Abstract
Toll-like receptors (TLRs) are involved in mediating cell activation on stimulation with microbial constituents. We investigated the role for TLRs in synovial fibroblast (SF) activation in rheumatoid arthritis (RA). We analyzed whether stimulation with interleukin-1β and tumor necrosis factor-α, cytokines present in RA synovium, influences expression of TLR genes in SFs. The effects were compared with those of treatment with lipopolysaccharide and a synthetic lipopeptide (sBLP). Gene expression was examined using quantitative polymerase chain reaction. TLR2-mediated cell activation was investigated by electromobility shift assay for nuclear factor-κB. To localize TLR2 expression in joint tissue sections of RA patients were stained using in situ hybridization. Expression of TLR2 in RA SFs was increased after treatment with interleukin-1β, tumor necrosis factor-α, lipopolysaccharide, and sBLP. Nuclear factor-κB translocation in SFs was triggered by TLR2-mediated cell stimulation. Synovial tissues from RA joints expressed TLR2 predominantly at sites of attachment and invasion into cartilage and bone. The observed elevated expression of TLR2 in RA SFs could be a consequence of direct exposure to microbial compounds or of the presence of inflammatory mediators in the joint. TLR-associated signaling pathways may contribute to the pathogenesis of RA, either by initiating or perpetuating activation of SFs.
Rheumatoid arthritis (RA) is a systemic debilitating disease whose primary symptoms consist of chronic inflammation, synovial hyperplasia, and destruction of cartilage and bone of numerous joints. The activation of synovial fibroblasts (SFs) has been found to be an important feature in the destructive processes of RA. However, the mechanisms leading to this activation are not clear.
Toll receptor proteins in Drosophila are involved in establishing the dorso-ventral axis during embryogenesis. They also play a fundamental role in the activation of the innate immune system on infections. The mammalian homologues belong to a family that currently consists of 10 members. Toll-like receptors (TLR) 1 to 10. 1-4 The cytoplasmic domains of the TLRs, beside their structural and functional similarities to Drosophila Toll, show homologies to the interleukin (IL)-1 receptor intracellular signaling domain (TIR domain). They are expressed differentially in certain tissues and cell types 1,4-6 and contribute to the specific recognition of various microbial constituents followed by the initiation of cellular activation. 7,8 TLR2 is involved in the recognition of various gram-positive bacterial compounds and leptospiral lipopolysaccharide (LPS), 9,10 whereas TLR4 and TLR9 have been shown to be involved in the innate response to LPS and to nonmethylated CG-rich DNA (CpG), respectively. TLRs have been proposed to act as pattern recognition receptors. 11,12 Pattern recognition receptors are germ line-encoded receptors, which are involved in the recognition of conserved microbial constituents such as LPS and zymosan. It remains unclear whether TLRs are only engaged by microbial constituents or if they can also bind to endogenous ligands, as it is the case for Drosophila Toll. There is some evidence for heat shock protein 60, which has been proposed to play a role in the induction of RA, being an endogenous ligand of TLR4. 13
TLRs have been shown to mediate activation of nuclear factor (NF)-κB and mitogen-activated protein kinase, 4,14 resulting in the production of mediators of the innate immune system such as IL-1, IL-6, IL-8, or tumor necrosis factor (TNF)-α. 15-17 Additionally, TLR2 has been shown to mediate induction of apoptosis. 18,19 Notably, an imbalance in the regulation of apoptosis in SFs is thought to be important for the induction of hyperplasia in the synovial lining. 20,21 Not much is known about the regulation of TLR expression. Recent studies have shown that expression of TLR2, in contrast to TLR4, is up-regulated after stimulation with TNF-α or other stimuli in myeloid 22,23 and muscle cells. 24
In the present study we investigated the expression of TLR2, TLR4, and TLR9 in cultured synovial cells after stimulation with IL-1β and TNF-α to examine the regulation of TLR-expression under RA typical conditions. Additionally, the cells were exposed to microbial compounds including LPS and a soluble synthetic bacterial lipopeptide (sBLP) that are known to act as ligands of TLRs and to influence the expression of TLR2 and TLR4. Activation of synovial cells by TLR2 was demonstrated with electrophoretic mobility shift assay (EMSA) for NF-κB. Further on, the expression pattern of TLR2 mRNA was assessed in synovial tissues by in situ hybridization.
Materials and Methods
Tissues
Normal synovial tissues were obtained from patients undergoing joint surgery after injury (Department of Surgery, University Hospital, Zurich, Switzerland). Synovial tissues from patients with RA and osteoarthritis (OA) were obtained from the Schulthess Clinic, Zurich, Switzerland. The tissues were either directly embedded in O.C.T. compound (Tissue-Tek TT 4583, Sakura Finetech, Torrance, CA) followed by snap-freezing in liquid nitrogen or fixed in 4% buffered formaldehyde for less than 24 hours at room temperature and embedded in paraffin.
