Abstract
Kruppel-like factors (KLFs) are a group of transcription factors that appear to be involved in different biological processes including carcinogenesis. In a recent study, KLF6 was reported as a tumor suppressor gene in prostate cancer because of its frequent loss of heterozygosity (LOH) and mutation as well as functional suppression of cell proliferation. Loss of chromosomal locus spanning KLF6 is relatively infrequent in other published studies of prostate cancer, however. To clarify the role of KLF6 in prostate cancers, particularly those that are high grade, we examined KLF6 for deletion, mutation, and loss of expression in 96 prostate cancer samples including 21 xenografts/cell lines. Loss of heterozygosity occurred in 4 (19%) of 21 xenografts/cell lines and 8 (28%) of 29 informative tumors. Fourteen of the 96 (15%) samples showed 15 somatic sequence changes in the KLF6 gene, including 7 that changed KLF6 peptide sequences, 4 that did not, and 4 that were located in untranslated regions. Expression levels of KLF6 were significantly lost in 4 of 20 (20%) xenografts/cell lines of prostate cancer, as detected by RT-PCR and Northern blot analysis. These findings indicate that significant genetic alterations of KLF6 occur in a minority of high-grade prostate cancers.
Deletion of the short arm of chromosome 10 (10p), suggesting the location of a tumor suppressor gene(s), has been demonstrated in several studies of prostate cancer. In a cytogenetic analysis, deletion at 10p13 was detected in 3 of 12 tumors. 1 Using the restriction fragment length polymorphism approach, loss of heterozygosity (LOH) at 10p was detected in 4 of 19 informative prostate cancers, although the region of deletion could not be defined in this study. 2 Examination of a larger number of prostate tumors with polymorphic microsatellite markers spanning chromosome 10 further showed relatively frequent LOH at 10p. 3-6 Loss of 10p was also detected in a cell line derived from a primary human prostate cancer. 7 In functional studies, a DNA fragment from 10p was able to suppress prostate cancer cell proliferation, 8,9 providing additional evidence for the presence of a tumor suppressor gene.
Recently, the KLF6 transcription factor located at 10p15 was reported as a candidate tumor suppressor gene in prostate cancer. 10 In this study, LOH at the KLF6 locus was detected in 17 of 22 (77%) tumors analyzed, which was more frequent compared to that reported in previous studies. The frequency of KLF6 mutation was also found to be high (55%). In addition, KLF6 was shown to reduce cell proliferation through up-regulation of p21 in a p53-independent manner. 10
To clarify the role of KLF6 in prostate cancer, we examined the gene for LOH, mutation, and expression change in 96 prostate cancer samples. We found that LOH and mutation of KLF6 occurred in a significant proportion but not the majority of prostate cancers, while the level of expression for KLF6 was greatly reduced in 4 of 20 of prostate cancer cell lines and xenografts.
Materials and Methods
Tumor Specimens, Cell Lines, and Xenografts
In total, we analyzed 96 samples from 93 patients with prostate cancer, including 75 tumor specimens and 21 cell lines and xenografts. Of the 75 tumor specimens, 43 had matched non-neoplastic cells and 32 did not. Thirty-one were primary tumors, 26 were hormone refractory cancers, and 18 were metastases. The patient age at diagnosis ranged from 51 to 75 years (average 66). Forty-seven (82%) of the primary tumors had Gleason scores of 8 to 10, whereas seven had a Gleason score of 7 and three had a Gleason score of 5 or 6. In most cases, tumor and non-neoplastic cells were collected from H&E stained sections of zinc formalin-fixed, paraffin-embedded tissues of manual microdissection, as described in our previous study. 11 Some cases were from prostate cancer patients who underwent autopsy. Use of human tissues was approved by each institution involved. Isolation of genomic DNA was performed as previously described. 11
We also analyzed seven prostate cancer cell lines (22Rv1, BRF-41T, DU145, LNCaP, MDA-Pca2b, NCI-H660, and PC-3) and 14 prostate cancer xenografts (CWR21, CWR91, LAPC-3, LAPC-4, LAPC-9, LUCaP23.1, LUCaP35, LUCaP41, LUCaP49, LUCaP58, LUCaP70, LUCaP73, LUCaP77, and PC-82,), which were described in detail in our previous studies. 12,13 Non-neoplastic controls included one total RNA sample from a pool of 19 normal human prostates (Clontech, Palo Alto, CA) and four immortalized non-neoplastic prostatic epithelial cell lines (PZ-HPV-7, PWR-1E, and RWPE-1 from ATCC (Manassas, VA), and BRF-55T from Biological Research Faculty & Facility (BRFF), Ijamsville, MD). Genomic DNA was isolated using the Blood & Cell Culture DNA Midi Kit from Qiagen (Valencia, CA), and cytoplasmic RNA was isolated using Qiagen’s RNeasy Maxi Kit.
