Abstract
The development and function of living tissues depends largely on interactions between cells that can vary in both time and space; however, temporal control of cell–cell interaction is experimentally challenging. By using a micromachined silicon substrate with moving parts, we demonstrate the dynamic regulation of cell–cell interactions via direct manipulation of adherent cells with micrometer-scale precision. We thereby achieve mechanical control of both tissue composition and spatial organization. As a case study, we demonstrate the utility of this tool in deconstructing the dynamics of intercellular communication between hepatocytes and supportive stromal cells in coculture. Our findings indicate that the maintenance of the hepatocellular phenotype by stroma requires direct contact for a limited time (≈hours) followed by a sustained soluble signal that has an effective range of <400 μm. This platform enables investigation of dynamic cell–cell interaction in a multitude of applications, spanning embryogenesis, homeostasis, and pathogenic processes.
Keywords: dynamic substrate, intercellular communication, microelectromechanical systems, microenvironment, microfabrication
Mammalian cells in vivo integrate and respond to cues in their microenvironment that vary in both time and space. In particular, interactions between neighboring cells can regulate both the fate and function of individual cells and govern the emergent properties of the resultant tissue. Because such cell–cell interactions occur primarily through direct contact or exchange of soluble factors, understanding the temporal and spatial aspects of these signals is of fundamental importance to tissue biology. Recent advances in cell “micropatterning” have already proven invaluable in increasing our understanding of the structure–function relationships of such multicellular communities (1–4); however, dynamic manipulation of tissue structure in vitro has remained largely out of reach.
Previous efforts toward spatiotemporal control of tissue organization at the cellular scale have focused on modulation of the adhesive properties of the culture substrate (5–7). Through the micropatterning of surface chemistries that can be dynamically altered, localized attachment and release of cells has been demonstrated (8, 9). Nonetheless, these manipulations are typically not reversible (i.e., nonadhesive surfaces are rendered adhesive just once), they do not allow the decoupling of processes associated with adhesion from those correlated with cell–cell interaction (i.e., attachment, spreading, and contact with neighboring cells have overlapping time scales), nor can these platforms accommodate serial manipulations to mimic key biological events (i.e., sequential exposure of a target cell population to different inducer populations). Manipulations of surface chemistry are also limited by the inability to precisely control tissue composition: (i) sequential seeding of different cell types can result in contamination of pure populations and (ii) maintaining μm-scale proximity of two different cell populations in the absence of contact over many days, important for decoupling the relative role of contact and paracrine signals (4), has not been achieved.
We introduce a different approach to this problem by leveraging tools from the field of microelectromechanical systems, which offers precise physical manipulation at a length scale comparable to that of many biological processes. In our approach, cells are grown on an array of micromachined plates that are physically rearranged to change the spatial organization of the culture, which will be referred to as micromechanical reconfigurable culture (μRC). Cells remain attached to the substrate throughout the repositioning process (10, 11). Using μRC, we are able to demonstrate dynamic regulation of cell–cell interactions via direct manipulation of cell positioning. Specifically, cell–cell contact between different cell populations is regulated by positioning plates together or apart. By imposing a small μm-scale separation between the plates, cell–cell contact can be abrogated while soluble signaling is maintained. By using larger separation distances, the extent of soluble signaling can also be modulated. In addition, by removing a plate and replacing it, one population of cells can be exchanged for another in a modular fashion. Thus, this micromechanical approach provides dynamic control of both tissue organization and composition.