Generation of TLR2-Specific RNA Probes (Riboprobes) for in Situ Hybridization
Digoxigenin-labeled anti-sense and sense probes specific for TLR2 mRNA were generated as follows: first strand complementary DNA (cDNA) from healthy donor blood mononuclear cells was synthesized by reverse transcription (Omniscript; Qiagen, Basel, Switzerland). A fragment corresponding to the 443 nucleotides ranging from position 2 to 445 of the human TLR2 cDNA sequence (accession no. U88878, National Center for Biotechnology Information; www.ncbi.nlm.nih.gov) was amplified by polymerase chain reaction (PCR) and ligated into pBluescript KS II (Stratagene, Heidelberg, Germany). The inserted DNA was sequenced (ABI Prism 310 Genetic analyzer; Applied Biosystems, Rotkreut, Switzerland). The riboprobes were synthesized by in vitro transcription using RNA polymerase (Stratagene) in the presence of digoxygenin-11-UTP (Boehringer-Mannheim, Rotkreuz, Switzerland).
Detection of TLR2 mRNA by in Situ Hybridization
In situ hybridization was performed according to a protocol reported earlier. 25 Briefly, after deparaffinization, slides were prehybridized in freshly prepared prehybridization solution (50% formamide, 1× Denhardt’s solution, 8× standard saline citrate, 0.2 g/ml dextran sulfate, 0.5 mg/ml herring sperm DNA, and 0.25 mg/ml yeast tRNA) for 1 hour at room temperature. Hybridization was performed using 0.2 μg of RNA probe (diluted 1:10 in prehybridization solution) per slide under a coverslip at 52°C overnight. Unbound RNA probe was digested at 37°C for 45 minutes with 10 μg/ml of RNase A (Boehringer-Mannheim). Sections then were washed with 50% formamide/2× standard saline citrate followed by three graded baths of sodium dodecyl sulfate/standard saline citrate (1×/0.5×/0.1×) for 18 minutes at 50°C. Unspecific binding was blocked with 2% normal horse serum in Tris-buffered saline (TBS) (pH 7.6) for 30 minutes at room temperature. Hybridized probes were detected using anti-digoxigenin Fab′ fragments coupled to alkaline phosphatase (Boehringer-Mannheim), diluted in TBS with 1% horse serum and nitro blue tetrazolim/5-bromo-4-chloro-3-indolyl phosphate substrate (Boehringer-Mannheim). Slices were counterstained with 0.1% nuclear fast red.
Immunohistochemical Detection of T Lymphocytes, Macrophages, and Fibroblasts
After in situ hybridization and detection as described above, immunohistochemistry was performed as follows: slices were blocked by treatment with 4% milk and 2% horse serum in TBS for 1 hour at room temperature. Monoclonal antibodies (mouse) against human CD3 (dilution 1:100; DAKO, Hamburg, Germany), antibodies (mouse) against human CD68 (dilution 1:200, DAKO), antibodies (mouse) against human prolyl-4-hydroxylase (1:25; Acris, Hiddenhausen, Germany), or antibodies (mouse) against human vimentin (dilution 1:200, V2258; Sigma, Buchs, Switzerland) in TBS with 2% milk powder were applied 1 hour at room temperature. Secondary goat anti-mouse antibodies coupled with horseradish peroxidase were diluted 1:200 in TBS with 2% milk powder and applied for 30 minutes at room temperature. Detection of the horseradish peroxidase was performed using a three-step alkaline phosphatase/anti-alkaline phosphatase kit (Biogenex, San Ramon, CA) according to the manufacturer’s instructions. For antibodies against CD3 only snap-frozen tissue sections were used to avoid antigen-unmasking procedures. The corresponding sections stained with the sense-control probes were negative (not shown).
Isolation and Culture of SFs
Synovial tissues were cut into small pieces and dissociated enzymatically for 1 hour with Dispase I (Boehringer-Mannheim). The resulting cell suspension was seeded into cell culture dishes and cultured in Dulbecco’s modified Eagle’s medium (DMEM/Nutrient Mix F12, 1:1; Life Technologies, Inc., Grand Island, NY), supplemented with 10% (v/v) fetal calf serum (Life Technologies, Inc.), 100 IU/ml penicillin, 100 μg/ml streptomycin (Life Technologies, Inc.), 1% l-Glutamine (Life Technologies, Inc.), 1% HEPES buffer (Life Technologies, Inc.) at 37°C in a 5% CO2 and 95% humidity. Cells were trypsinized and mRNA extracted. In all experiments, cells were used in passages 3 to 8.