LOH Analyses
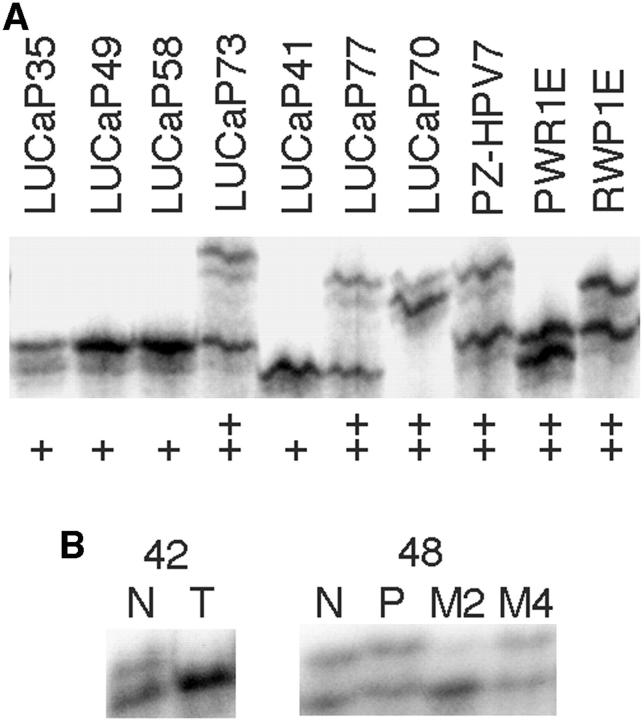
Seven well-mapped microsatellite markers in or near the KLF6 locus were used to detect LOH in prostate cancer xenografts/cell lines (Figure 1A) ▶ by the method of homozygosity-mapping-of-deletion (HOMOD), in which an LOH is defined when 5 or more consecutive polymorphic markers with a heterozygosity value of 0.75 or greater show a homozygous genotype. 14 Primer sequences, sizes of PCR products, and locations of these markers are available from The Genome Database at http://gdbwww.gdb.org. The average distance between these markers was 800 kb, and the KLF6 gene is located between D10S533 and D10S591. The heterozygosity scores for these markers were 0.7 for D10S1716, 0.85 for D10S1745, 0.22 for D10S533, 0.71 for D10S591, 0.53 for D10S552, 0.72 for D10S1729, and 0.73 for D10S189, according to The Genome Database. It was unknown, however, what populations were used to obtain these scores thus they may not completely match the scores for the population used in this study.
Figure 1.
Detection of allelic status at the KLF6 locus in prostate cancer cell lines and xenografts (A) and LOH in prostate cancer specimens (B) using microsatellite markers. Markers D10S189 and D10S591 were used for panel A and B respectively. Sample names are shown at the top of each panel. In panel A, ++ denotes two different alleles whereas + denotes one allele. N, normal; T, primary tumor, P, primary tumor from patients whose metastases were also used; M, metastasis.
Four of the seven markers were also used for LOH analysis of prostate cancer specimens (Figure 1B) ▶ . PCR, gel electrophoresis, and determination of LOH were performed as previously described. 12,15 Briefly, tumor samples were microdissected to ensure a minimum number of 70% neoplastic cells. Non-neoplastic cells from lymph nodes or seminal vesicles (or spleen for autopsy specimens) in almost 90% of the cases, and from normal prostate in the remainder, were obtained from paraffin blocks that contained no neoplastic cells. LOH was determined when the signal for one allele in the tumor was reduced significantly when compared to that for the non-neoplastic cells. All autoradiograms were analyzed independently by two of the investigators, and any conflicts in interpretation were resolved by repetition of the experiment.
Mutation Detection
Both single-strand conformational polymorphism (SSCP) analysis and direct DNA sequencing were used to determine the mutation status of KLF6 in prostate cancer cells. Briefly, PCR was performed in a volume of 10 μl using GeneAmp PCR System 9600 or 9700 (Applied Biosystems Inc., Foster City, CA), with 5 to 25 ng of genomic DNA, 1X PCR buffer (1.5 mmol/L MgCl2), 0.2 mmol/L of each dNTPs, 0.6 unit of AmpliTaqGold DNA polymerase (Applied Biosystems), 0.4 μmol/L of each primer, and 0.4 μCi of α[33P]dATP (>2500 Ci/mmol). After initial incubation at 95°C for 5 minutes, a total of 35 cycles were performed, each including 94°C for 30 seconds, 55–60°C for 30 seconds, and 72°C for 45 seconds. The FastStart DNA polymerase kit (Roche) was used for exon 1. Primer sequences for SSCP analysis were: 5′-ATGAAACTTTCACCTGCGCTCC-3′/5′-TCTGAACCCC AAACAGCCGA-3′ (exon 1), 5′-TTTGGTCGTCATGGCAATCAC-3′/5′-TTCCCGAG CTGACCAAA ACTTC-3′ (exon 2a), 5′-GTTTACCTCCGACCCCATTGGC-3′/5′-TCCAGGG CTGGTGCAATGCA-3′ (exon 2b), 5′-CTGCGGGTCAGTGAAGTCATGGG-3′/5′-CATTCCCAAGGCCCCACGCT-3′(exon 3), and 5′-AACACTGGCAAGGGATGGGAACC-3′/5′-GCTATGCCGCTTCTTACAGGACC-3′ (exon 4). PCR fragments were subsequently heat-denatured and run for 15 to 24 hours at 800 V in both 0.5 × and 0.8 × mutation detection enhancement (MDE) gels (BMA, Rockland, ME) containing 1% glycerol. After electrophoresis, the gels were dried and exposed to Kodak BioMax MR film overnight. When a band shift was observed in a sample, the SSCP procedure including PCR was repeated for that sample to verify the band shift. Once verified, the PCR products were subject to DNA sequencing using an ABI 377 sequencing unit (Applied Biosystems). All potential mutations were sequenced on both DNA strands.
