Abstract
Hepatoma cell lines can produce a massive amount of chemokines in response to various stimuli including hepatitis viruses and their products. However, it remains elusive on the types of chemokine receptor(s) expressed in the hepatoma tissues and its roles in hepatoma development. To clarify these points, we examined the chemokine receptor expression in six human hepatoma cell lines. All of the hepatoma cell lines constitutively and exclusively expressed CCR1 mRNA and its protein on their cell surface. CCR1 expression was also detected on hepatoma cells and to a lesser degree, on endothelial cells in hepatoma tissues but not in normal liver tissues. Furthermore, CCL3 expression was detected in hepatoma cells, endothelial cells, and to a lesser degree, fibroblast-like cells in hepatoma tissue, whereas only occasional vascular endothelial cells and inflammatory cells in normal liver tissues were weakly positive for CCL3. Moreover, the forskolin-mediated increases in intracellular cAMP concentrations were inhibited by the ligands for CCR1, CCL3, CCL4, and CCL5, suggesting that the expressed CCR1 was functional. Four hepatoma cell lines produced CCL3 only in response to interleukin (IL)-1α and IL-1β. Finally, IL-1α and IL-1β were detected abundantly in hepatoma tissues but not in normal liver tissues. Thus, IL-1 may enhance the local production of CCL3, which may interact with CCR1 expressed on hepatoma cells, in an autocrine and/or paracrine manner.
Chemokines are a family of chemoattractant cytokines that can be classified into four groups based on the position of the first two cysteines adjacent to the amino-terminus, CXC, CC, C, and CX3C. 1-4 The biological effects of chemokines seem to be mediated through their interaction with a family of specific G protein-coupled seven-transmembrane domain receptors. 1-4 Based on their specific chemokine ligands, the receptor proteins are categorized as CC chemokine receptors (CCR), CXC chemokine receptors (CXCR), C chemokine receptor (CR), and CX3C chemokine receptor (CX3CR). 1-4 Until the present, nearly 20 chemokine receptors have been identified and each receptor mediates the functions of a corresponding chemokine(s) (http://cytokine.medic.kumamoto-u.ac.jp). A wide variety of tumor cells and normal cells can produce various chemokines under various conditions. Moreover, several lines of evidence suggest that produced chemokines have a crucial role in the progression of tumor cells by recruiting leukocyte infiltration and modulating tumor cell motility. 5-8
Persistent chronic infection with hepatitis B virus or hepatitis C virus frequently resulted in the development of hepatocellular carcinoma, which ranks as the eighth cause of death among human cancers and is endemic in Asia, Africa, and southern Europe. 9 We previously observed that the X protein of hepatitis B virus and the hepatitis C virus NS5A gene product can induce in vitro gene expression and protein production of interleukin (IL)-8/CXCL8, 10,11 one of CXC chemokines. Moreover, IL-8/CXCL8 protein was detected in hepatoma cells in some hepatoma tissues. 12 However, it still remains elusive whether receptors for IL-8/CXCL8 are expressed in hepatoma tissues. Moreover, little is currently known about the expression of other chemokine receptors in hepatoma tissues.
Hence, we examined chemokine receptor expression in hepatoma cell lines. CCR1 was the only chemokine receptor expressed consistently in all hepatoma cell lines that we investigated. Moreover, CCR1 and its ligand, CCL3/macrophage inflammatory protein-1α, were expressed abundantly in hepatoma cells in human hepatoma tissues. Furthermore, we provided evidence that a proinflammatory cytokine, IL-1, may regulate the expression of CCL3 in hepatoma tissues.
Materials and Methods
Reagents
Human recombinant CCL3/macrophage inflammatory protein-1α and an anti-human CCL3 monoclonal antibody (clone ANOC801) were generously provided by Ohtsuka Pharmaceutical Co., Ltd. (Tokushima, Japan). Human IL-1α and tumor necrosis factor (TNF)-α were kindly provided by Dainippon Pharmaceutical Company (Osaka, Japan). Rabbit anti-human CCR1 antibodies were prepared as described previously. 13-16 Human recombinant CCL4/macrophage inflammatory protein-1β, CCL5/regulated on activation normal T lymphocyte expressed and secreted (RANTES), CCL11/eotaxin, and IL-1β were purchased from Pepro Tech EC (London, UK). Monoclonal mouse anti-human IL-1α and IL-1β antibodies were purchased from Genzyme (Cambridge, MA) and R&D Systems, Inc. (Minneapolis, MN), respectively. Rabbit anti-human CCL4 antibodies were prepared by immunizing rabbits with recombinant human CCL4.
Cell Culture
Six human hepatoma-derived cell lines including HuH7 (differentiated hepatoma cell line), HepG2 (epithelial-like cell line), HLE and HLF (nondifferentiated epithelial cell lines), SK-Hep1 (endothelial cell origin), and Hep3B (hepatitis B virus-positive hepatocellular carcinoma cell line) were used in the experiments. 17-21 These cell lines were maintained in Dulbecco’s modified essential medium (DMEM; Sigma Chemical Co., St. Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Norcross, GA) in a humidified incubator at 37°C in 5% CO2.
Liver Samples
Liver specimens were surgically obtained from 15 individuals with their informed consent. Twelve of them were confirmed to be positive for antibodies against hepatitis C virus and presented with histological diagnosis of cirrhosis with hepatocellular carcinoma. The remaining three specimens were obtained from the surrounding hepatic parenchyma of metastatic liver cancers, in which no evidence of metastasis or chronic liver disease was diagnosed by at least two pathologists specializing in liver disease. These three specimens were also enrolled as controls. Study protocols were approved by the Ethical Committee of Kanazawa University, Graduate School of Medicine.
Reverse Transcriptase (RT)-Polymerase Chain Reaction (PCR)
Total RNAs were extracted from cultured human hepatoma cell lines at a subconfluence with the use of RNAzol (Tel-Test Inc., TX) according to the manufacturer’s instructions. Then the RNA preparations were treated with ribonuclease-free deoxyribonuclease (DNase) I (Life Technologies, Inc., Gaithersburg, MD) to remove residual genomic DNA. Two μg of total RNA were reverse-transcribed at 42°C for 1 hour in 20 μl of reaction mixture containing mouse Moloney leukemia virus reverse transcriptase (Toyobo, Osaka, Japan) and hexanucleotide random primers (Amersham Bioscience, Tokyo, Japan). Serially twofold diluted cDNA products were amplified for β-actin using the specific set of the primers (Table 1) ▶ with 27 cycles of 94°C for 30 seconds, 55°C for 1 minute, and 72°C for 1 minute, in 20 μl of reaction mixture containing Taq polymerase (Takara Shuzo, Kyoto, Japan) to evaluate the amount of the transcribed cDNA. Thereafter, equal amounts of cDNA products were amplified for chemokine and chemokine receptor genes using the specific sets of primers based on the reported sequences (Table 1) ▶ . As a negative control, PCRs were performed without reverse transcriptase treatment (no-RT-PCR). Total RNAs extracted from human peripheral blood mononuclear cells were used as a positive control for chemokine receptor mRNA expression. After the amplification, the resultant PCR products were fractionated on a 1.2% agarose gel and visualized by ethidium bromide staining under ultraviolet light transillumination.
Table 1.