Stimulation of SFs
SFs from joints affected from RA (n = 6) or OA (n = 7) were cultured to 80 to 90% confluency in Petri dishes, washed with phosphate-buffered saline (PBS), and incubated with 2 ng/ml of IL-1β (R&D Systems, Minneapolis, MN) in PBS with 0.1% bovine serum albumin, 100 U/ml recombinant human TNF-α (Roche, Rotkreuz, Switzerland), 100 ng/ml of LPS from Escherichia coli or Salmonella minnesota (List Biologicals, CA) or 100 ng/ml of synthetic bacterial lipopeptide (sBLP, Boehringer-Mannheim) with 0.05% human serum albumin. All reagents were diluted in complete culture medium. Cells were collected and mRNA extracted after 24 hours of incubation.
Reverse Transcription-PCR
Total RNA from cultured SFs was isolated with RNeasy total RNA isolation system (Qiagen) including treatment with RNase-free DNase (Qiagen). Reverse transcription of each 1 μg of RNA was performed using MuLV reverse transcriptase (Applied Biosystems) according to the manufacturer’s protocol.
Real-Time Quantitative Reverse Transcriptase-PCR (TaqMan)
Primers and probes for 18S RNA, TLR2, TLR4, and TLR9 are listed with their corresponding optimal concentrations in Table 1 ▶ . The method of relative quantification was based on the fluorogenic 5-minute nuclease assay using the ABI Prism 7700 Sequence Detection System (Applied Biosystems). Successful DNase treatment of the RNA samples was ensured by comparing the Ct values of 18S rRNA amplified from the equal amount of RNA sample that was used for cDNA transcription. The amounts of target mRNA (TLR2, TLR4, and TLR9) and of the endogenous reference (18S rRNA) were determined by calculation based on the appropriate standard curve as established in preliminary experiments according to the manufacturer’s instructions (Applied Biosystems). The amount of the corresponding target mRNA divided by the amount of mRNA of the endogenous control results in a normalized target value. All normalized target values are compared to the untreated controls.
Table 1.
Primers and Probes Used for Real-Time RT-PCR (TaqMan)
| Primer/probe | Sequence | Final concentration |
|---|---|---|
| 18-S rRNA forward | 5′-AGTCCCTGCCCTTTGTACACA-3′ | 400 nmol/L |
| 18-S rRNA reverse | 5′-GATCCGAGGGCCTCACTAAAC-3′ | 400 nmol/L |
| 18-S rRNA probe | 5′-CGCCCGTCGCTACTACCGATTGG-3′ | 250 nmol/L |
| TLR2 forward | 5′-GGCCAGCAAATTACCTGTGTG-3′ | 200 nmol/L |
| TLR2 reverse | 5′-AGGCGGACATCCTGAACCT-3′ | 200 nmol/L |
| TLR2 probe | 5′-CTCCATCCCATGTGTGCGTGGCC-3′ | 250 nmol/L |
| TLR4 forward | 5′-GTCCTGCAGAAGGTGGAGAAGA-3′ | 250 nmol/L |
| TLR4 reverse | 5′-GGTAAGTGTTCCTGCTGAGAAGG-3′ | 250 nmol/L |
| TLR4 probe | 5′-CAGCAGGTGGAGCTGTACCGCCTTCT-3′ | 900 nmol/L |
| TLR9 forward | 5′-TGAAGACTTCAGGCCCAACTG-3′ | 300 nmol/L |
| TLR9 reverse | 5′-TGCACGGTCACCAGGTTGT-3′ | 100 nmol/L |
| TLR9 probe | 5′-AGCACCCTCAACTTCACCTTGGATCTGTC | 175 nmol/L |
Activation of NF-κB and EMSA
RA SFs (n = 3) were cultured to 80 to 90% confluency in culture flasks (225 cm2), and incubated with 100 U/ml of recombinant human TNF-α (Roche), 10 μg/ml of synthetic bacterial lipopeptide (sBLP, Boehringer-Mannheim) with 0.05% human serum albumin, or as a negative control for TLR2-specific stimulation with 10 μg/ml of the biologically inactive analogue of sBLP, Pam3Cys (Novabiochem, Laufelfingen, Switzerland) with 0.05% human serum albumin. All reagents were diluted in complete culture medium. Cells were collected by scratching them in ice-cold PBS after stimulation at different time points (0 minutes/20 minutes/40 minutes/60 minutes) after stimulation with sBLP or after 30 minutes after stimulation with TNF-α or Pam3Cys. Nuclear extracts were either prepared according to the method of Andrews and Faller 26 or by using NE-PER nuclear and cytoplasmic extraction reagents (Pierce, Rockford, IL). Nuclear protein concentrations were determined using the BCA Protein Assay Reagent kit (Pierce). Nonradioactive EMSA was performed using an EMSA kit according to the manufacturer’s instructions (Panomics, Inc., Redwood City, CA). Six μg of nuclear protein were used to bind biotinylated oligonucleotides containing the NF-κB-binding site for 30 minutes at room temperature. The samples were separated in a nondenaturing polyacrylamide gel (6%, with 2.5% glycerol) and blotted on a Biodyne B (0.45 μm) positively charged nylon membrane (Pall Schweiz AG, Basel, Switzerland). The biotin was labeled with alkaline phosphatase-conjugated streptavidin (1:1000, DAKO) and alkaline phosphatase was detected with CDP-Star substrate (Applied Biosystems) according to the manufacturer’s instructions.
