Abstract
Homologs of the chromatin-bound yeast silent information regulator 2 (SIR2) protein are found in organisms from all biological kingdoms. SIR2 itself was originally discovered to influence mating-type control in haploid cells by locus-specific transcriptional silencing. Since then, SIR2 and its homologs have been suggested to play additional roles in suppression of recombination, chromosomal stability, metabolic regulation, meiosis, and aging. Considering the far-ranging nature of these functions, a major experimental goal has been to understand the molecular mechanism(s) by which this family of proteins acts. We report here that members of the SIR2 family catalyze an NAD–nicotinamide exchange reaction that requires the presence of acetylated lysines such as those found in the N termini of histones. Significantly, these enzymes also catalyze histone deacetylation in a reaction that absolutely requires NAD, thereby distinguishing them from previously characterized deacetylases. The enzymes are active on histone substrates that have been acetylated by both chromatin assembly-linked and transcription-related acetyltransferases. Contrary to a recent report, we find no evidence that these proteins ADP-ribosylate histones. Discovery of an intrinsic deacetylation activity for the conserved SIR2 family provides a mechanism for modifying histones and other proteins to regulate transcription and diverse biological processes.
Yeast silent information regulator 2 (SIR2) protein functions in transcriptional silencing of the silent mating loci, telomeres, and rDNA (1–3). It is found in a chromatin-bound complex with SIR3 and SIR4 at the silent mating loci and telomeres, and in a different complex at rDNA (4–6). Four additional SIR2 homologs exist in yeast (HST1–4), and related proteins are found from archaeabacteria to eubacteria to mammals (7). Until recently, very little was known about the in vivo activity of this family of proteins. An important breakthrough came with the identification of the Salmonella typhimurium CobB protein as a SIR2 homolog (8). CobB can partially fulfill the requirement for CobT in vitamin B12 synthesis. Because CobT protein was known to transfer ribose 5′-phosphate from nicotinic acid mononucleotide to a precursor of vitamin B12, it prompted tests of Sir2-like proteins for phosphoribosyltransferase activity. Indeed, Frye (9) found that Escherichia coli CobB had NAD-dependent ADP-ribosyltransferase activity. He also reported that both CobB and a human SIR2-like protein could transfer radioactivity from [32P]NAD to albumin. Very recently, another group (10) reported that yeast SIR2 can ADP-ribosylate itself as well as histones and albumin.
Here we show that members of the SIR2 family of enzymes catalyze an NAD–nicotinamide exchange reaction that requires the presence of acetylated lysines such as are found in the N termini of histones. Furthermore, these enzymes also catalyze histone deacetylation in a reaction that absolutely depends on NAD, thereby distinguishing them from previously known deacetylases. Contrary to the recent report (10), we see no evidence that these proteins ADP-ribosylate histones. Very recently, an independent report (11) described NAD-dependent deacetylase activity on peptide substrates for yeast and mouse SIR2, in agreement with the findings reported here.
Materials and Methods
Construction of E. coli Expression Plasmids.
The HST2 E. coli expression plasmid was constructed as follows: the HST2 coding sequence was amplified from yeast genomic DNA by using primers HST2–NcoI (5′-TATACCATGGCTGTTTCTACCGCCTCTACA-3′) and HST2–XhoI (5′-GCAGTCGCTCGAGTTCTTTAGCGGCTTTTTG-3′). Amplified product was digested with NcoI and XhoI. The NcoI–NcoI and NcoI–XhoI fragments were purified and inserted into the expression vector, pET28b, in two steps to create pJWL03. The entire HST2 coding region was sequenced in both directions to confirm that there were no amplification-induced mutations. The cobB E. coli expression plasmid, pHEX-2TCobB, was a gift from R. Frye (9). The SIR2 E. coli expression plasmid, pDM111a, was a gift from D. Moazed (10).
Protein Overexpression.
HST2 was expressed from pJWL03 in E. coli BL21(DE3) after 3-h induction with 0.35 mM isopropyl β-d-thiogalactoside (IPTG) at room temperature. The protein was purified by using Novagen His Bind Affinity Resin according to the manufacturer's instructions. CobB from pHEX-2TCobB and SIR2 from pDM111a were expressed in BL21(DE3) and purified as described (9, 10). Purified proteins were dialyzed against 50 mM sodium phosphate (pH 7.2) and frozen at −80°C in 50 mM sodium phosphate (pH 7.2)/0.5 mM DTT/10% glycerol. Protein concentrations were estimated by comparing Coomassie brilliant blue staining of samples with BSA standards, analyzed by SDS/PAGE.