Specific Sets of Primers and Conditions of PCR
| Primers | Nucleotide sequence (5′ → 3′) sense/anti-sense | Annealing temperature, ° | PCR Cycles |
|---|---|---|---|
| β-actin | CACTGTGTTGGCGTACAGGT | 55 | 27 |
| TCATCACCATTGGCAATGAG | |||
| CXCR1 | CATGTCAAATATTACAGATCCA | 55 | 35 |
| CCAGCAGCCAAGACAAACAAAC | |||
| CXCR2 | CATGGAGAGTGACAGCTTTG | 58 | 35 |
| GCTGGGCTAACATTGGATGA | |||
| CXCR3 | CCACCCACTGCCAATACAAC | 60 | 35 |
| CGGAACTTGACCCCTACAAA | |||
| CXCR4 | GGTGGTCTATGTTGGCGTCT | 57 | 35 |
| TCGATGCTGATCCCAATGTA | |||
| CXCR5 | GAGAGAACCTCACGCACCTC | 57 | 35 |
| TGCTTGGTCAAGATGACTGC | |||
| CCR1 | CTCTTCCTGTTCACGCTTCC | 57 | 35 |
| GCCTGAAACAGCTTCCACTC | |||
| CCR2 | ATGCTGTCCACATCTCGTTCTC | 55 | 35 |
| GGACAGAAGCAAACACAGCCAC | |||
| CCR3 | GTCATCACCAGCATCGTCAC | 57 | 35 |
| TCATGCAGCAGTGGGAGTAG | |||
| CCR5 | GGGATAGCACTGAGCAAAGC | 57 | 35 |
| GTTTTAGCCATCCCCCAAAT | |||
| CCL3/MIP-1α | CAGGTCTCCACTGCTGCC | 59 | 35 |
| CACTCAGCTCCAGGTCACT | |||
| CCL4/MIP-1β | CCAAACCAAAAGAAGCAAGC | 57 | 35 |
| ACAGTGGACCATCCCCATAG | |||
| CCL5/RANTES | ACCCAGCAGTCGTCTTTGTC | 57 | 35 |
| CTCCCAAAGTGCTGGGATTAC |
Immunocytochemical and Immunohistochemical Analysis
For an immunocytochemical analysis of hepatoma cell lines, cultured hepatoma cells were seeded into the wells of a Lab-Tec chamber slide with eight wells (Nalge Nunc International Corp., Naperville, IL) at 2 × 104 cells/well. The cells were cultured for 1 to 2 days in a 37°C incubator with 5% CO2 and then subjected to immunocytochemical study. The cells in the wells were fixed by sequential treatment with 4% paraformaldehyde for 15 minutes at room temperature and with a freshly prepared mixture of methanol/acetone (1:1) for 15 minutes. For an immunohistochemical analysis of tissue samples, paraffin-embedded sections were deparaffinized with xylene and graded concentrations of ethanol. After treatment with 0.3% (v/v) hydrogen peroxide in methanol, sections were incubated sequentially with an avidin/biotin blocking kit (Vector Laboratories Inc., Burlingame, CA) and with goat normal serum. For the detection of CCR1 protein expression, the slides were incubated sequentially with primary rabbit anti-human CCR1 antibody (3 μg/ml) at 4°C overnight and with goat anti-rabbit IgG (Vector Laboratories, Inc.) for 30 minutes at room temperature. Immune complexes were detected with the use of the Vectastain Elite ABC kit (Vector Laboratories, Inc.) according to the manufacturer’s instructions. For the detection of CCL3, IL-1α, and IL-1β proteins, the slides were sequentially incubated with mouse anti-human CCL3 monoclonal antibody (0.5 μg/ml) for 1 hour at room temperature, mouse anti-human IL-1α (3 μg/ml), and mouse anti-human IL-1β (2 μg/ml) antibodies overnight, respectively. Then, the slides were incubated with Envision (goat anti-mouse IgG; DAKO, Tokyo, Japan) for 30 minutes at room temperature. The resultant immune complexes were detected with the use of DAB kit (DAKO). Before being mounted, the slides were counterstained with hematoxylin. Negative and isotype controls were also performed in the same condition. An observer without a prior knowledge on the histological diagnosis examined at least randomly chosen five fields of the tissue sections at ×400 magnification and scored the immune reactions as follows: −, less than 10% cells were positive; +, 10 to 20% cells exhibited positive reactions; ++, 20 to 50% cells showed positive reactions; +++, more than 50% showed positive reactions.
Flow Cytometric Analysis of the Hepatoma Cell Lines Cell-Surface CCR1 Expression
Cultured hepatoma cells were harvested and washed two times in washing buffer (phosphate-buffered saline, pH 7.4, 0.1% w/v bovine serum albumin, 0.1% w/v sodium azide) and were first incubated in a washing buffer containing 5% normal goat serum to block nonspecific binding. For antibody staining, 5 to 10 × 105 cells were incubated with 20 μg/ml of rabbit anti-human CCR1 or the same concentration of normal rabbit IgG as a control. After being washed twice with washing buffer, the cells were incubated with fluorescein isothiocyanate-conjugated F(ab′)2 goat anti-rabbit IgG (5 μg/ml; Caltag Laboratories, Burlingame, CA). The cells were filtered thorough nylon mesh after being washed twice. All incubations were done for 20 minutes on ice followed by two washings in a washing buffer. Cells were gated based on forward and side scatter and propidium-iodine-positive dead cells were excluded from the analysis. A total of 1 × 104-gated events was collected from each flow cytometry sample.
CCL3 Protein Production and mRNA Expression by Hepatoma Cell Lines
Hepatoma cell lines were suspended in DMEM supplemented with 10% fetal bovine serum at a cell concentration of 1 × 105 cells/ml. One ml of cell suspension was incubated in the absence or presence of the designated concentrations of human IL-1α, IL-1β, TNF-α, and lipopolysaccharide for 24 hours. Supernatants were collected after centrifugation at 1,500 × g to remove particles, and aliquots were stored frozen at −70°C until use in enzyme-linked immunosorbent assay. CCL3 concentrations in the supernatants were determined with the use of human CCL3 assay kit (Genzyme) according to the manufacturer’s instructions. Total RNAs were extracted from hepatoma cell lines cultured for 6 hours in the presence or the absence of either IL-1α or IL-1β (100 ng/ml) and subjected to RT-PCR as described above.
Determination of Intracellular cAMP Levels
Approximately 1 × 105 Hep3B cells were suspended in 1 ml of DMEM supplemented with 10% fetal bovine serum and seeded into each well of 24-multiwell plates. After incubation overnight, the medium was changed to serum-free DMEM. After the incubation for additional 24 hours, the medium was again removed. Then, 0.5 ml of serum-free DMEM was added to each well in the presence or the absence of 10 μmol of forskolin (Wako Pure Chemical Industries, Osaka, Japan) in the presence of the indicated concentrations of CCR1 ligands or CCR1 ligands with 10 μg/ml of anti-CCR1. The effects of CCL11/eotaxin were also examined in parallel. After incubation at 37°C for 20 minutes, the cells were lysed and intracellular cAMP levels were determined with the use of Biotrak cAMP enzyme immunoassay system (Amersham Bioscience) according to the manufacturer’s instructions.
Statistical Analysis
All data were expressed as mean ± SD and were analyzed statistically with analysis of variance. P < 0.05 was considered to be significant.