To verify the efficiency of SSCP in detecting gene mutations, we also performed direct DNA sequencing of the coding region for KLF6 in most of the cell lines and xenografts. Briefly, first strand cDNA was synthesized from total RNA samples from one normal prostate, four immortalized nonneoplastic prostate epithelial cell lines, and 20 prostate cancer xenografts/cell lines as described. 16 Primers 5′-GCCTCGCCGCCCTCGCAAC-3′ (forward) and 5′-CTCTCAGCCTGGAAGCCTTTTAGCCTAC-3′ (reverse) were used to amplify KLF6 coding region in a volume of 50 μl. The PC-3 cell line has a mutation in the forward primer sequence, thus another primer was used (5′-GACTTTCGGCTCCGGCTCCCA-3′). Resultant PCR products were purified using the Qiaquick PCR Purification Kit (Qiagen) and were subject to DNA sequence determination using an ABI 377 sequencing unit with two nested primers (5′-ACAGCTCCGAGGAACTTTCTCCCA-3′ and 5′-CCCGAGCTGACCAAAACTTCGCCA-3′).
RT-PCR and Northern Blot
RT-PCR was performed on each of the xenografts and cell lines. RNA samples were treated with DNase I to remove contaminating genomic DNA. The Superscript cDNA synthesis system (Invitrogen, Carlsbad, CA) was used to synthesize first-strand cDNA from 2 μg of total RNA with oligo(dT) primers. Following cDNA synthesis, ribonuclease H was added to the reaction mixture, which was then heated to 37°C for 15 minutes. Released cDNA was purified using the Qiaquick PCR Purification Kit (Qiagen) and was resuspended in 100 μl of water. For each PCR, 2 μl of purified cDNA were used. Primer sequences for the KLF6 genes and β-actin are as follows: 5′-CTCTCAG CCTGGAAGCTTTTAGCCTAC-3′/5′-ACAGCTCCGAGGAACTTTCTCCCA-3′(KLF6) and 5′-GAAATCGTGCGTGACATTAAG-3′/5′-CTAGAAGCATTTGCGGTGGACG ATGAGGGGCC-3′ (β-actin). Duplex PCR was performed with platinum Taq polymerase (Invitrogen) in a volume of 20 μl and 30 cycles, each with 94°C for 15 seconds, 60°C for 15 seconds, and 72°C for 1 minute.
For the Northern blot analysis, purified PCR products of KLF6 coding region from mutation detection experiments were used as the probe, which was labeled with α[32P]dCTP using a random primer DNA labeling system (Invitrogen). Only the 11 samples with sufficient RNA available were used. In the Northern blot, 15 μg of total RNA was loaded in each lane. Hybridization was performed at 68°C for 3 hours in QuikHyb solution (Stratagene, La Jolla, CA). The membrane was washed following the manufacturer’s instructions. After exposure to X-ray film, the KLF6 probe was stripped from the blot and the membrane was re-hybridized with the β-actin probe.
Results and Discussion
We first examined the frequency of LOH at the KLF6 locus in tumors, cell lines, and xenografts of prostate cancer. For the 21 prostate cancer xenografts/cell lines, homozygosity was detected at each of the seven microsatellite markers in four samples, ie, PC-3, LUCaP23.1, LUCaP35, and LUCaP41 (Figure 1A ▶ , Figure 2 ▶ ). Based on the heterozygosity value for each marker, the possibility of being homozygous for these markers in a normal genome is 3.6 × 10−4, which meets the criteria for the definition of an LOH in a sample without matched normal control. 14 Therefore, LOH occurred in 4 of the 21 (19%) prostate cancer xenografts/cell lines. With this method, however, there is the possibility of underestimating the frequency of LOH, because a smaller region of loss not covered by each of the defining markers cannot be detected. Based on the HOMOD method, none of the five non-neoplastic samples had LOH at the KLF6 locus (Figure 2) ▶ . To determine the LOH frequency at KLF6 in prostate cancer from patients, we then examined four markers in 43 prostate cancers that had matched nonneoplastic cells. Of the 29 cases that were informative (showing two different alleles) at one or more of the markers, eight (28%) showed LOH at one or more markers examined (Figure 1B ▶ and Figure 3 ▶ ). This frequency of LOH may be slightly higher than that seen in xenografts/cell lines (Figure 2) ▶ but is lower than previously reported. 10 We noticed that Narla et al identified a number of patients with small regions of LOH using tightly flanking markers. 10 Such losses would not be detectable in this study. Therefore, we may have underestimated the frequency of LOH at the KLF6 locus.