Exchange Reactions.
Exchange reactions were performed in 20-μl volumes with 5 mM NAD (Sigma N-1511), ≈50 ng of enzyme, 0.1 mM [14C]nicotinamide (Sigma N-2142, 53.1 mCi/mmol), 50 mM sodium phosphate (pH 7.2), and 0.5 mM DTT. Chicken erythrocyte histones were prepared as described previously (12) and used at a concentration of 0.2 mg/ml. H4 or H3 peptides, when present, were at a concentration of 25 μM. The H4 peptide corresponded to the first 28 amino acids of H4, and the H3 peptide corresponded to the first 20 amino acids of H3. Peptides were acetylated as described below. Exchange reactions were incubated at 37°C for 60 min. After incubation, 8 μl of each reaction was spotted to Whatman HPKF Silica Gel 60 A TLC plates. The plates were developed in a preequilibrated chamber with 80:20 ethanol:2.5 M ammonium acetate. After chromatography, the plates were air dried and exposed to film (Kodak X-Omat AR) for 36–48 h.
Deacetylation Reactions.
Deacetylation reactions were carried out in a two-step process. Histones/peptides were first acetylated with [3H]acetyl-CoA in 50-μl reactions with 75 mM Tris⋅Cl (pH 8.8), 135 mM NaCl, 0.2 mg/ml chicken erythrocyte histones, or 50 μM H4 or H3 peptide, 0.875 μCi of [3H]acetyl-CoA (Amersham TRK688, 3.80 Ci/mmol), and 40 ng of enzyme (HAT1, ESA1, or HPA2). Reactions were incubated at 37°C for 30 min and then heated at 55°C for 30 min to inactivate the histone acetyltransferase. Tests showed that this inactivated HAT1 and ESA1 but not HPA2. Acetylated histones/peptides were assayed for deacetylase activity in 25-μl reactions including 0.5 mM NAD, 50 mM sodium phosphate (pH 7.2), 0.5 mM DTT, 50 ng of enzyme to be tested, and 20 μl of a histone acetylation reaction as the substrate. Reactions were incubated at 37°C for 60 min. After incubation, the reactions were spotted to Whatman P81 cation-exchange paper and air dried. The dried papers were washed for 5 min three times with 1 M sodium carbonate (pH 9.0), followed by a wash in acetone. Papers were air dried, and radioactivity was quantitated in a liquid scintillation counter.
SDS/PAGE Analysis of Deacetylated Histones.
Chicken erythrocyte histones (0.15 mg) were acetylated with HAT1 or ESA1, and deacetylated with SIR2 or HST2, as described above. The histones then were precipitated by the addition of Triton X-100 and trichloroacetic acid to a final concentration of 1% and 10%, respectively. After 30 min on ice, the samples were centrifuged at 16,000 × g for 5 min. The pellet was washed twice with 1 ml of −80°C acetone and then air dried. Dried proteins were redissolved in 20 μl of protein loading buffer and resolved on a 15% SDS/polyacrylamide gel. Proteins were stained with Coomassie blue R-250 and destained. The destained gel was saturated with EN3HANCE (NEN catalog no. NEF981), dried under vacuum, and exposed to film (Kodak X-Omat AR) at −80°C for 24 h.
Triton–Acid–Urea Gel Electrophoresis.
Triton–acid–urea gel electrophoresis was carried out as described previously (13). Histone acetylation and deacetylation were done as described above with the exception that 0.24 mM nonradioactive acetyl-CoA replaced the [3H]acetyl-CoA. A total of 400 μg of histones in 2.1 ml was acetylated and deacetylated for each reaction. After deacetylation, Triton X-100 and trichloroacetic acid were added to a final concentration of 1% and 8.5%, respectively. Samples were chilled on ice for 30 min and centrifuged at 16,000 × g for 2 min. The precipitate was washed 2 times with −80°C acetone, air dried, and dissolved in 40 μl of sample buffer.