Results
Chemokine Receptor Gene Expression of Hepatoma Cell Lines
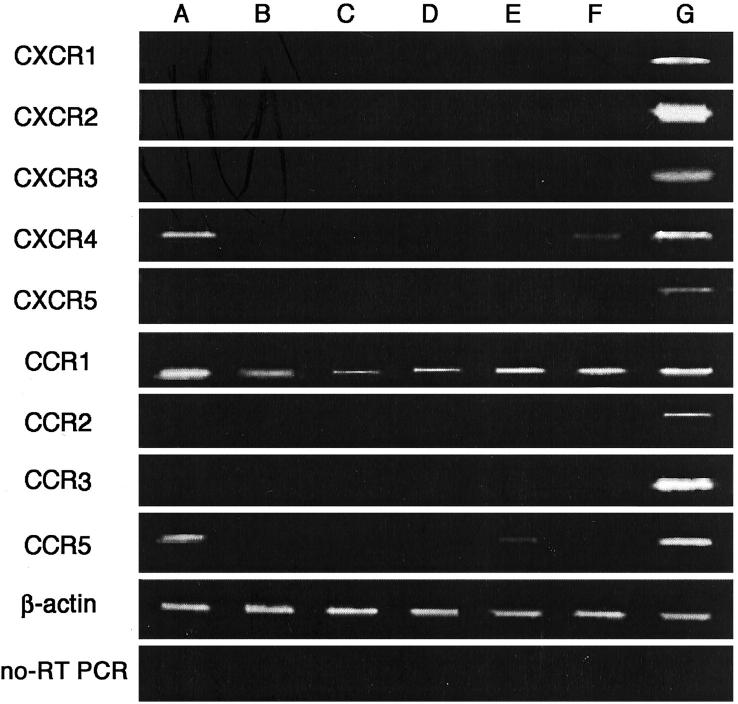
Several lines of evidence suggest the potential involvement of chemokines and their receptors in the invasion and metastatic processes of tumors. Hence, we first examined the gene expression of chemokine receptors in several hepatoma cell lines with the use of RT-PCR. Among the chemokine receptor genes that we examined, CCR1 mRNA was detected in all hepatoma cell lines (Figure 1) ▶ . In addition, two hepatoma cell lines (HuH7 and Hep3B) expressed CXCR4 mRNA, while HuH7 and HLF cells expressed CCR5 mRNA. To exclude the possibility that contaminated genomic DNA gave rise to the generation of the amplified bands, we used total RNA samples that were treated with DNase. Moreover, we performed RT-PCR under the same conditions except that RT was omitted from the reaction and did not detect any bands at all (Figure 1 ▶ , bottom). Thus, although the coding region of most chemokine receptor genes consists of one exon, the amplified bands derived from the expressed transcript but not the contaminated genomic DNA.
Figure 1.
Expression of chemokine receptor mRNAs of human hepatoma cell lines by RT-PCR. Total RNA was extracted from HuH7 (A), HepG2 (B), HLE (C), SK-Hep1 (D), HLF (E), and Hep3B (F) hepatoma cell lines and human peripheral blood mononuclear cells (G) and amplified by RT-PCR to detect chemokine receptor mRNAs as described in Materials and Methods. As a positive control, RNA from peripheral blood mononuclear cells was used. Analysis for CCR1 expression was performed without reverse transcriptase treatment and the results are shown as no-RT-PCR in the lowest panel. β-actin served as a control for sample loading.
CCR1 Protein Expression by Hepatoma Cell Lines
We next examined by immunocytochemical analysis whether hepatoma cell lines expressed CCR1 protein. We detected positive immune reactions in all of the cell lines that we examined (Figure 2A) ▶ . The use of control antibodies did not give rise to positive reactions at all (Figure 2A ▶ ; a, b, and c). Moreover, positive reactions were abolished completely when the primary antibodies were absorbed with the peptide used for the immunization (Figure 2A ▶ ; d, e, and f). These results implied that the observed immunoreactivities were specific. Furthermore, flow cytometric analysis also detected CCR1 expression on all hepatoma cell lines (Figure 2B) ▶ , indicating that CCR1 was expressed on the cell surface of these hepatoma cell lines.
Figure 2.
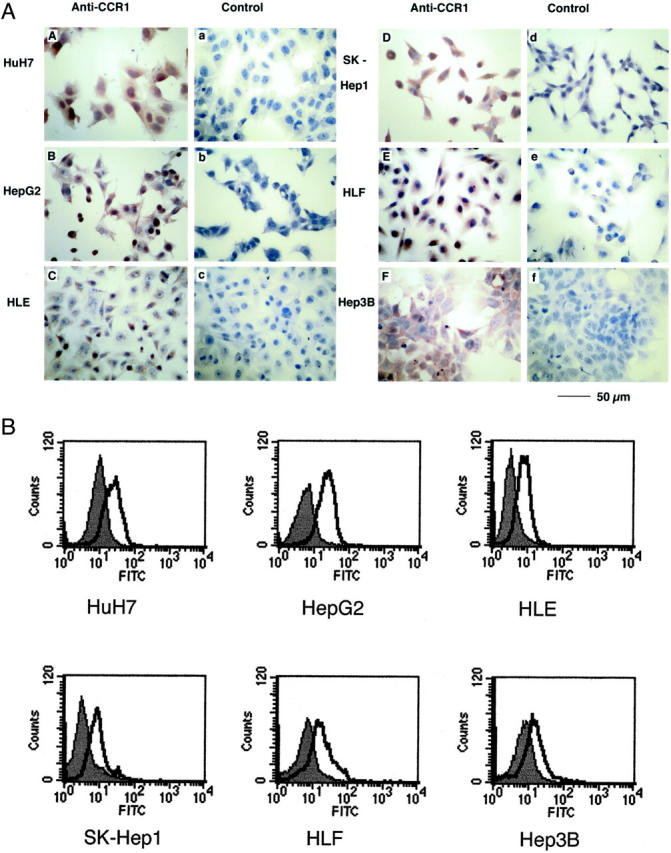
CCR1 protein expression of hepatoma cell lines. A: An immunocytochemical analysis of CCR1 protein expression in hepatoma cell lines. Localization of CCR1 in HuH7 (A), HepG2 (B), HLE (C), SK-Hep1 (D), HLF (E), and Hep3B (F) were determined immunocytochemically using specific polyclonal rabbit anti-human CCR1 antibodies as described in Materials and Methods. As a control, the first antibody was replaced by normal rabbit IgG (a, b, and c) or by primary antibodies absorbed with respective peptide antigen (d, e, and f). Scale bar, 50 μm; original magnifications, ×400. B: Flow cytometric analysis of cell-surface CCR1 expression by hepatoma cell lines. Cells (5 to 10 × 105) were incubated with rabbit anti-human CCR1 or normal rabbit IgG as an isotype control. Subsequently, cells were stained with fluorescein isothiocyanate-labeled goat anti-rabbit IgG. Open heavy-lined and filled histograms represent those with the test antibody (anti-CCR1) and the control IgG, respectively. The representative results from three independent experiments are shown.
Functionality of CCR1 Expressed on a Hepatoma Cell Line
Because most chemokine receptors are coupled with the Gα subunit and adenylyl cyclase, we examined the effects of chemokines on the forskolin-induced increase in intracellular cAMP levels. Forskolin induced a marked increase in intracellular cAMP levels in Hep3B cells (Figure 3A) ▶ . All three ligands for CCR1, CCL3, CCL4, and CCL5, reduced the forskolin-induced increases in intracellular cAMP levels (Figure 3A) ▶ . Because CCL5 can bind CCR3, we also examined the effect of CCL11/eotaxin, another specific ligand for CCR3. However, CCL11/eotaxin failed to decrease forskolin-induced cAMP accumulation (Figure 3B) ▶ . Furthermore, anti-CCR1 antibodies abrogated the effects of CCL3 (Figure 3B) ▶ . Similar results were obtained when HuH7 cells were used instead of Hep3B cells (data not shown). Because CCR1 but not CCR5 mRNA was detected in Hep3B cells, these results would indicate that Hep3B cells responded to CCL3, CCL4, and CCL5 through functional CCR1 on their surface.
Figure 3.