Figure 2.
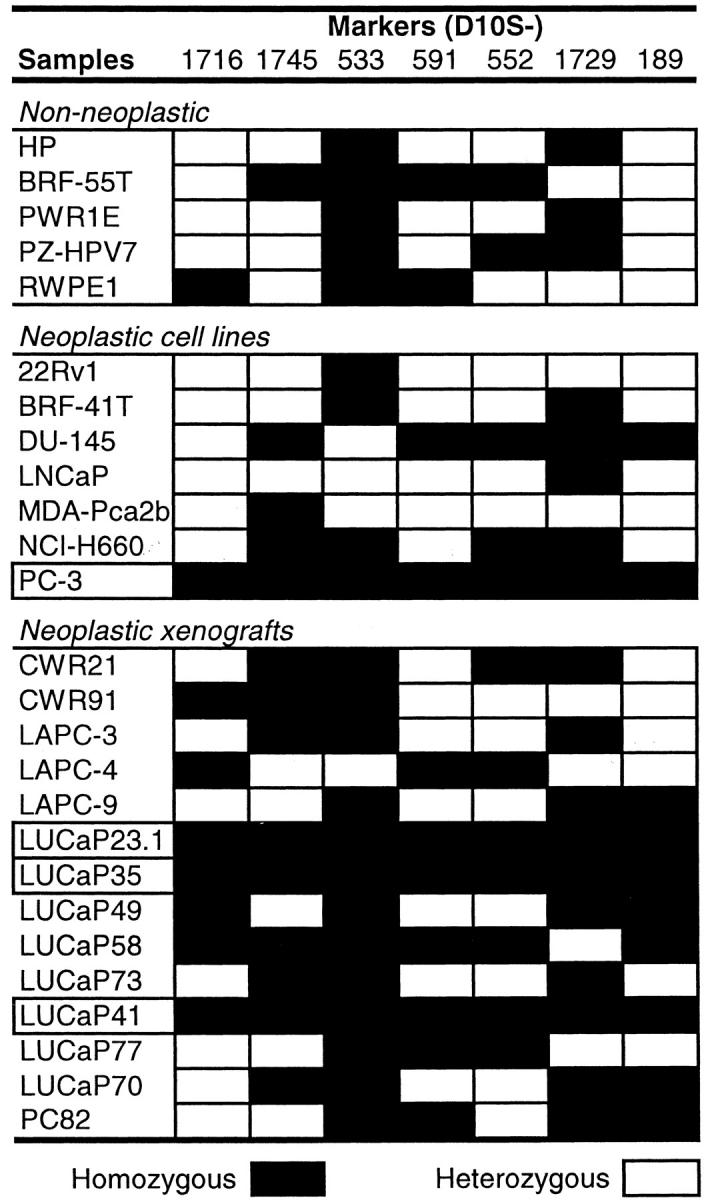
Detection of LOH at the KLF6 locus in cell lines and xenografts of prostate cancer by the method of HOMOD analysis. Marker names are listed at the top with the order from telomere to centromere. Cases with LOH are boxed. Black boxes indicate one allele and white boxes indicate two alleles.
Figure 3.