HPLC.
Reverse-phase HPLC analysis was performed on H4 peptides, either untreated, or acetylated with HAT1 and acetyl-CoA, or acetylated by HAT1 and then treated with HST2, as described above. Reactions were centrifuged for 5 min at 16,000 × g. Supernatants (≈50 μl) were added to 450 μl of 0.1% trifluoroacetic acid (TFA), and samples were resolved with a Vydac protein/peptide C18 column (catalog no. 218 TP54). Peptides were eluted with a 30-min gradient from 0% to 80% acetonitrile plus 0.1% TFA throughout. Peptides were detected at 214 nm, and data were collected and processed by using Rainin Instruments dynamax software version 1.4.
Results
Exchange Reaction.
Based on the report by Frye (9) that members of the SIR2 family had ADP-ribosyltransferase activity, we hypothesized that these proteins might cleave the glycosidic bond between nicotinamide and ADP-ribose in NAD. To test this hypothesis, we used an exchange reaction, in which the enzyme is incubated with radioactive nicotinamide and nonradioactive NAD. After incubation, the nicotinamide is separated from NAD by TLC, and the chromatogram is subjected to autoradiography. If the enzyme is able to cleave the glycosidic bond in NAD and form an enzyme-ADP-ribose intermediate, then the following equilibrium will be established: NAD + enzyme ⇌ enzyme-ADP-ribose + nicotinamide. Thus, NAD should become radioactively labeled in an enzyme and time-dependent manner.
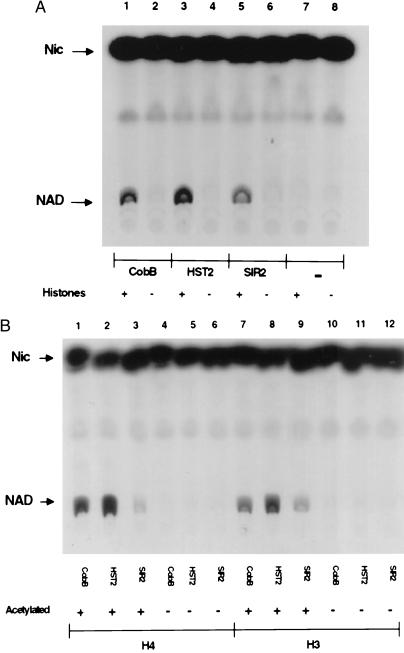
We expressed epitope-tagged versions of E. coli CobB as well as yeast SIR2 and the SIR2 homolog, HST2, in E. coli and purified them by affinity chromatography. Initially none of the proteins was found to catalyze the NAD–nicotinamide exchange reaction described above. But when histones isolated from chicken erythrocytes were added, each of the three proteins was able to promote significant exchange (Fig. 1A). Next we tested whether chemically synthesized peptides corresponding to N termini of histones H4 and H3 could promote exchange. These peptides did not promote exchange, but if the peptides were first acetylated by histone acetyltransferases, HAT1 (14) for H4 and HPA2 (15) for H3, then significant exchange occurred (Fig. 1B). This result suggested that the SIR2-like enzymes require acetylated lysine for the exchange reaction, and that chicken histones could promote exchange because they had been at least partially acetylated in vivo. Two other results support this conclusion. First, two related acetyltransferases, HPA2 and HPA3, known to acetylate themselves (S.T.T. and R.S., unpublished results) could replace histones to support the exchange reaction, but only after they became autoacetylated by incubation with acetyl-CoA. Second, even monomeric Nɛ-acetyllysine at a concentration of 25 mM supported the exchange reaction, whereas lysine itself and Nα-acetylserine at the same concentration did not (data not shown).
Figure 1.
NAD–nicotinamide exchange reactions with CobB, HST2, or SIR2. (A) Reactions were performed with (lanes 1, 3, 5, and 7) and without (lanes 2, 4, 6, and 8) histones, as indicated. (B) Reactions with acetylated (lanes 1–3, 7–9) or unacetylated (lanes 4–6, 10–12) H4 or H3 peptides. Peptides were acetylated as described in Materials and Methods.
Deacetylation.