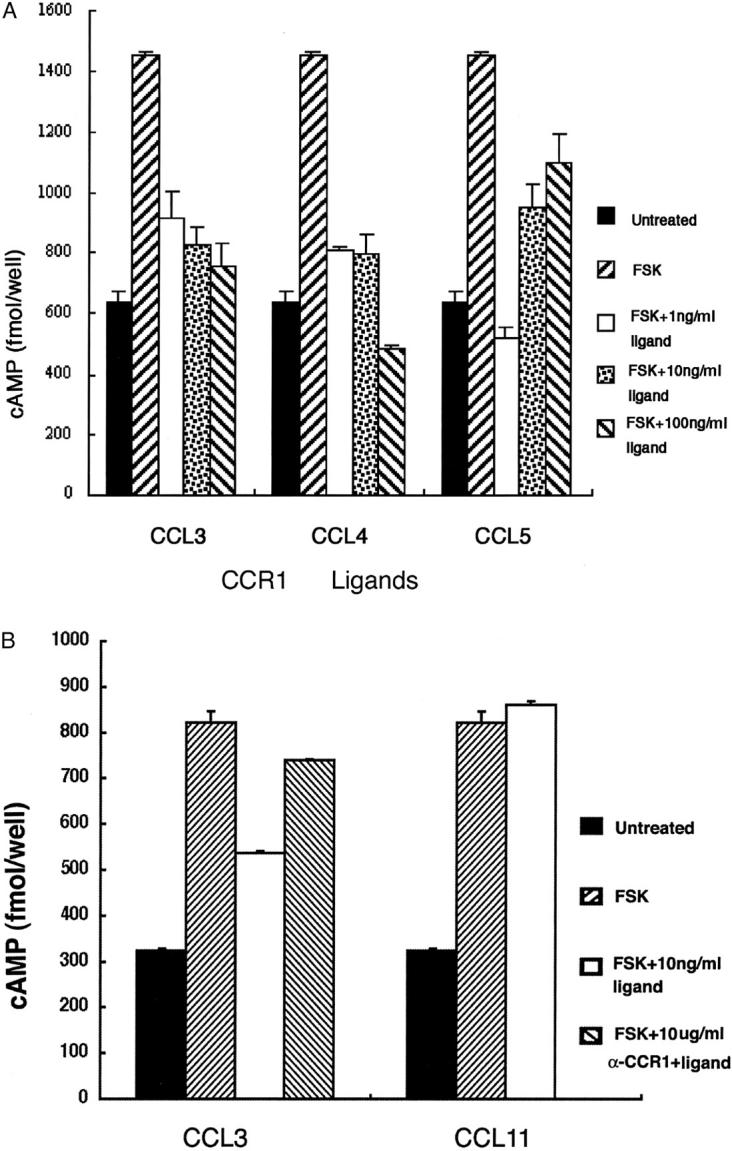
Effects of ligands for CCR1 on forskolin-stimulated cAMP production in a hepatoma cell line. A: Hep3B hepatoma cells were incubated for 20 minutes without (untreated) or with 10 μmol of forskolin at the indicated concentrations of human CCL3, CCL4, and CCL5. Then, intracellular cAMP levels were measured and are expressed in fmol/well. One representative result from three independent experiments is shown. Data are expressed as mean ± SD of data in triplicates. B: Hep3B cells were incubated for 20 minutes without (untreated) or with 10 μmol of forskolin in the absence or presence of 10 ng/ml of human CCL11 and CCL3 with or without preincubation with anti-CCR1 antibodies (10 μg/ml). Then, intracellular cAMP levels were measured and are expressed in fmol/well. One representative result from three independent experiments is shown. Data are expressed as mean ± SD of data in triplicates.
CCR1 Expression in Hepatoma Tissues
We next examined CCR1 protein expression in hepatoma tissues by an immunohistochemical analysis. We could not detect any CCR1 immunoreactivities in normal liver tissues (Figure 4D ▶ and Table 2 ▶ ). On the contrary, CCR1 immunoreactivities were detected on hepatoma tissues (Figure 4, A to C ▶ ; and Table 2 ▶ ). The observation at a higher magnification demonstrated the immunoreactivities in hepatoma cells and to a lesser degree, vascular endothelial cells and occasionally small bile duct epithelial cells (Figure 4, B and C ▶ ; and Table 2 ▶ ). However, only in a small number of cases (3 of 12), fibroblast-like cells were also weakly positive for CCR1.
Figure 4.
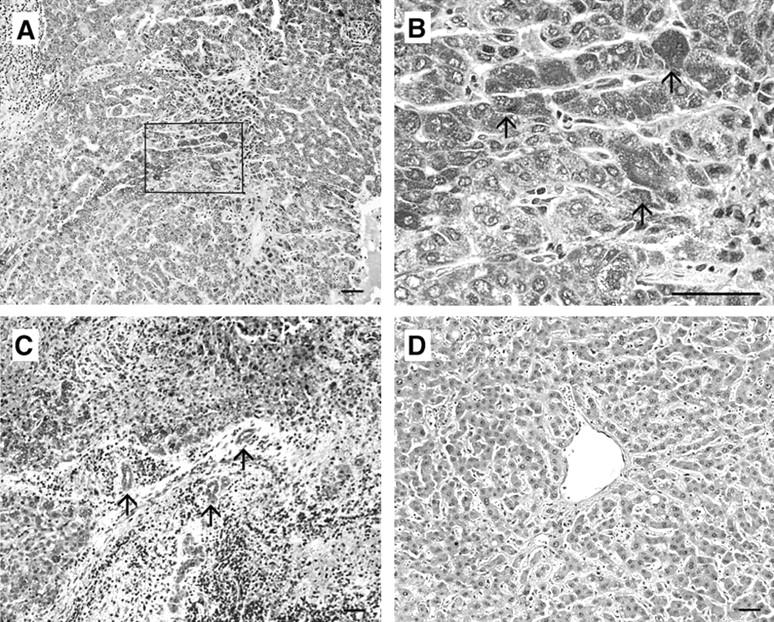
Immunohistochemical analysis to identify CCR1. Human hepatoma tissues (A to C) or normal liver tissues (D) were immunostained with anti-CCR1 antibodies as described in Materials and Methods. B represents the square in A at the higher magnification. Arrows in B indicate the positively stained hepatoma cells. Arrows in C indicate the positively stained vascular endothelial cells and small bile duct epithelial cells. Representative results shown in Table 2 ▶ are shown. Scale bars, 50 μm. Original magnifications: ×100 (A, C, and D); ×400 (B).
Table 2.
CCR1 and CCL3 Expression in Normal and Hepatoma Tissue Samples by Immunohistochemical Analysis
| Sample no. | Histology | CCR1 | CCL3 | ||||
|---|---|---|---|---|---|---|---|
| Hepatocyte or hepatoma | Endothelium | Fibroblast | Hepatocyte or hepatoma | Endothelium | Fibroblast | ||
| 1 | Normal | − | − | − | − | − | − |
| 2 | Normal | − | − | − | − | − | − |
| 3 | Normal | − | − | − | − | + | − |
| 4 | HCC | ++ | + | − | ++ | + | − |
| 5 | HCC | ++ | ++ | + | ++ | ++ | − |
| 6 | HCC | ++ | + | − | ++ | ++ | + |
| 7 | HCC | + | + | + | + | + | + |
| 8 | HCC | + | + | − | + | + | + |
| 9 | HCC | ++ | ++ | − | ++ | ++ | + |
| 10 | HCC | ++ | ++ | + | ++ | ++ | + |
| 11 | HCC | + | − | − | + | + | − |
| 12 | HCC | − | − | − | + | − | − |
| 13 | HCC | ++ | + | − | ++ | ++ | + |
| 14 | HCC | + | + | − | + | + | − |
| 15 | HCC | + | + | − | + | + | − |
HCC, Hepatocellular carcinoma.
−, <10% of the cells are positive; +, 10–20% of the cells are positive; ++, 20 to 50% of the cells are positive; +++, >50% of the cells are positive.
