of Epstein-Barr virus infection of human thymocytes: expression of viral genome and impact on cellular receptor expression in the T-lymphoblastic cell line, HPB-ALL. Blood 1995, 85:456-464 [PubMed] [Google Scholar]
- 56.Paterson RL, Kelleher CA, Streib JE, Amankonah TD, Xu JW, Jones JF, Gelfand EW: Activation of human thymocytes after infection by EBV. J Immunol 1995, 154:1440-1449 [PubMed] [Google Scholar]
- 57.Klein G, Purtilo D: Summary: symposium on Epstein-Barr virus-induced lymphoproliferative diseases in immunodeficient patients. Cancer Res 1981, 41:4302-4304 [PubMed] [Google Scholar]
- 58.Hanto DW, Gajl-Peczalska KJ, Frizzera G, Arthur DC, Balfour HH, Jr, McClain K, Simmons RL, Najarian JS: Epstein-Barr virus (EBV) induced polyclonal and monoclonal B-cell lymphoproliferative diseases occurring after renal transplantation. Clinical, pathologic, and virologic findings and implications for therapy. Ann Surg 1983, 198:356-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shearer WT, Ritz J, Finegold MJ, Guerra IC, Rosenblatt HM, Lewis DE, Pollack MS, Taber LH, Sumaya CV, Grumet FC: Epstein-Barr virus-associated B-cell proliferations of diverse clonal origins after bone marrow transplantation in a 12-year-old patient with severe combined immunodeficiency. N Engl J Med 1985, 312:1151-1159 [DOI] [PubMed] [Google Scholar]
- 60.Cleary ML, Nalesnik MA, Shearer WT, Sklar J: Clonal analysis of transplant-associated lymphoproliferations based on the structure of the genomic termini of the Epstein-Barr virus. Blood 1988, 72:349-352 [PubMed] [Google Scholar]
- 61.Knowles DM, Cesarman E, Chadburn A, Frizzera G, Chen J, Rose EA, Michler RE: Correlative morphologic and molecular genetic analysis demonstrates three distinct categories of posttransplantation lymphoproliferative disorders. Blood 1995, 85:552-565 [PubMed] [Google Scholar]