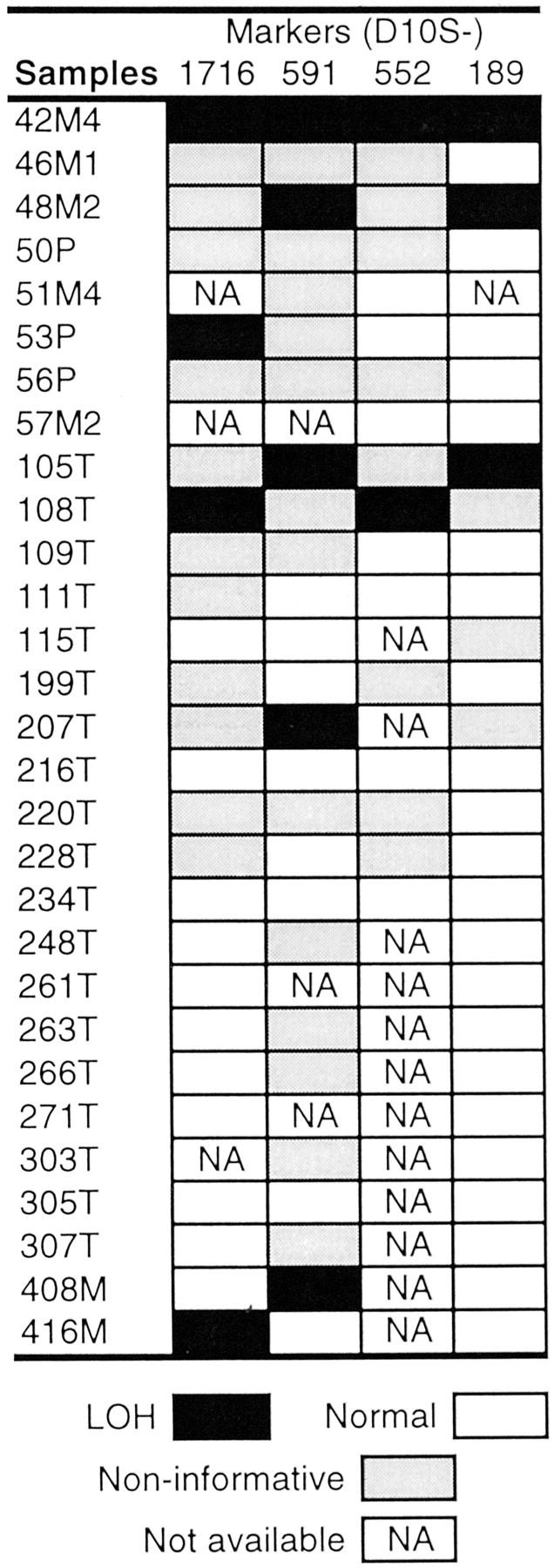
LOH of the KLF6 locus in prostate cancer specimens. Only cases informative for at least one marker are shown. Markers are shown at the top, and samples at the left. N, normal; T and P, primary tumor; M, metastasis.
To determine whether KLF6 is inactivated by “two-hits” as for some other tumor suppressor genes, ie, one allele is lost and the other allele is mutated, 17 we examined the gene for point mutations in all samples using different methodologies. We first used SSCP analysis combined with DNA sequence determination to analyze the four exons and their adjacent intron nucleotides (Figure 4A ▶ , Table 1 ▶ ). A mutation of C>A occurred in exon 1, four nucleotides upstream to the ATG start codon, in the LAPC-3 and LUCaP49 xenografts (Table 1 ▶ , Figure 4 ▶ ). In addition, a polymorphism G/A was detected in intron 1, at nucleotide 24 upstream to exon 2. The prostate cancer xenograft LUCaP35 showed a heterozygous genotype at this single nucleotide polymorphism (SNP) (G/A), while the non-neoplastic cell line RWPE1 was homozygous A/A, and the rest of the samples including four non-neoplastic controls were homozygous G/G. Therefore, SSCP analysis revealed mutations of KLF6 in 2 of the 21 prostate cancer xenografts/cell lines.
Figure 4.
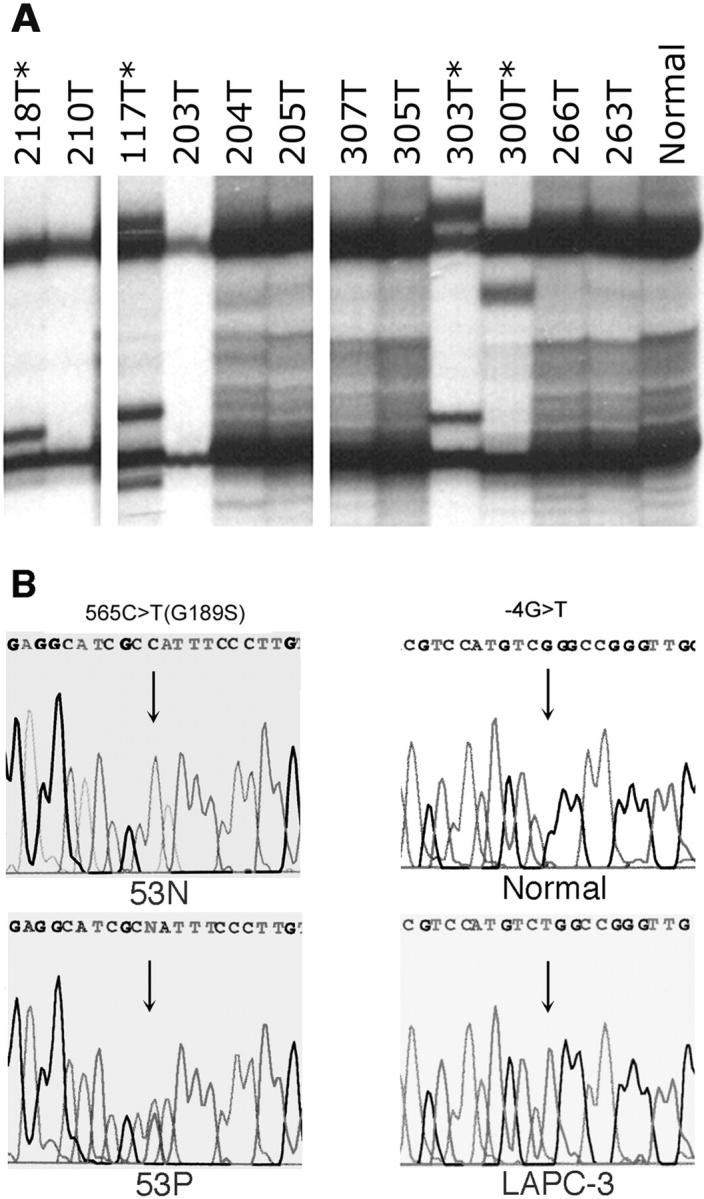
Detection of KLF6 mutations in prostate cancer samples. A: Examples of SSCP analysis. Asterisks denote samples with shifted bands. B: DNA sequence determination in representative samples with mutations. Case 53 has a mutation of 565C>T(G189S) and xenograft LAPC-3 has a mutation of –4G>T.