CCL3 Expression by Hepatoma Tissues but Not Hepatoma Cell Lines under a Resting Condition
We next examined whether hepatoma cell lines expressed ligands for CCR1. However, we detected neither CCL3 (Figure 5, A and B) ▶ nor CCL4 immunoreactivities in any hepatoma cell lines (data not shown). In normal liver tissues, CCL3 immunoreactivities were weakly detected in a few inflammatory cells and endothelium cells (Figure 5F ▶ and Table 2 ▶ ). Unexpectedly, CCL3 immunoreactivities were detected in hepatoma cells, vascular endothelial cells, and small bile duct epithelial cells, and to a lesser degree, fibroblast-like cells in hepatoma tissues (Figure 5, C to E ▶ ; and Table 2 ▶ ). The immunoreactivities were not detected when control antibodies or the antibodies preabsorbed with the corresponding antigens were used as a primary antibody (data not shown), indicating the specificity of the reactions. However, we did not detect immunoreactive CCL4 expression in hepatoma tissues (data not shown).
Figure 5.
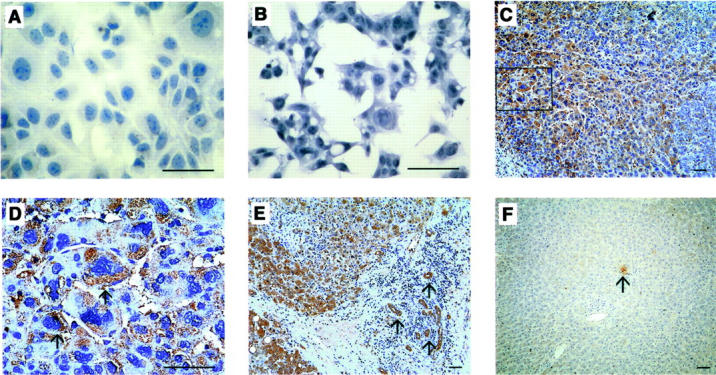
Immunohistochemical analysis to identify CCL3. HuH7 (A), HepG2 hepatoma cell lines (B), human hepatoma tissues (C to E), or normal liver tissues (F) were immunostained with anti-CCL3 antibodies as described in Materials and Methods. D represents the square in C at the higher magnification. Arrows in D indicate the positively stained hepatoma cells. Arrows in E indicate the positively stained vascular endothelial cells and small bile duct epithelial cells. Representative results shown in Table 2 ▶ are shown. Scale bars, 50 μm. Original magnifications: ×400 (A, B, and D); ×100 (C, E, and F).
Induction of CCL3 Expression by Hepatoma Cell Lines in Response to Proinflammatory Cytokines
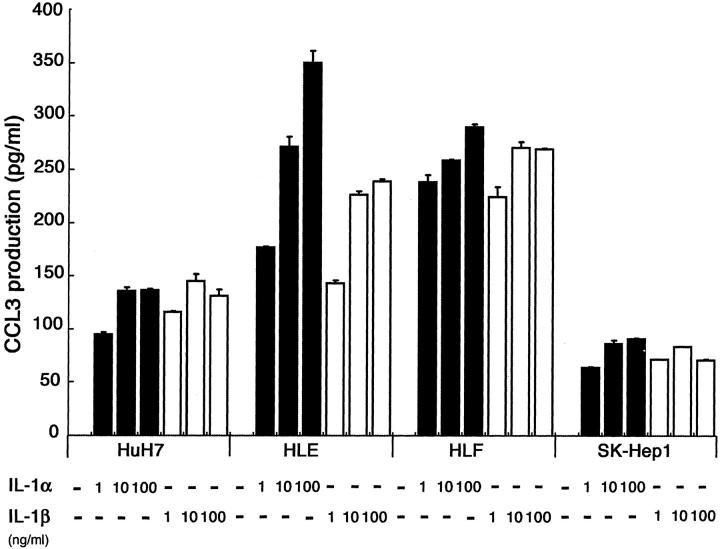
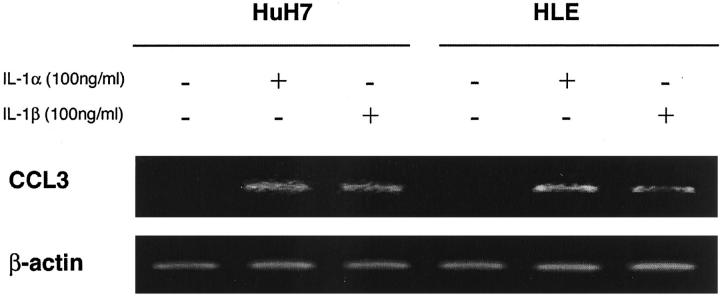
Hepatoma cells were positive for CCL3 although hepatoma cell lines did not express CCL3 at all without any stimulation. These observations prompted us to assume that hepatoma cells in tissues produce CCL3 in response to various stimuli such as proinflammatory cytokines and lipopolysaccharide, which may be present in hepatoma tissues. To explore this possibility, we measured CCL3 production by hepatoma cell lines stimulated with IL-1α, IL-1β, TNF-α, and lipopolysaccharide. The treatment with human IL-1α or IL-1β enhanced markedly CCL3 secretion into the supernatants in a dose-dependent manner in four cell lines among six cell lines that we investigated (Figure 6) ▶ . SK-Hep1 cell line but not other cell lines secreted CCL3 in a dose-dependent manner in the stimuli of TNF-α and lipopolysaccharide (data not shown). Only the HuH7 cell line secreted CCL4 in response to IL-1α in a dose-dependent manner (data not shown). The induction of CCL3 production was at pretranslational levels because CCL3 mRNA expression was induced in HuH7 and HLE cells when they were stimulated with either IL-1α or IL-1β (Figure 7) ▶ .
Figure 6.
CCL3 production of hepatoma cell lines in response to IL-1α or IL-1β. Hepatoma cell lines were incubated at a concentration of 1 × 105 cells in 1 ml of DMEM with 10% fetal bovine serum after 24 hours in the absence or presence of the indicated concentration of human IL-1α or IL-1β. Then, the supernatants were collected to measure CCL3 contents as described in Materials and Methods. A representative result from three independent experiments is shown. Mean and SD are shown.
Figure 7.
Total RNAs were extracted from HuH7 or HLE cell lines, which were incubated in the presence or the absence of IL-1α (100 ng/ml) or IL-1β (100 ng/ml) for 6 hours. RT-PCR was performed on the obtained total RNAs to detect CCL3 as described in Materials and Methods. Amplification of β-actin is shown to ensure the same amount of used cDNA. A representative result from three independent experiments is shown.
IL-1α and IL-1β Expression of Hepatoma Tissues
To further support our hypothesis that hepatoma cells in tissue produce CCL3 in response to proinflammatory cytokines, we examined IL-1α and IL-1β expression in hepatoma and normal liver tissue by an immunohistochemical analysis. In normal liver tissue, IL-1α (Figure 8D) ▶ and IL-1β (data not shown) were scarcely detected in hepatocytes. In hepatoma tissues, IL-1α was detected abundantly in hepatoma cells, vascular endothelial cells, and small bile duct epithelial cells (Figure 8, A to C) ▶ whereas IL-1β was detected in hepatoma cells and infiltrating cells (data not shown). The immunoreactivities were not detected when the primary antibody was omitted or the isotype-matched control antibody was used as the primary one (data not shown), indicating the specificity of the reactions. These results suggest that endogenously produced IL-1 induces CCL3 expression in hepatoma tissues that express the specific receptor for CCL3, CCR1.
Figure 8.