Results
TaqMan Analysis
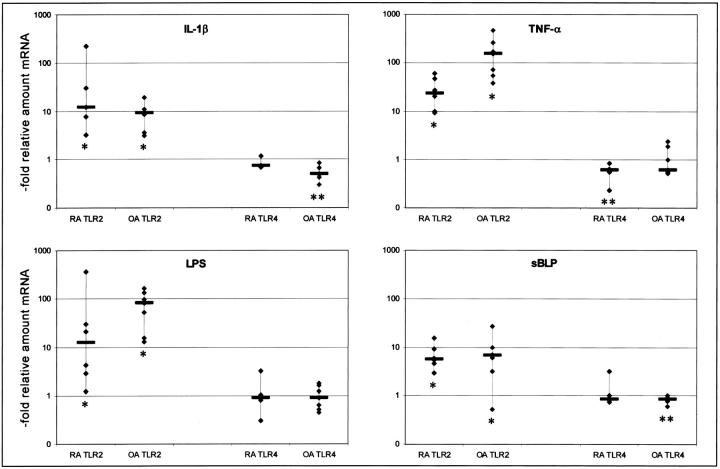
We initially examined the expression of TLR mRNAs in SFs derived from joints affected with RA, OA, or nonarthritic joints. No differences in the base line expression of TLR2, TLR4, or TLR9 were detectable between these different tissues (data not shown). For the following experiments, we used SFs from RA-affected joints and cells from joints affected with OA. After stimulation of SFs with sBLP or LPS we found a marked statistically significant increase in the expression of TLR2 (Wilcoxon paired-sample test, P < 0.05) after exposure to both stimuli in RA SFs as well as in OA SFs (Figure 1) ▶ . Stimulation of SFs with the proinflammatory cytokines IL-1β and TNF-α also led to an increase of TLR2 mRNA. Neither TLR4 (Figure 1) ▶ not TLR9 (data not shown) gene expression was up-regulated by any of the stimuli used. A slight, but reproducible decrease in TLR4 gene expression was observed after stimulation with IL-1β, TNF-α, and sBLP, partially reaching statistical significance (Wilcoxon paired-sample test, P < 0.05) (Figure 1) ▶ . Expression of TLR9 mRNA remained undetectable by quantitative PCR.
Figure 1.
Effects of stimulation on the expression of TLR2 and TLR4 mRNA in RA (n = 6) and OA (n = 7) SFs. The ordinate shows the relative increase of specific mRNA compared to nonstimulated cells. Each symbol indicates one cell culture from one RA patient examined. The bars indicate the medians. The increase of TLR2 mRNA in response to all four stimuli reached statistical significance (*, Wilcoxon paired-sample test; tied Z values from −2.37 to −2.02, P values from 0.02 to 0.04). Levels of TLR4 mRNA expression of stimulated cells did not differ significantly from nonstimulated cells apart from stimulation of RA SFs with TNF-α, and from stimulation of OA SFs with IL-1β or sBLP (**, Wilcoxon paired-sample test; tied Z values from−2.2 to −2.36, P values from 0.02 to 0.04).
In Situ Hybridization and Immunohistochemical Double Labeling
We investigated whether the increase in the expression of TLR2 mRNA could also be detected in synovial tissues from RA patients. In situ hybridization revealed that tissues from joints affected by RA showed a pronounced expression of TLR2 mRNA in the synovial lining and at sites of attachment and invasion into cartilage or bone tissue (Figure 2A) ▶ . Labeling was also observed in areas around small vessels (Figure 2C) ▶ and in areas of infiltrating lymphocytes (Figure 2E) ▶ . Figure 2, B, D, and F ▶ , shows the corresponding control hybridizations with the sense probes for TLR2 mRNA. Double labeling for CD3 and TLR2 mRNA revealed intense expression of TLR2 mRNA at sites of infiltrating lymphocytes (Figure 3C) ▶ , but the synovial cells expressing high levels of TLR2 mRNA were not CD3-positive T cells. Tissue sections double-stained for TLR2 and CD68 to detect macrophages (Figure 3 ▶ ; D to F) showed that the majority of the cells expressing TLR2 were also not positive for CD68. Finally, sections double-stained for TLR2 and vimentin or prolyl-4-hydroxylase (Figure 3 ▶ ; G to I) showed that a large percentage of cells expressing TLR2 were also positive for these fibroblast markers suggesting that the expression of TLR2 mRNA is mainly confined to SFs. Representative examples of tissue sections derived from nonarthritic joints (Figure 3B) ▶ or from patients affected by OA (Figure 3A) ▶ showed an overall weak expression of TLR2 mRNA compared to tissues from joints affected with RA. No sites of strong local expression of TLR2 mRNA were found in these non-RA tissues.