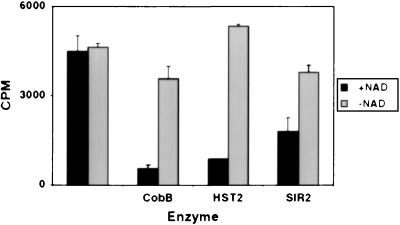
The reports that SIR2 and related proteins could ADP-ribosylate BSA or histones prompted us to test HST2 for this activity. Using [32P]NAD, we could not detect enzyme-dependent transfer of radioactivity to various histone substrates. But in one experiment, we first acetylated histones with the enzyme HAT1 and [3H]acetyl-CoA, and then incubated those histones with HST2 and [32P]NAD, the rationale being that perhaps the putative ADP-ribosylation would occur on acetylated substrates. Rather than observing ADP-ribosylation, we instead observed a decrease in 3H on the histones, indicative of deacetylation. Subsequent experiments verified that this decrease was indeed deacetylation and that it depended absolutely on NAD, as depicted for all three enzymes in Fig. 2. Similar deacetylation was observed for histones acetylated by HPA2 or SAS3, which act mainly on H3 (data not shown).
Figure 2.
Histone deacetylation by CobB, HST2, and SIR2 depends on NAD. The cpm present in the histones after enzyme incubation are depicted. Error bars show the variation between two independent experiments.
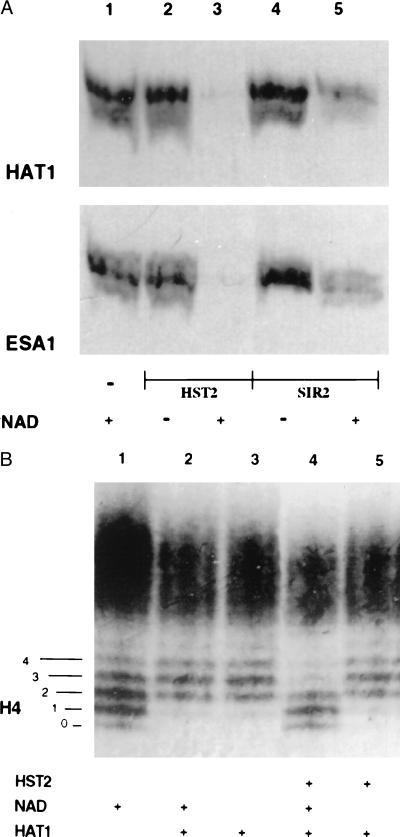
Additional evidence for deacetylation is shown in Fig. 3A. Histones, 3H-labeled by HAT1 or by another histone acetyltransferase, ESA1 (16, 17), were electrophoresed on an SDS/PAGE gel, before and after treatment with HST2 or SIR2, and the gel was subjected to fluorography. Radioactivity was no longer present on the histones treated with HST2 and was greatly reduced in those treated with SIR2. This deacetylation absolutely depended on NAD (Fig. 3A).
Figure 3.
Histone deacetylation analyzed by gel electrophoresis. (A) An SDS/PAGE gel of histones acetylated with [3H]acetyl-CoA by HAT1 or by ESA1, as indicated, and then treated with no enzyme (lane 1), with HST2 (lanes 2 and 3), or with SIR2 (lanes 4 and 5). A fluorogram of the gel is depicted. (B) A Triton–acid–urea gel of chicken erythrocyte histones acetylated with HAT1 followed by deacetylation by HST2. The gel was stained with Coomassie blue to visualize the histone isoforms. The numbers 0–4 on the left refer to isoforms of H4 differing in charge, with 0 corresponding to the most positively charged isoform and 4 corresponding to the least positively charged form. Lane 1, histones incubated without HAT1; lanes 2 and 3, histones acetylated with HAT1, in the presence or absence of NAD; lanes 4 and 5, histones acetylated by HAT1, incubated with HST2, with and without NAD.