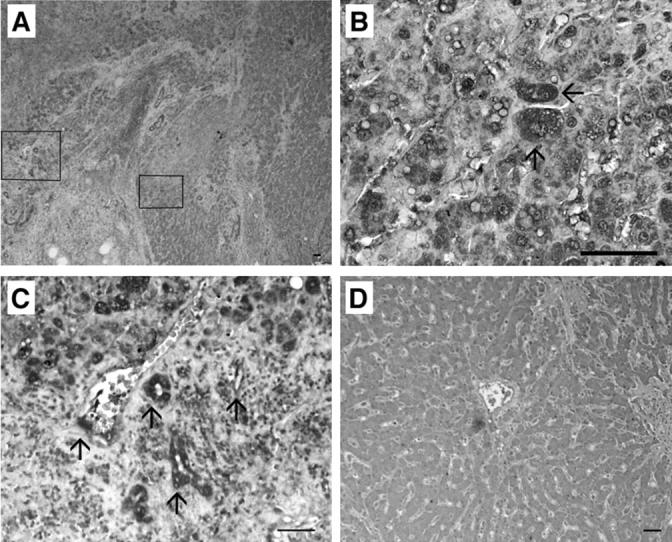
Immunohistochemical analysis of IL-1α. Human hepatoma tissues (A to C) and normal liver tissues (D) were immunostained with anti-IL-1α antibodies as described in Materials and Methods. B and C represent the squares in A at the higher magnification. Arrows in B indicate the positively stained hepatoma cells. Arrows in C indicate the positively stained vascular endothelial cells and small bile duct epithelial cells. Representative results are shown here. Scale bars, 50 μm. Original magnifications: ×40 (A); ×400 (B); ×200 (C); ×100 (D).
Discussion
Chemokines were originally discovered as chemotactic factors for specific types of leukocytes. Subsequent studies have demonstrated that chemokines exert a wide variety of actions also on nonleukocytic cells. Of interest is that some CXC chemokines such as IL-8/CXCL8 is presumed to be involved in neovascularization in some types of tumors through their potent angiogenic activities. 7,22-25 Moreover, several lines of evidence indicate that chemokine receptors expressed on tumor cells may determine the destination of metastasis of several types of tumors such as breast cancer and melanoma. 26-33 However, it remains elusive which chemokine receptor(s) is expressed by hepatoma. Hence, we examined chemokine receptor gene and protein expression by hepatoma cell lines. We observed that hepatoma cell lines constitutively expressed CCR1.
Accumulating evidence indicates that several chemokines have important roles in tumor progression. 6-8,34-36 We previously observed that gastric cancer-derived CXCL8 acted on some gastric carcinoma cell lines through interacting with its specific receptors, suggesting the presence of an autocrine amplifying loop. 37 We have previously observed that the X protein of hepatitis B virus and NS5A protein of hepatitis C virus induced the transcription of another chemokine, CXCL8. 10,11 Moreover, CXCL8 protein was detected immunohistochemically in hepatoma tissues. 12,38,39 However, we could not detect CXCR1 and CXCR2 mRNA in any hepatoma cell lines under our present experiment conditions. CXCL8 is a potent angiogenic factor and hepatoma tissue is characterized by abundant neovascularization. Thus, it is probable that hepatoma-derived CXCL8 might act on normal cells, particularly endothelium cells, thereby inducing neovascularization.
CCR1 binds a particular set of CC chemokines including CCL3, CCL4, and CCL5, and is expressed by a variety of cells, including lymphocytes, monocytes, basophils, neutrophils, and bone marrow progenitor cells. 13,15,16 CCR1 expression was detected in ovarian cancer tissues and ascites. 32,33 However, the expression was restricted to infiltrating macrophages and T lymphocytes. CCR1 expression was detected in malignant plasma cells by cDNA microarray analysis, 40 although its protein expression and function remain to be investigated. In hepatoma tissues, CCR5, another specific receptor for CCL3, CCL4, and CCL5, was detected also on infiltrating lymphocytes but not hepatoma cells. 39 Here, we provided definitive evidence that hepatoma cells express CCR1 in vitro and in vivo. Because hepatoma cell lines express functional CCR1 in terms of adenylate-mediated cAMP changes, it is probable that CCR1-mediated signals may have some effects on hepatoma cells. In our preliminary experiments, neither CCL3 nor anti-CCR1 antibodies have consistent effects on hepatoma cell proliferation (data not shown). However, because some of chemokine receptors can induce survival signals, 41 CCR1 may induce survival signals in hepatoma cells.
CCL3 can induce the infiltration of various types of leukocytes including monocytes/macrophages and neutrophils. 1-3 Because these cells are a major source of CXCL8, 1-3 a potent angiogenic factor, CCL3 may indirectly induce abundant neovascularization, which is frequently observed in hepatoma tissues. Moreover, intravenous injection of CCL3 can induce the rapid migration of bone marrow progenitor cells into peripheral blood. 42 Bone marrow progenitor cell population overlaps the population of endothelial cell progenitor cells and can direct the migration of endothelial progenitor cells in some instances. 43,44 Endothelial cells were positive for CCR1 in hepatoma tissues. Thus, it is tempting to speculate that locally produced CCL3 may induce neovascularization also by inducing the migration of bone marrow progenitor cells.
CCL3 expression by hepatoma cells in vivo but not hepatoma cell lines, prompted us to investigate the effect of IL-1 and TNF on CC chemokine expression by hepatoma cell lines because these proinflammatory cytokines can induce various types of cells to produce CC chemokine. 45-50 IL-1 and TNF induced CCL3 and CCL4 production by several hepatoma cell lines and an immunohistochemical analysis demonstrated the presence of IL-1α and IL-1β in hepatoma tissues. Thus, it is reasonable to speculate that endogenously produced IL-1 induces CCL3 production in an autocrine and/or paracrine manner. The IL-1 receptor antagonist gene was selectively expressed in liver during the course of diethylnitrosamine-induced hepatoma development. 51 Thus, it is tempting to speculate that aberration in IL-1 axis resulted in aberrant production of chemokines including CCL3 and CXCL8.
Kupffer cells belong to the similar lineage of monocytes/macrophages, which have an abundant ability to produce proinflammatory cytokines. 52,53 Thus, it is presumed that Kupffer cells are a major source of proinflammatory cytokines. However, hepatoma cells and other resident cells but not Kupffer cells were immunostained with anti-IL-1 antibodies. Similarly, hepatocytes but not Kupffer cells were a major source of proinflammatory cytokines in liver of Propionibacterium acnes-primed mice after lipopolysaccharide challenge. 54 These observations suggest that proinflammatory cytokines can be produced by other types of cells in liver than Kupffer cells under some conditions.
Several lines of evidence suggest that gene transfer of CCL3 or CCL4 into tumor was effective against several types of cancers under some instances. 55,56 However, the present study suggests potential involvement of these CC chemokines in tumor progression. Hence, it is necessary to elucidate the roles of the CCR1-CCL3 axis in tumor progression to advance an immune gene therapy using these CC chemokines.
Acknowledgments
We thank Dr. Haruhiro Higashida (Department of Biophysical Genetics, Kanazawa University, Graduate School of Medicine) for his invaluable suggestions, Dr. Toshikazu Kondo for his photographical assistance, and Ms. Akemi Nakano for her excellent technical assistance.
Footnotes
Address reprint requests to Dr. Naofumi Mukaida, Division of Molecular Bioregulation, Cancer Research Institute, Kanazawa University, 13-1, Takara-machi, Kanazawa 920-0934, Japan. E-mail: naofumim@kenroku.kanazawa-u.ac.jp.
Supported in part by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan.