Table 1.
Mutations of KLF6 in Prostate Cancer
| Samples | Mutation | Location in gene | Gleason score | Metastasis |
|---|---|---|---|---|
| Tumors | ||||
| 7T | 359C>T (S120F) | Exon 2 | 10 | |
| 53P | 565G>A (G189S) | Exon 2 | 9 | Yes |
| 117T | 262G>A (D88N) | Exon 2 | 9 | Yes |
| 216T | 924G>A | 3′-UTR | 7 | No |
| 218T | 127C>T (L43F) | Exon 2 | 9 | Yes |
| 232T | 104C>T (T35I) | Exon 2 | 10 | No |
| 244T | 371A>T (K124M) | Exon 2 | 6 | |
| 300T | 366G>A | Exon 2 | 7 | No |
| 303T | 277C>T (P93S) | Exon 2 | 9 | No |
| 360T>C | ||||
| 404M | 201C>T | Exon 2 | 10 | Yes |
| 410M | 207G>A | Exon 2 | 10 | Yes |
| Xenografts/cell lines | ||||
| LAPC-3 | −4C>A | 5′-UTR | No | |
| LUCaP49 | −4C>A | 5′-UTR | ||
| PC-3 | −29A>G | 5′-UTR | Yes |
For 18 of the 21 prostate cancer xenografts/cell lines and each of the five non-neoplastic controls, we also amplified the coding region of KLF6 by RT-PCR and determined the DNA sequences of these PCR products to verify the SSCP results. Using this approach, the mutations in LAPC-3 and LUCaP49 were also detected, but no additional mutations were found. Two of the samples, ie, PC-3 and LUCaP41, however, failed to generate PCR products for sequencing analysis. With another set of primers that flank the earlier PCR fragment of the KLF6 coding region, we were able to amplify the KLF6 coding region in PC-3 cells and identify a mutation in its 5′-untranslated region (-29A>G). These findings indicate the sensitivity and reliability of SSCP analysis in detecting point mutations. In total, three of 21 (14%) xenografts/cell lines had point mutations at KLF6, yet none of them resulted in a change in amino acid sequence of the gene. We also noticed two sequence discrepancies between the KLF6 mRNA reported in GenBank (accession number AF001461) and the KLF6 coding region that we sequenced in 22 samples. The AF001461 sequence may have acquired two sequence changes (827C>G and 860G>A).
We then screened 75 prostate cancer specimens from 72 patients for KLF6 mutation using SSCP and DNA sequencing. As shown in Figure 4 ▶ and Table 1 ▶ , we detected 12 mutations in 11 of the 75 (15%) tumors. Seven of the mutations changed amino acid codons, but none of them resulted in a stop codon. Three were samesense (silent) mutations, while one mutation occurred in the 3′-untranslated region. Tumor 303T had two mutations, one of which changed the amino acid sequence. None of these mutations were detected in the study by Narla et al. 10
According to available human genome sequences in GenBank, as seen in contig NT 008705 of the draft sequence of chromosome 10, the KLF6 gene has four exons. The size for exon 1 to 4 is 218 bp, 574 bp, 124 bp, and 525 bp, respectively. Whereas none of the 12 mutations occurred in the same codon twice or more, 10 (83%) of them occurred in exon 2. Clustering of mutations in exon 2 was also reported in the previous study. 10 The majority of exon 2 encodes for the transactivation domain of the KLF6 protein, which varies greatly among different KLF members. 18 None of the mutations occurred in the zinc finger domains, which are encoded by exons 2 to 4 and are more evolutionary conserved.
In total, mutation at KLF6 was detected in 14 of 96 (15%) samples from 93 patients. The majority of the tumors were high grade and many were from metastases of prostate cancer. Half of the mutations changed amino acid sequences, but it is unknown whether such changes have an impact on the function of the KLF6 protein. The allelic status of KLF6 was available for only three of the 11 tumors with mutations due to the lack of non-neoplastic control DNA. One of these three tumors (53P) could have LOH at the KLF6 locus (Figure 3) ▶ . Among the three xenografts/cell lines with mutations, one (PC-3) also had LOH. Therefore, although we were unable to determine exactly how many tumors with mutations also had LOH, at least two may be inactivated by “two-hits”. We are aware of the limit of SSCP in mutation detection, which may be partially responsible for the lower frequency of mutations in this study compared to that reported by Narla et al, 10 in which PCR products were sequenced to determine a mutation. In addition, most of the mutations in the study by Narla et al were detected in samples with LOH, which was less frequent in this study and could also contribute to the lower mutation frequency of KLF6.