Using HST2, the most active of the enzymes under our conditions, two other experiments were done to confirm that the loss of [3H]acetyl groups from histones was NAD-dependent deacetylation. Histones again were acetylated by HAT1, which predominantly acetylates H4 on Lys-5 and -12 (14), and then incubated with HST2 with and without NAD. The histones were then separated on a Triton–acid–urea gel that separates histones not only by size but also by charge (13). On such a gel, the various modified species of H4 can be separated clearly (Fig. 3B). The untreated chicken histones are shown in Fig. 3B, lane 1; five prominent isoforms of H4 can be seen. Lanes 2 and 3 depict H4 after treatment with HAT1; the center of the distribution shifts by 1 to 2 steps, consistent with the known mono- and diacetylation of H4 on Lys-5 and -12 (14). (The result also suggests that the majority of native chicken erythrocyte H4 is not acetylated on those lysines.) Lane 4 shows that HST2 treatment returns H4 to its original state before acetylation by HAT1. Lane 5 shows that this deacetylation absolutely requires NAD. The fact that HST2 does not shift the H4 distribution to the most positively charged state, labeled “0” in Fig. 3B, means either that modifications other than acetylation (such as phosphorylation) contribute to the original distribution shown in lane 1 or that HST2 shows specificity in which lysines it deacetylates. This issue has not yet been resolved. It should be emphasized that the histones in the gel in Fig. 3B are visualized with Coomassie blue, not radioactivity, and thus represent significant quantities of acetylation by HAT1 and deacetylation by HST2.
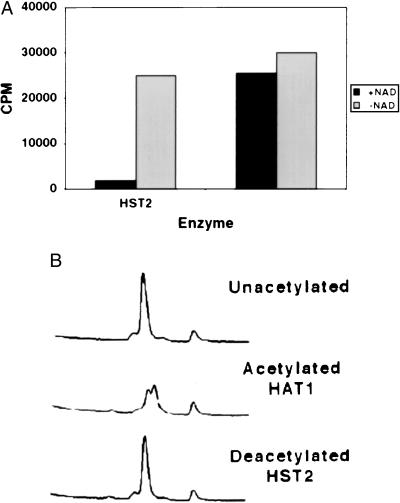
Another experiment to evaluate deacetylation used peptides corresponding to the N termini of H4 and H3. When these peptides were [3H]acetyl-labeled with HAT1 or HPA2, and then treated with HST2 or SIR2, the radioactivity was removed from the peptides in an NAD-dependent reaction (Fig. 4A and Table 1). Reverse-phase HPLC analysis was performed on an H4 peptide that had been acetylated on Lys-5 and -12 by HAT1 and then deacetylated by HST2. As shown in Fig. 4B, the chromatogram shows that the peptide changes retention time after acetylation and then returns to its original state after deacetylation by HST2. The two peaks seen after acetylation are most likely due to mono- and diacetylation by HAT1, consistent with previous results (14). Notably, the chromatogram reveals no significant ADP-ribosylation of the H4 peptide from the NAD that was present in the reaction. If there had been such a modification, the absorption at 214 nm of the peptide would have increased after HST2 treatment, and the retention time would most likely have decreased. Neither was observed (Fig. 4B).
Figure 4.
Deacetylation of an H4 peptide by HST2. The peptide was initially acetylated by HAT1. (A) H4 peptide deacetylation by HST2 depends on NAD. The cpm present in the peptide after enzyme incubation are depicted. (B) HPLC traces of the input unacetylated H4 peptide, the peptide after acetylation by HAT1, and the peptide after subsequent deacetylation by HST2.
Table 1.
Deacetylation of acetylated H3 and H4 peptides by SIR2
| Peptide | Acetylated by | cpm*
|
|
|---|---|---|---|
| −NAD | +NAD | ||
| H3 | HPA2 | 6,900 | 3,400 |
| H4 | HAT1 | 40,800 | 20,700 |
cpm remaining on acetylated peptides after incubation with SIR2, with (+) and without (−) NAD, as described in Materials and Methods.
Discussion
The exchange reaction we describe demonstrates that the SIR2-like proteins form an enzyme-ADP-ribose intermediate, in agreement with a recent report (10). Our data show that this reaction occurs only in the presence of proteins containing acetyllysine. The enzymes also deacetylate the lysines on these proteins, and this deacetylation is, in turn, dependent on NAD. The exact mechanism by which these reactions occur is not clear. The enzymes must have at least two recognition sites, one for NAD and one for acetyllysine, and two activities, auto-ADP-ribosylation and deacetylation. The crystal structure of these enzymes should prove enlightening because previously characterized deacetylases do not exhibit the dependence on NAD described here. A major unanswered question is whether NAD turns over during the deacetylation. In other words, is an ADP-ribose released for every acetyl group removed, or is a stably ADP-ribosylated enzyme the active deacetylase? Future studies defining enzyme and cofactor turnover numbers should resolve this question.