Figure 2.
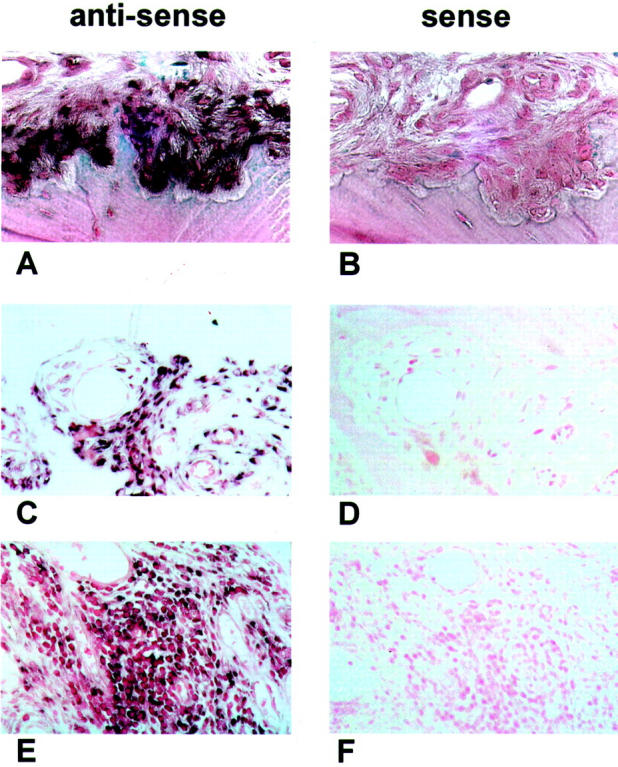
TLR2 in situ hybridizations of RA synovial tissues. One representative section of RA synovial tissue of seven patients, hybridized in situ with specific RNA probes for TLR2 mRNA (A, C, E). Cells positive for TLR2 mRNA are dark purple. As negative controls, corresponding tissue sections were hybridized with the sense probes. All tissues hybridized with the sense control probes show no specific signals (B, D, F). Cells expressing detectable levels of TLR2 mRNA are located in areas of adhesion and invasion into bone or cartilage (A; negative control, B), around small vessels (C; negative control, D) and in areas of lymphocyte infiltrations (E; negative control, F). Original magnifications: ×200 (A, B, C, E); ×100 (D, F).
Figure 3.
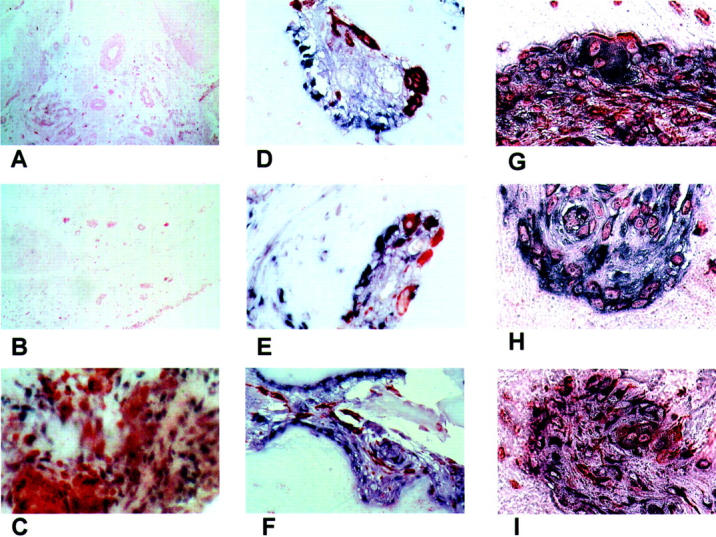
Control and double stainings. A: One representative section from OA tissues of four patients stained for TLR2 mRNA. B: One representative tissue section from one normal subject of two stained for TLR2 mRNA. Representative tissue section derived from RA-synovium, stained with anti-CD3 antibodies (specific for T cells) after in situ hybridization with a probe specific for TLR2 mRNA. TLR2 mRNA presents as purple/brown color, T cells are red. C: Expression of TLR2 mRNA and CD3 is detectable in distinct cells. Representative tissue sections derived from RA-synovium at sites of synovial invasion into cartilage. The sections are stained with anti-CD68 antibodies (specific for macrophages) after in situ hybridization with probes specific for TLR2 mRNA. TLR2 mRNA presents as purple color, macrophages are red. D–F: Most of the cells expressing TLR2 mRNA are not expressing CD68. Representative tissue section derived from RA synovium at sites of synovial invasion into cartilage. The sections are stained with antibodies against vimentin (G–H) or prolyl-4-hydroxylase (I) after in situ hybridization with probes specific for TLR2 mRNA. TLR2 presents as purple/black color, antibodies are visible as orange. A great portion of cells expressing TLR2 is positive for the fibroblast markers. Original magnifications: ×100 (A, B); ×200 (C); ×630 (D, E, G, I); ×400 (F).