References
- 1.Premack BA, Schall TJ: Chemokine receptors: gateways to inflammation and infection. Nat Med 1996, 2:1174-1178 [DOI] [PubMed] [Google Scholar]
- 2.Rollins BJ: Chemokines. Blood 1997, 90:909-928 [PubMed] [Google Scholar]
- 3.Rossi D, Zlotnik A: The biology of chemokines and their receptors. Annu Rev Immunol 2000, 18:217-242 [DOI] [PubMed] [Google Scholar]
- 4.Mellado M, Rodriguez-Frade JM, Manes S, Martinez AC: Chemokine signaling and functional responses: the role of receptor dimerization and TK pathway activation. Annu Rev Immunol 2001, 19:397-421 [DOI] [PubMed] [Google Scholar]
- 5.Gerard C, Rollins BJ: Chemokines and disease. Nat Immunol 2001, 2:108-115 [DOI] [PubMed] [Google Scholar]
- 6.Desbaillets I, Diserens AC, Tribolet N, Hamou MF, Van Meir EG: Upregulation of interleukin 8 by oxygen-deprived cells in glioblastoma suggests a role in leukocyte activation, chemotaxis, and angiogenesis. J Exp Med 1997, 186:1201-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunz M, Hartmann A, Flory E, Toksoy A, Koczan D, Thiesen HJ, Mukaida N, Neumann M, Rapp UR, Brocker EB, Gillitzer R: Anoxia-induced up-regulation of interleukin-8 in human malignant melanoma. A potential mechanism for high tumor aggressiveness. Am J Pathol 1999, 155:753-763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukaida N: Interleukin-8: an expanding universe beyond neutrophil chemotaxis and activation. Int J Hematol 2000, 72:391-398 [PubMed] [Google Scholar]
- 9.Schafer DF, Sorrell MF: Hepatocellular carcinoma. Lancet 1999, 353:1253-1257 [DOI] [PubMed] [Google Scholar]
- 10.Mahe Y, Mukaida N, Kuno K, Akiyama M, Ikeda N, Matsushima K, Murakami S: Hepatitis B virus X protein transactivates human interleukin-8 gene through acting on nuclear factor kB and CCAAT/enhancer-binding protein-like cis-elements. J Biol Chem 1991, 266:13759-13763 [PubMed] [Google Scholar]
- 11.Polyak SJ, Khabar KS, Paschal DM, Ezelle HJ, Duverlie G, Barber GN, Levy DE, Mukaida N, Gretch DR: Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J Virol 2001, 75:6095-6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iguchi A, Kitajima I, Yamakuchi M, Ueno S, Aikou T, Kubo T, Matsushima K, Mukaida N, Maruyama I: PEA3 and AP-1 are required for constitutive IL-8 gene expression in hepatoma cells. Biochem Biophys Res Commun 2000, 279:166-171 [DOI] [PubMed] [Google Scholar]
- 13.Su SB, Mukaida N, Wang J, Nomura H, Matsushima K: Preparation of specific polyclonal antibodies to a C-C chemokine receptor, CCR1, and determination of CCR1 expression on various types of leukocytes. J Leukoc Biol 1996, 60:658-666 [DOI] [PubMed] [Google Scholar]
- 14.Furuichi K, Wada T, Sakai N, Iwata Y, Yoshimoto K, Shimizu M, Kobayashi K, Takasawa K, Kida H, Takeda SI, Mukaida N, Matsushima K, Yokoyama H: Distinct expression of CCR1 and CCR5 in glomerular and interstitial lesions of human glomerular diseases. Am J Nephrol 2000, 20:291-299 [DOI] [PubMed] [Google Scholar]
- 15.de Wynter EA, Heyworth CM, Mukaida N, Jaworska E, Weffort-Santos A, Matushima K, Testa NG: CCR1 chemokine receptor expression isolates erythroid from granulocyte-macrophage progenitors. J Leukoc Biol 2001, 70:455-460 [PubMed] [Google Scholar]
- 16.Su S, Mukaida N, Wang J, Zhang Y, Takami A, Nakao: Inhibition of immature erythroid progenitor cell proliferation by macrophage inflammatory protein-1alpha by interacting mainly with a C-C chemokine receptor, CCR1. Blood 1997, 90:605-611 [PubMed] [Google Scholar]
- 17.Pugh JC, Yaginuma K, Koike K, Summers J: Duck hepatitis B virus (DHBV) particles produced by transient expression of DHBV DNA in a human hepatoma cell line are infectious in vitro. J Virol 1988, 62:3513-3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diamond L, Kruszewski F, Aden DP, Knowles BB, Baird WM: Metabolic activation of benzo[a]pyrene by a human hepatoma cell line. Carcinogenesis 1980, 1:871-875 [DOI] [PubMed] [Google Scholar]
- 19.Dor I, Namba M, Sato J: Establishment and some biological characteristics of human hepatoma cell lines. Gann 1975, 66:385-392 [PubMed] [Google Scholar]
- 20.Heffelfinger SC, Hawkins HH, Barrish J, Taylor L, Darlington GJ: SK HEP-1: a human cell line of endothelial origin. In Vitro Cell Dev Biol 1992, 28A:136-142 [DOI] [PubMed] [Google Scholar]
- 21.Knowles BB, Howe CC, Aden DP: Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980, 209:497-499 [DOI] [PubMed] [Google Scholar]
- 22.Huang S, Mills L, Mian B, Tellez C, McCarty M, Yang XD, Gudas JM, Bar-Eli M: Fully humanized neutralizing antibodies to interleukin-8 (ABX-IL8) inhibit angiogenesis, tumor growth, and metastasis of human melanoma. Am J Pathol 2002, 161:125-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshizaki T, Horikawa T, Qing-Chun R, Wakisaka N, Takeshita H, Sheen TS, Lee SY, Sato H, Furukawa M: Induction of interleukin-8 by Epstein-Barr virus latent membrane protein-1 and its correlation to angiogenesis in nasopharyngeal carcinoma. Clin Cancer Res 2001, 7:1946-1951 [PubMed] [Google Scholar]
- 24.Addison CL, Daniel TO, Burdick MD, Liu H, Ehlert JE, Xue YY, Buechi L, Walz A, Richmond A, Strieter RM: The CXC chemokine receptor 2, CXCR2, is the putative receptor for ELR+ CXC chemokine-induced angiogenic activity. J Immunol 2000, 165:5269-5277 [DOI] [PubMed] [Google Scholar]
- 25.Inoue K, Slaton JW, Kim SJ, Perrotte P, Eve BY, Bar-Eli M, Radinsky R, Dinney CP: Interleukin 8 expression regulates tumorigenicity and metastasis in human bladder cancer. Cancer Res 2000, 60:2290-2299 [PubMed] [Google Scholar]
- 26.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegui E, Zlotnik A: Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410:50-56 [DOI] [PubMed] [Google Scholar]
- 27.Mashino K, Sadanaga N, Yamaguchi H, Tanaka F, Ohta M, Shibuta K, Inoue H, Mori M: Expression of chemokine receptor CCR7 is associated with lymph node metastasis of gastric carcinoma. Cancer Res 2002, 62:2937-2941 [PubMed] [Google Scholar]
- 28.Li A, Varney ML, Singh RK: Expression of interleukin 8 and its receptors in human colon carcinoma cells with different metastatic potentials. Clin Cancer Res 2001, 7:3298-3304 [PubMed] [Google Scholar]
- 29.Loukinova E, Dong G, Enamorado-Ayalya I, Thomas GR, Chen Z, Schreiber H, Van Waes C: Growth regulated oncogene-alpha expression by murine squamous cell carcinoma promotes tumor growth, metastasis, leukocyte infiltration and angiogenesis by a host CXC receptor-2 dependent mechanism. Oncogene 2000, 19:3477-3486 [DOI] [PubMed] [Google Scholar]
- 30.Robledo MM, Bartolome RA, Longo N, Rodriguez-Frade JM, Mellado M, Longo I, van Muijen GN, Sanchez-Mateos P, Teixido J: Expression of functional chemokine receptors CXCR3 and CXCR4 on human melanoma cells. J Biol Chem 2001, 276:45098-45105 [DOI] [PubMed] [Google Scholar]