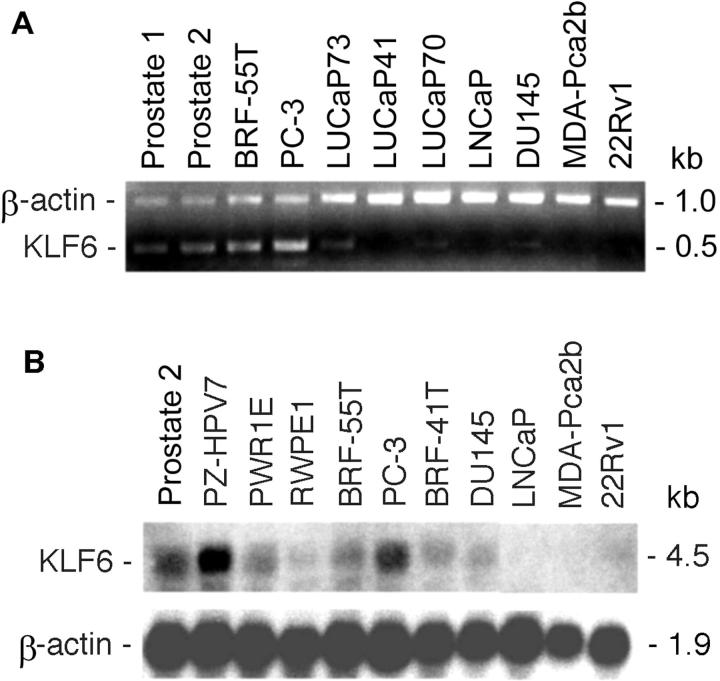
Several members of the KLF family appear to be down-regulated in different cancers, including KLF4 in colon carcinoma 19 and KLF5 and KLF10 in breast cancer. 16,20 We thus determined expression levels of KLF6 in 20 xenografts/cell lines of prostate cancer by duplex RT-PCR, in which two pairs of primers were added to the same tube, and Northern blot analysis. Whereas the majority of the 20 samples analyzed showed similar levels of expression as in normal prostate samples, four, ie, 22Rv1, LNCaP, LUCaP41, and MDA-Pca2b, showed loss of KLF6 expression significantly (Figure 5) ▶ . None of them had mutations and only the LUCaP41 xenograft had LOH. The PC-3 cancer cell line, on the other hand, had LOH and a mutation in the 5′-UTR of KLF6 but showed a high level of expression for KLF6. Two prostate cancer xenografts also had mutations in the 5′-UTR but retained KLF6 expression.
Figure 5.
Loss of expression for KLF6 in some prostate cancer cell lines or xenografts, as detected by duplex RT-PCR (A) and Northern blot analysis (B). In each panel, samples are listed at the top, gene names at the left, and DNA/RNA sizes at the right. Prostate 1 and prostate 2 were normal prostates from different individuals; PZ-HPV-7, PWR1E, and RWPE1 are immortalized cell lines from normal prostate epithelium; and BRF-55T is an immortalized cell line from a benign prostatic hyperplasia.
LOH and mutation are the common “two-hits” that inactivate a tumor suppressor gene. 17 Loss of expression is another mechanism to functionally inactivate a tumor suppressor gene, as demonstrated for the PTEN gene. 21 Detection of LOH, point mutations, and loss of expression of KLF6 in a minority of high-grade prostate cancers in this study suggests that KLF6 plays a role in some prostate cancers. Additional studies may better clarify the role of KLF6 as a potential tumor suppressor gene in prostate cancer.
Footnotes
Address reprint requests to Jin-Tang Dong, Ph.D., Winship Cancer Institute, Emory University School of Medicine, 1365-B Clifton Road, Suite B4100, Atlanta, GA 30322. E-mail: jdong2@emory.edu.
Supported by grants from the National Institutes of Health (CA85560 and CA87921 to J.T.D), the Medical Research Fund of Tampere University Hospital, and the Finnish Cancer Institute (to P.A.K.).