We see no evidence for transfer of ADP-ribose from the enzymes to proteins such as albumin or histones, contrary to recent reports (9, 10). Although we cannot exclude the possibility that a small amount of ADP-ribosylation might occur, three experiments argue against significant transfer. First, we see no [32P]NAD transfer to recipient proteins by either CobB or HST2. The groups that reported such transfer used very high concentrations of enzyme to observe it. We see significant deacetylation at a 100-fold lower enzyme concentration. This result suggests that deacetylation is the physiologically significant reaction. Second, the H4 visualized in the Triton–acid–urea gel in Fig. 3B shows no sign of ADP-ribosylation after HST2 treatment. If that had occurred, the H4 isoforms would be expected to migrate more slowly in the gel because of extra negative charges. Instead they moved more rapidly, consistent with deacetylation. Finally, the H4 peptide shows no evidence for ADP-ribosylation in the HPLC chromatogram (Fig. 4B), as argued above.
Overproduction of SIR2 in yeast was earlier found to cause bulk deacetylation of histones, leading to the suggestion that SIR2 was a histone deacetylase or stimulated such an activity (18). The work presented here demonstrates that SIR2 can deacetylate histones, and is the likely explanation for the overexpression phenotype observed previously. At normal levels, SIR2 presumably deacetylates histones in chromatin where it is bound, including the silent mating loci, the telomeres, and within the rDNA. When SIR2 is removed by mutation, these loci are aberrantly activated, and recombination is observed to increase within the repetitive rDNA (reviewed in refs. 2 and 3).
We observed that HST2 and SIR2 can both deacetylate histones that have been acetylated by HAT1 or ESA1 (Fig. 3A). These two HATs have somewhat different substrate specificities and different biological roles. HAT1 is likely to function in chromatin assembly by modifying newly synthesized histones, thereby promoting their deposition in the nucleosomal octamer of histones (14, 19). In contrast, ESA1 is the only known essential HAT in yeast and may contribute to both transcriptional regulation and other critical biological processes (16, 17, 20). The fact that the recombinant SIR2-like enzymes can deacetylate histones modified by either class of HATs raises several possibilities. One is that the enzymes have very broad specificities in vivo, and may in fact have the capacity to modulate chromatin structure under many different circumstances to yield different biological consequences. A second possibility is that the functions of the family members are far more restricted in vivo, and the activities that we observe here reflect some relaxation of enzyme specificity. This would not be unprecedented and introduces the possibility for significant control mechanisms. For example, because SIR2 is targeted primarily to silenced loci, perhaps its activity is restricted to these loci and modulated by other components of silencing complexes. The recent observation that SIR2's localization changes during mitosis (5) raises the possibility that its activity may also be dynamic. Less is known about HST2, but because it does not function in mating type or telomeric silencing (7), it presumably functions elsewhere in the cell, perhaps in the context of a complex of proteins that contribute to its specificity. Indeed, reports of cytoplasmic localization for human and trypanosome homologs (21, 22) that are closely related to HST2 emphasize the necessity of thinking broadly about the roles of these enzymes.
The functions of the homologs of SIR2 in other organisms have not yet been well established. The fact that E. coli CobB has comparable catalytic activity to two eukaryotic enzymes suggests that not all homologs will function to deacetylate histones, and that there may be other significant in vivo substrates. For example, diverse proteins such as myoD, p53, and tubulin are known to be acetylated on specific lysine residues (23–25). Perhaps these proteins are deacetylated by SIR2-like proteins to modulate their biological activities. Indeed, mutation of a fission yeast SIR2 homolog results in sensitivity to microtubule-depolymerizing drugs and chromosome loss (26). Likewise, in the fungus Candida albicans, mutation of a SIR2 homolog leads to genomic instability, which is correlated with clinical pathology (27). Defects in acetylation status of the mitotic spindle, centromeric histones, or other protein substrates would be consistent with these phenotypes.