- 31.Schrader AJ, Lechner O, Templin M, Dittmar KE, Machtens S, Mengel M, Probst-Kepper M, Franzke A, Wollensak T, Gatzlaff P, Atzpodien J, Buer J, Lauber J: CXCR4/CXCL12 expression and signalling in kidney cancer. Br J Cancer 2002, 86:1250-1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scotton C, Milliken D, Wilson J, Raju S, Balkwill F: Analysis of CC chemokine and chemokine receptor expression in solid ovarian tumours. Br J Cancer 2001, 85:891-897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milliken D, Scotton C, Raju S, Balkwill F, Wilson J: Analysis of chemokines and chemokine receptor expression in ovarian cancer ascites. Clin Cancer Res 2002, 8:1108-1114 [PubMed] [Google Scholar]
- 34.Ugurel S, Rappl G, Tilgen W, Reinhold U: Increased serum concentration of angiogenic factors in malignant melanoma patients correlates with tumor progression and survival. J Clin Oncol 2001, 19:577-583 [DOI] [PubMed] [Google Scholar]
- 35.Koshiba T, Hosotani R, Miyamoto Y, Ida J, Tsuji S, Nakajima S, Kawaguchi M, Kobayashi H, Doi R, Hori T, Fujii N, Imamura M: Expression of stromal cell-derived factor 1 and CXCR4 ligand receptor system in pancreatic cancer: a possible role for tumor progression. Clin Cancer Res 2000, 6:3530-3535 [PubMed] [Google Scholar]
- 36.Salcedo R, Ponce ML, Young HA, Wasserman K, Ward JM, Kleinman HK, Oppenheim JJ, Murphy WJ: Human endothelial cells express CCR2 and respond to MCP-1: direct role of MCP-1 in angiogenesis and tumor progression. Blood 2000, 96:34-40 [PubMed] [Google Scholar]
- 37.Kitadai Y, Haruma K, Mukaida N, Ohmoto Y, Matsutani N, Yasui W, Yamamoto S, Sumii K, Kajiyama G, Fidler IJ, Tahara E: Regulation of disease-progression genes in human gastric carcinoma cells by interleukin 8. Clin Cancer Res 2000, 6:2735-2740 [PubMed] [Google Scholar]
- 38.Akiba J, Yano H, Ogasawara S, Higaki K, Kojiro M: Expression and function of interleukin-8 in human hepatocellular carcinoma. Int J Oncol 2001, 18:257-264 [DOI] [PubMed] [Google Scholar]
- 39.Yoong KF, Afford SC, Jones R, Aujla P, Qin S, Price K, Hubscher SG, Adams DH: Expression and function of CXC and CC chemokines in human malignant liver tumors: a role for human monokine induced by gamma-interferon in lymphocyte recruitment to hepatocellular carcinoma. Hepatology 1999, 30:100-111 [DOI] [PubMed] [Google Scholar]
- 40.De Vos J, Couderc G, Tarte K, Jourdan M, Requirand G, Delteil MC, Rossi JF, Mechti N, Klein B: Identifying intercellular signaling genes expressed in malignant plasma cells by using complementary DNA arrays. Blood 2001, 98:771-780 [DOI] [PubMed] [Google Scholar]
- 41.Vlahakis SR, Villasis-Keever A, Gomez T, Vanegas M, Vlahakis N, Paya CV: G protein-coupled chemokine receptors induce both survival and apoptotic signaling pathway. J Immunol 2002, 169:5546-5554 [DOI] [PubMed] [Google Scholar]
- 42.Hunter MG, Bawden L, Brotherton D, Craig S, Cribbes S, Czaplewski LG, Dexter TM, Drummond AH, Gearing AH, Heyworth CM, et al: BB-10010: an active variant of human macrophage inflammatory protein-1 α with improved pharmaceutical properties. Blood 1995, 86:4400-4408 [PubMed] [Google Scholar]
- 43.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner JM: Isolation of putative progenitor endothelial cells for angiogenesis. Science 1997, 275:964-967 [DOI] [PubMed] [Google Scholar]
- 44.Takakura N, Watanabe T, Suenobu S, Yamada Y, Noda T, Ito Y, Satake M, Suda T: A role for hematopoietic stem cells in promoting angiogenesis. Cell 2000, 102:199-209 [DOI] [PubMed] [Google Scholar]
- 45.Chabaud M, Page G, Miossec P: Enhancing effect of IL-1, IL-17, and TNF-α on macrophage inflammatory protein-3α production in rheumatoid arthritis: regulation by soluble receptors and Th2 cytokines. J Immunol 2001, 167:6015-6020 [DOI] [PubMed] [Google Scholar]
- 46.Mustafa M, Wondimu B, Bakhiet M, Modeer T: Production of Rantes/CCL5 in human gingival fibroblasts challenged with tumor necrosis factor α. Eur J Oral Sci 2001, 109:44-49 [DOI] [PubMed] [Google Scholar]
- 47.Saji F, Nonaka M, Pawankar R: Expression of RANTES by IL-1 β and TNF-α stimulated nasal polyp fibroblasts. Auris Nasus Larynx 2000, 27:247-252 [DOI] [PubMed] [Google Scholar]
- 48.Bian ZM, Elner SG, Strieter RM, Kunkel SL, Lukacs NW, Elner VM: IL-4 potentiates IL-1β- and TNF-α-stimulated IL-8 and MCP-1 protein production in human retinal pigment epithelial cells. Curr Eye Res 1999, 18:349-357 [DOI] [PubMed] [Google Scholar]
- 49.Marra F, Valente AJ, Pinzani M, Abboud HE: Cultured human liver fat-storing cells produce monocyte chemotactic protein-1. Regulation by proinflammatory cytokines. J Clin Invest 1993, 92:1674-1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiura TS, Kempiak SJ, Nel AE: Activation of the human RANTES gene promoter in a macrophage cell line by lipopolysaccharide is dependent on stress-activated protein kinases and the IκB kinase cascade: implications for exacerbation of allergic inflammation by environmental pollutants. Clin Immunol 1999, 90:287-301 [DOI] [PubMed] [Google Scholar]
- 51.Yamada Y, Karasaki H, Matsushima K, Lee GH, Ogawa K: Expression of an IL-1 receptor antagonist during mouse hepatocarcinogenesis demonstrated by differential display analysis. Lab Invest 1999, 79:1059-1067 [PubMed] [Google Scholar]
- 52.Kurosaka K, Watanabe N, Kobayashi Y: Production of proinflammatory cytokines by resident tissue macrophages after phagocytosis of apoptotic cells. Cell Immunol 2001, 211:1-7 [DOI] [PubMed] [Google Scholar]
- 53.Lichtman SN, Wang J, Schwab JH, Lemasters JJ: Comparison of peptidoglycan-polysaccharide and lipopolysaccharide stimulation of Kupffer cells to produce tumor necrosis factor and interleukin-1. Hepatology 1994, 19:1013-1022 [PubMed] [Google Scholar]
- 54.Fujioka N, Mukaida N, Harada A, Akiyama M, Kasahara T, Kuno K, Ooi A, Mai M, Matsushima K: Preparation of specific antibodies against murine IL-1ra and the establishment of IL-1ra as an endogenous regulator of bacteria-induced fulminant hepatitis in mice. J Leukoc Biol 1995, 58:90-98 [DOI] [PubMed] [Google Scholar]
- 55.Miyata T, Yamamoto S, Sakamoto K, Morishita R, Kaneda Y: Novel immunotherapy for peritoneal dissemination of murine colon cancer with macrophage inflammatory protein-1beta mediated by a tumor-specific vector, HVJ cationic liposomes. Cancer Gene Ther 2001, 8:852-860 [DOI] [PubMed] [Google Scholar]
- 56.Van Deventer HW, Serody JS, McKinnon KP, Clements C, Brickey WJ, Ting JP: Transfection of macrophage inflammatory protein 1alpha into B16 F10 melanoma cells inhibits growth of pulmonary metastases but not subcutaneous tumors. J Immunol 2002, 169:1634-1639 [DOI] [PubMed] [Google Scholar]