EMSA (NF-κB)
We examined the functional potential of TLR2 expression on cultured SFs using nonradioactive EMSA for NF-κB after stimulation with specific ligands of TLR2. Stimulation with 10 μg/ml of sBLP led to a strong increase of NF-κB translocation into the nucleus at levels comparable to those observed after stimulation with 20 ng/ml of TNF-α. As controls we used nonstimulated cells from the same culture or cells treated with a biologically inactive analogue of sBLP (Pam3Cys) (Figure 4) ▶ .
Figure 4.
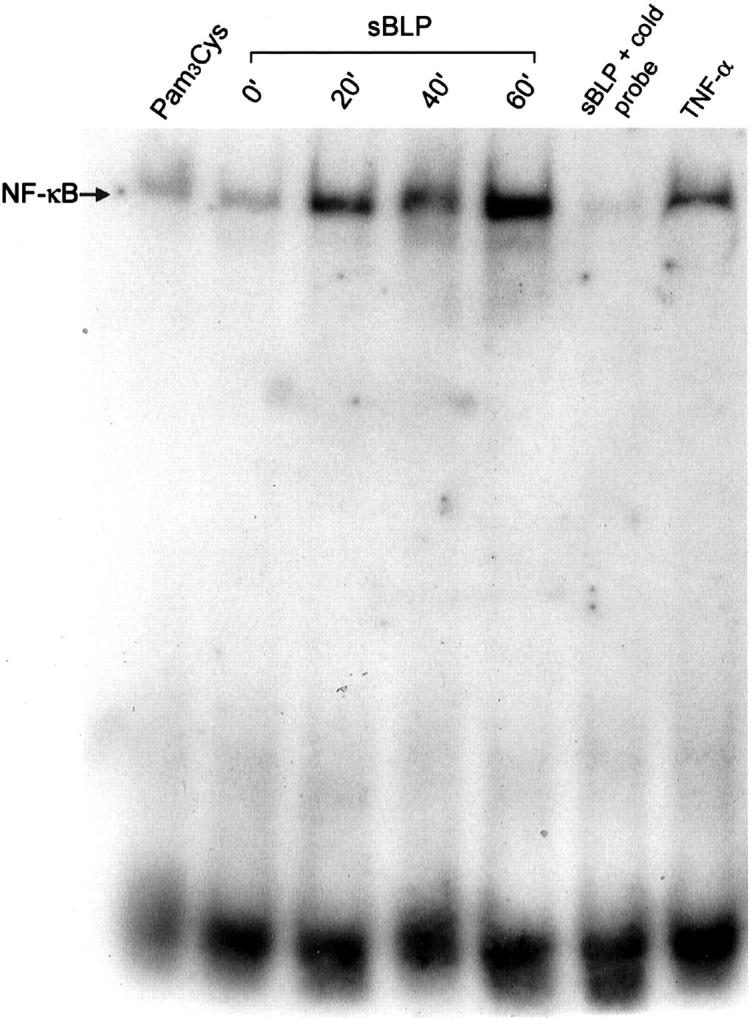
Nuclear translocation of NF-κB after TLR2-dependent stimulation. Representative EMSA for examining the effects of stimulation with sBLP on the nuclear translocation of NF-κB in RA SFs. Three different time points (20 minutes, 40 minutes, 60 minutes) after stimulation are shown compared to nonstimulated cells (0 minutes), cells exposed to Pam3Cys, or stimulated with TNF-α. The specificity of NF-κB binding to the oligonucleotides was confirmed by the addition of unlabeled probe (cold probe) to the binding reaction. The data show one representative experiment of three.
Discussion
Activation of the innate immune system is an important feature in the pathogenesis of RA. The direct involvement of a specific pathogen for RA has to date not been demonstrated. However, many studies still follow the assumption that microbial triggers are involved in the pathogenesis of various autoimmune diseases and RA, either through direct activation of synovial cells or through molecular mimicry processes turning an infection into an autoimmune disease.
TLRs are directly involved in signaling the presence of various microbial constituents including LPS and in mediating proinflammatory reactions through activating NF-κB and other proinflammatory signaling pathways. Hsp60, an agent possibly contributing to RA, 27 has recently been proposed to be an endogenous ligand for TLR4 in mouse macrophages. 14 TLRs are not only up-regulating the expression of agents inducing inflammation such as IL-1β, IL-6, IL-8, and TNF-α, 28 but also initiating the activation of the adaptive immune responses through an elevation of the expression of co-stimulatory factors such as B7.1. 28,29 A recent report provides evidence that innate immune responses mediated by TLRs to bacterial DNA could be involved in triggering B cells to produce the autoreactive rheumatoid factor in a T-cell-independent manner. 30,31
It has been shown that TLR2 can induce apoptosis when constitutively overexpressed in THP-1 cells, 18 whereas in muscle cells it mediates anti-apoptotic effects. 19 Indeed, TLR2 can mediate activation of both caspase 8, a key element in inducing apoptosis, as well as translocation of NF-κB and anti-apoptotic signaling. 18 Because a misbalance in the regulation of apoptosis has been proposed to play an important role in the hyperplasia of RA synovium, 21 such effects of TLR2 related to apoptosis have to be taken into account in further investigations.