Recent studies indicate that, in human cells, hyperacetylation may be a determinant for productive V(D)J recombination that ultimately yields appropriate Ag receptor gene expression in differentiated B and T lymphocytes (28). Because human homologs of the SIR2 family of deacetylases are expressed in multiple tissues, including those of lymphocytic and erythropoietic lineages (9, 29), it seems possible that these enzymes may contribute to processes as diverse as suppression of recombination, maintenance of genome stability, and transcriptional regulation. Understanding the unique mechanism and targets of these NAD-dependent deacetylase activities will be critical goals for future studies.
Acknowledgments
We thank B. Olivera for suggesting the exchange reaction, R. Haltiwanger for advice, R. Dutnall for HAT1, and R. Frye and D. Moazed for plasmids. This work was supported by National Institutes of Health Grants GM28220 and GM55641 to R.S. and GM54778 to L.P.
Abbreviation
- SIR2
silent information regulator 2
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110148297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110148297
References
- 1.Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 2.Sherman J M, Pillus L. Trends Genet. 1997;13:308–313. doi: 10.1016/s0168-9525(97)01198-0. [DOI] [PubMed] [Google Scholar]
- 3.Guarente L. Nat Genet. 1999;23:281–285. doi: 10.1038/15458. [DOI] [PubMed] [Google Scholar]
- 4.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 5.Straight A F, Shou W, Dowd G J, Turck C W, Deshaies R J, Johnson A D, Moazed D. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 6.Shou W, Seol J H, Shevchenko A, Baskerville C, Moazed D, Chen S Z W, Jang J, Shevchenko A, Charbonneau H, Deshaies R J. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 7.Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 8.Tsang A W, Escalante-Semerena J C. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 9.Frye R A. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 10.Tanny J, Dowd G J, Huang J, Hilz H, Moazed D. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 11.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 12.Graziano V, Gerchman S E, Ramakrishnan V. J Mol Biol. 1988;203:997–1007. doi: 10.1016/0022-2836(88)90124-6. [DOI] [PubMed] [Google Scholar]
- 13.Alfageme C, Zweidler A, Mahowald A, Cohen L. J Biol Chem. 1974;249:3729–3733. [PubMed] [Google Scholar]
- 14.Kleff S, Andrulis E D, Anderson C W, Sternglanz R. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 15.Angus-Hill M L, Dutnall R N, Tafrov S T, Sternglanz R, Ramakrishnan V. J Mol Biol. 1999;294:1311–1325. doi: 10.1006/jmbi.1999.3338. [DOI] [PubMed] [Google Scholar]
- 16.Smith E R, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook R G, Lucchesi J C, Allis C D. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke A S, Lowell J E, Jacobson S J, Pillus L. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braunstein M, Rose A B, Holmes S C, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 19.Parthun M R, Widom J, Gottschling D E. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 20.Allard S, Utley R T, Savard J, Clarke A, Grant P, Brandl C J, Pillus L, Workman J L, Cote J. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zemzoumi K, Sereno D, Francois C, Guilvard E, Lemesre J L, Ouaissi A. Biol Cell. 1998;90:239–245. doi: 10.1016/s0248-4900(98)80020-8. [DOI] [PubMed] [Google Scholar]
- 22.Afshar G, Murnane J P. Gene. 1999;234:161–168. doi: 10.1016/s0378-1119(99)00162-6. [DOI] [PubMed] [Google Scholar]
- 23.Sartorelli V, Puri P L, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang J Y J, Kedes L. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 24.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan K F. Annu Rev Cell Biol. 1988;4:687–716. doi: 10.1146/annurev.cb.04.110188.003351. [DOI] [PubMed] [Google Scholar]
- 26.Freeman-Cook L L, Sherman J M, Brachmann C B, Allshire R C, Boeke J D, Pillus L. Mol Biol Cell. 1999;10:3171–3186. doi: 10.1091/mbc.10.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Martin J, Uria J A, Johnson A D. EMBO J. 1999;18:2580–2592. doi: 10.1093/emboj/18.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMurry M T, Krangel M S. Science. 2000;287:495–498. [PubMed] [Google Scholar]
- 29.Sherman J M, Stone E M, Freeman-Cook L L, Brachmann C B, Boeke J D, Pillus L. Mol Biol Cell. 1999;10:3045–3059. doi: 10.1091/mbc.10.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]