References
- 1.Teixeira MR, Waehre H, Lothe RA, Stenwig AE, Pandis N, Giercksky KE, Heim S: High frequency of clonal chromosome abnormalities in prostatic neoplasms sampled by prostatectomy or ultrasound-guided needle biopsy. Genes Chromosomes Cancer 2000, 28:211-219 [PubMed] [Google Scholar]
- 2.Phillips SM, Morton DG, Lee SJ, Wallace DM, Neoptolemos JP: Loss of heterozygosity of the retinoblastoma and adenomatous polyposis susceptibility gene loci and in chromosomes 10p, 10q and 16q in human prostate cancer. Br J Urol 1994, 73:390-395 [DOI] [PubMed] [Google Scholar]
- 3.Kunimi K, Bergerheim US, Larsson IL, Ekman P, Collins VP: Allelotyping of human prostatic adenocarcinoma. Genomics 1991, 11:530-536 [DOI] [PubMed] [Google Scholar]
- 4.Ittmann M: Allelic loss on chromosome 10 in prostate adenocarcinoma. Cancer Res 1996, 56:2143-2147 [PubMed] [Google Scholar]
- 5.Komiya A, Suzuki H, Ueda T, Yatani R, Emi M, Shimazaki J: Allelic losses at loci on chromosome 10 are associated with metastasis and progression of human prostate cancer. Genes Chromosomes Cancer 1996, 17:245-253 [DOI] [PubMed] [Google Scholar]
- 6.Trybus TM, Burgess AC, Wojno KJ, Glover TW, Macoska JA: Distinct areas of allelic loss on chromosomal regions 10p and 10q in human prostate cancer. Cancer Res 1996, 56:2263-2267 [PubMed] [Google Scholar]
- 7.Yasunaga Y, Nakamura K, Ko D, Srivastava S, Moul JW, Sesterhenn IA, McLeod DG, Rhim JS: A novel human cancer culture model for the study of prostate cancer. Oncogene 2001, 20:8036-8041 [DOI] [PubMed] [Google Scholar]
- 8.Murakami YS, Albertsen H, Brothman AR, Leach RJ, White RL: Suppression of the malignant phenotype of human prostate cancer cell line PPC-1 by introduction of normal fragments of human chromosome 10. Cancer Res 1996, 56:2157-2160 [PubMed] [Google Scholar]
- 9.Fukuhara H, Maruyama T, Nomura S, Oshimura M, Kitamura T, Sekiya T, Murakami Y: Functional evidence for the presence of tumor suppressor gene on chromosome 10p15 in human prostate cancers. Oncogene 2001, 20:314-319 [DOI] [PubMed] [Google Scholar]
- 10.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, Ferrari AC, Martignetti JA, Friedman SL: KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 2001, 294:2563-2566 [DOI] [PubMed] [Google Scholar]
- 11.Dong JT, Chen C, Stultz BG, Isaacs JT, Frierson HF, Jr: Deletion at 13q21 is associated with aggressive prostate cancers. Cancer Res 2000, 60:3880-3883 [PubMed] [Google Scholar]
- 12.Chen C, Frierson HF, Jr, Haggerty PF, Theodorescu D, Gregory CW, Dong JT: An 800 kb region of deletion at 13q14 in human prostate and other carcinomas. Genomics 2001, 77:135-144 [DOI] [PubMed] [Google Scholar]
- 13.Chen C, Brabham WW, Stultz BG, Frierson HFJ, Barrett JC, Sawyers CL, Isaacs JT, Dong JT: Defining a common region of deletion at 13q21 in human cancers. Genes Chromosomes Cancer 2001, 31:333-344 [DOI] [PubMed] [Google Scholar]
- 14.Goldberg EK, Glendening JM, Karanjawala Z, Sridhar A, Walker GJ, Hayward NK, Rice AJ, Kurera D, Tebha Y, Fountain JW: Localization of multiple melanoma tumor-suppressor genes on chromosome 11 by use of homozygosity mapping-of-deletions analysis. Am J Hum Genet 2000, 67:417-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyytinen ER, Frierson HF, Boyd JC, Chung LWK, Dong JT: Three distinct regions of allelic loss at 13q14, 13q21–22, and 13q33 in prostate cancer. Genes Chromosomes Cancer 1999, 25:108-114 [PubMed] [Google Scholar]
- 16.Chen C, Bhalala HV, Qiao H, Dong JT: A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene 2002, 21:6567-6572 [DOI] [PubMed] [Google Scholar]
- 17.Knudson AG, Jr: Genetics and etiology of human cancer. Adv Hum Genet 1977, 8:1-66 [DOI] [PubMed] [Google Scholar]
- 18.Philipsen S, Suske G: A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res 1999, 27:2991-3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang DT, Bachman KE, Mahatan CS, Dang LH, Giardiello FM, Yang VW: Decreased expression of the gut-enriched Kruppel-like factor gene in intestinal adenomas of multiple intestinal neoplasia mice and in colonic adenomas of familial adenomatous polyposis patients. FEBS Lett 2000, 476:203-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramaniam M, Hefferan TE, Tau K, Peus D, Pittelkow M, Jalal S, Riggs BL, Roche P, Spelsberg TC: Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. J Cell Biochem 1998, 68:226-236 [PubMed] [Google Scholar]
- 21.Whang YE, Wu X, Suzuki H, Reiter RE, Tran C, Vessella RL, Said JW, Isaacs WB, Sawyers CL: Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci USA 1998, 95:5246-5250 [DOI] [PMC free article] [PubMed] [Google Scholar]