TLR2 may induce the expression of pro-IL-1β and activate caspase 1, an enzyme activating IL-1β, and thus might be directly involved in the inflammatory responses caused by stimulation with synthetic soluble bacterial lipopeptide. 18
To further investigate the potential role of TLRs in the pathogenesis of RA, we stimulated cultured SFs with the proinflammatory cytokines IL-1β and TNF-α as well as with LPS and sBLP. Stimulation with all of the four substances resulted in a significant increase in TLR2 mRNA expression. By contrast, levels of TLR4 remained unchanged or, after stimulation with TNF-α, showed a slight decrease. TLR9 expression was not detectable by quantitative PCR in both nonstimulated and stimulated cells. These effects seem not to be specific features of RA SFs because cells derived from joints affected with OA responded similarly or even stronger (TNF-α, LPS) to the same stimuli. Similar changes in expression of TLR2 and TLR4 after stimulation with inflammatory stimuli have been observed in myeloid cells. 22,23 Nevertheless, recent literature is controversial whether this change in expression of TLR2 is associated on a functional level by an increased or decreased responsiveness to microbial compounds.
In summary, based on the increased expression of TLR2 mRNA in synovial tissues of patients with RA and on the in vitro experiments with cell cultures, we propose that the up-regulation of TLR2 corresponds either to a response to an exposure to microbial compounds or is secondary to the inflammatory milieu present in the rheumatoid synovium. At the current time, we cannot discriminate between these two possibilities. However, based on our finding that activation of NF-κB is strongly enhanced after TLR2-specific cell stimulation, we suggest that TLR2-dependent mechanisms may contribute to the activation of synovial cells, possibly leading to the destruction of cartilage and bone in the pathogenesis of RA.
Footnotes
Address reprint requests to Roger P. Lauener, M.D., Div. for Immunology, Zurich University Children’s Hospital, Steinwiesstrasse 75, CH-8032 Zürich, Switzerland. E-mail: rlauener@kispi.unizh.ch.
Supported by the Swiss National Research Foundation (grants 32-41649.94 and 32-50878.97) and the Bonizzi Theler Foundation.
References
- 1.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF: A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA 1998, 95:588-593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeuchi O, Kawai T, Sanjo H, Copeland NG, Gilbert DJ, Jenkins NA, Takeda K, Akira S: TLR6: a novel member of an expanding Toll-like receptor family. Gene 1999, 231:59-65 [DOI] [PubMed] [Google Scholar]
- 3.Du X, Poltorak A, Wei Y, Beutler B: Three novel mammalian Toll-like receptors: gene structure, expression, and evolution. Eur Cytokine Netw 2000, 11:362-371 [PubMed] [Google Scholar]
- 4.Aderem A, Ulevitch RJ: Toll-like receptors in the induction of the innate immune response. Nature 2000, 406:782-787 [DOI] [PubMed] [Google Scholar]
- 5.Muzio M, Bosisio D, Polentarutti N, D’Amico G, Stoppacciaro A, Mancinelli R, van’t Veer C, Penton-Rol G, Ruco LP, Allavena P, Mantovani A: Differential expression and regulation of Toll-like receptors (TLR) in human leukocytes: selective expression of TLR3 in dendritic cells. J Immunol 2000, 164:5998-6004 [DOI] [PubMed] [Google Scholar]
- 6.Daun JM, Fenton MJ: Interleukin-1/Toll receptor family members: receptor structure and signal transduction pathways. J Interferon Cytokine Res 2000, 20:843-855 [DOI] [PubMed] [Google Scholar]
- 7.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S: Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 1999, 11:443-451 [DOI] [PubMed] [Google Scholar]
- 8.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S: A Toll-like receptor recognizes bacterial DNA. Nature 2000, 408:740-745 [DOI] [PubMed] [Google Scholar]
- 9.Iwaki D, Mitsuzawa H, Murakami S, Sano H, Konishi M, Akino T, Kuroki Y: The extracellular Toll-like receptor 2 domain directly binds peptidoglycan derived from Staphylococcus aureus. J Biol Chem 2002, 277:24315-24320 [DOI] [PubMed] [Google Scholar]
- 10.Werts C, Tapping RI, Mathison JC, Chuang TH, Kravchenko V, Saint Girons I, Haake DA, Godowski PJ, Hayashi F, Ozinsky A, Underhill DM, Kirschning CJ, Wagner H, Aderem A, Tobias PS, Ulevitch RJ: Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat Immunol 2001, 2:346-352 [DOI] [PubMed] [Google Scholar]
- 11.Janeway CA, Jr: The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol Today 1992, 13:11-16 [DOI] [PubMed] [Google Scholar]
- 12.Medzhitov R, Janeway CAJ: On the semantics of immune recognition. Res Immunol 1996, 147:208-214 [DOI] [PubMed] [Google Scholar]
- 13.Ohashi K, Burkart V, Flohe S, Kolb H: Heat shock protein 60 is a putative endogenous ligand of the Toll-like receptor-4 complex. J Immunol 2000, 164:558-561 [DOI] [PubMed] [Google Scholar]
- 14.Kaisho T, Akira S: Dendritic-cell function in Toll-like receptor- and MyD88-knockout mice. Trends Immunol 2001, 22:78-83 [DOI] [PubMed] [Google Scholar]
- 15.Wang JE, Warris A, Ellingsen EA, Jorgensen PF, Flo TH, Espevik T, Solberg R, Verweij PE, Aasen AO: Involvement of CD14 and Toll-like receptors in activation of human monocytes by Aspergillus fumigatus hyphae. Infect Immun 2001, 69:2402-2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi O, Hoshino K, Akira S: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J Immunol 2000, 165:5392-5396 [DOI] [PubMed] [Google Scholar]
- 17.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F: Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem 1999, 274:10689-10692 [DOI] [PubMed] [Google Scholar]
- 18.Aliprantis AO, Yang RB, Weiss DS, Godowski P, Zychlinsky A: The apoptotic signaling pathway activated by Toll-like receptor-2. EMBO J 2000, 19:3325-3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frantz S, Kelly RA, Bourcier T: Role of TLR-2 in the activation of nuclear factor kappa B by oxidative stress in cardiac myocytes. J Biol Chem 2001, 276:5197-5203 [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto S, Muller-Ladner U, Gay RE, Nishioka K, Gay S: Ultrastructural demonstration of apoptosis, Fas and Bcl-2 expression of rheumatoid synovial fibroblasts. J Rheumatol 1996, 23:1345-1352 [PubMed] [Google Scholar]
- 21.Franz JK, Pap T, Hummel KM, Nawrath M, Aicher WK, Shigeyama Y, Muller-Ladner U, Gay RE, Gay S: Expression of sentrin, a novel antiapoptotic molecule, at sites of synovial invasion in rheumatoid arthritis. Arthritis Rheum 2000, 43:599-607 [DOI] [PubMed] [Google Scholar]
- 22.Matsuguchi T, Musikacharoen T, Ogawa T, Yoshikai Y: Gene expressions of Toll-like receptor 2, but not Toll-like receptor 4, is induced by LPS and inflammatory cytokines in mouse macrophages. J Immunol 2000, 165:5767-5772 [DOI] [PubMed] [Google Scholar]
- 23.Flo TH, Halaas O, Torp S, Ryan L, Lien E, Dybdahl B, Sundan A, Espevik T: Differential expression of Toll-like receptor 2 in human cells. J Leukoc Biol 2001, 69:474-481 [PubMed] [Google Scholar]
- 24.Watari M, Watari H, Nachamkin I, Strauss JF: Lipopolysaccharide induces expression of genes encoding pro-inflammatory cytokines and the elastin-degrading enzyme, cathepsin S, in human cervical smooth-muscle cells. J Soc Gynecol Invest 2000, 7:190-198 [DOI] [PubMed] [Google Scholar]
- 25.Hummel KM, Petrow PK, Franz JK, Muller-Ladner U, Aicher WK, Gay RE, Bromme D, Gay S: Cysteine proteinase cathepsin K mRNA is expressed in synovium of patients with rheumatoid arthritis and is detected at sites of synovial bone destruction. J Rheumatol 1998, 25:1887-1894 [PubMed] [Google Scholar]
- 26.Andrews NC, Faller DV: A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res 1991, 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirata D, Hirai I, Iwamoto M, Yoshio T, Takeda A, Masuyama JI, Mimori A, Kano S, Minota S: Preferential binding with Escherichia coli hsp60 of antibodies prevalent in sera from patients with rheumatoid arthritis. Clin Immunol Immunopathol 1997, 82:141-148 [DOI] [PubMed] [Google Scholar]
- 28.Medzhitov R, Preston-Hurlburt P, Janeway CAJ: A human homologue of the drosophila Toll protein signals activation of adaptive immunity. Nature 1997, 388:394-397 [DOI] [PubMed] [Google Scholar]
- 29.Ogata H, Su I, Miyake K, Nagai Y, Akashi S, Mecklenbrauker I, Rajewsky K, Kimoto M, Tarakhovsky A: The Toll-like receptor protein RP105 regulates lipopolysaccharide signaling in B cells. J Exp Med 2000, 192:23-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A: Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature 2002, 416:603-607 [DOI] [PubMed] [Google Scholar]
- 31.Krieg AM: A role for Toll in autoimmunity. Nat Immunol 2002, 3:423-424 [DOI] [PubMed] [Google Scholar]