Results and Discussion
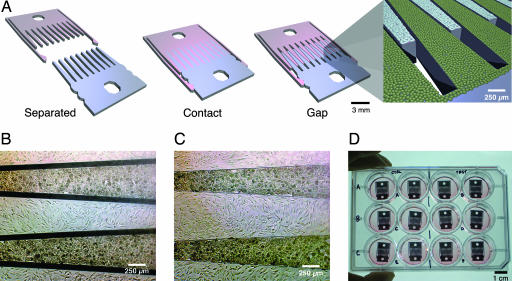
The μRC device consists of two parts with interlocking comb fingers and an integrated snap-lock mechanism. The parts can be fully separated, locked together with the fingers in contact, or locked together with a fixed gap between the comb fingers (Fig. 1A). We will refer to these configurations as the separated, contact, and gap modes, respectively. Cells are cultured on the top surface of the fingers (Fig. 1 B and C). The silicon parts are spin-coated with polystyrene, resulting in a surface comparable to tissue culture plastic. Parts can be separated into individual wells of a 12-well plate for coating of extracellular matrix proteins and seeding of cells, so as to avoid cross-contamination. After cell attachment, the two parts can be assembled in a fresh well (Fig. 1D), where cell culture and functional assays can be performed in a standard manner. The actuation strategy to switch between modes was designed for simplicity and compatibility with standard aseptic cell-culture technique. The matching V-shaped latches and notches are self-centering, allowing the parts to be accurately and reproducibly positioned by using only tweezers, without the need for microscopic visualization or micromanipulation machinery [supporting information (SI) Movie 1]. The extent of finger separation in the gap mode can be tuned via notch positioning; multiple sets of notches could also be used to allow variable spacing.
Fig. 1.
Micromechanical substrates enable micrometer-resolution cell positioning. (A) Microfabricated silicon parts can be fully separated (Left), locked together with comb fingers in contact (Center), or slightly separated (Right). Cells are cultured on the top surfaces; manual scraping can be used to restrict cells to the comb fingers only (Inset). The slope of the tapered comb fingers results in a 20:1 mechanical transmission ratio; that is, sliding the parts 1.6 mm changes the gap between the fingers by only 80 μm. Together with the integrated snap-lock mechanism, it is thereby possible to control separation with repeatable micrometer-scale precision by using unassisted manual actuation. (B and C) Bright-field images of hepatocytes (darker cells) and 3T3 fibroblasts cultured on the comb fingers. The silicon is first functionalized by spin-coating with polystyrene followed by plasma treatment, resulting in a surface comparable to tissue culture plastic. Devices can be reused multiple times (>20). (D) Devices in a standard 12-well plate. Cell culture and functional assays were performed with standard methods. Actuation is also performed directly on the plate with sterile tweezers.
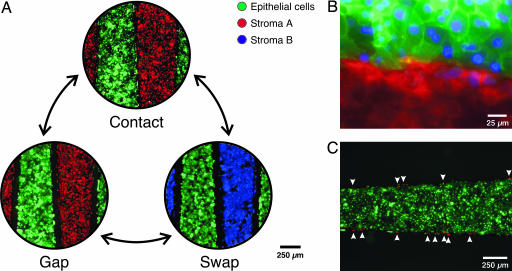
The μRC system can be reversibly switched between a number of configurations (Fig. 2A). In the contact mode, cells on opposing fingers can engage in direct cell–cell contact. In the gap mode, the cells are held at a small uniform separation that is just large enough to prevent contact, yet maintains cells within the range of short-range soluble factors (≈100 μm, demonstrated below). Finally, cell populations can be exchanged in a modular fashion by removing one set of fingers and snapping in a replacement seeded with another cell type. Thus, contact-mediated signaling can be dynamically regulated by switching between the contact and gap modes. Likewise, soluble signaling can be dynamically regulated by swapping between various cell types in the gap mode. To provide the necessary mechanical precision, silicon parts were fabricated in a single-mask, through-wafer, deep reactive ion etching process (12, 13). A separation of 6 μm or less was measured in the contact mode, and a separation of 79 ± 1 μm was measured in the gap mode. Fluorescence microscopy with membrane dyes showed that cells on opposing fingers form intimate contacts in contact mode (Fig. 2B). In addition, contamination of cells between adjacent fingers after 18 h of contact was minimal (Fig. 2C).
Fig. 2.
Reconfigurable cell culture. Cultures can be reversibly switched to initiate or to eliminate contact between two cell populations; individual populations can also be removed and replaced. (A) Fluorescent images illustrating possible device manipulations. Each cell type was prelabeled with an individual dye color. (B) Fluorescent image showing intimate contact between hepatocytes (green) and stroma (red, 3T3 fibroblasts) at the interface between neighboring comb fingers. The image was taken 18 h after initiation of contact. Cell nuclei are counterstained in blue. (C) Cross-migration of cells is minimal for moderate durations of contact. Representative fluorescent image showing small numbers of stromal cells (red, arrows indicate selected cells) remaining behind on a hepatocyte finger (green) after combs were separated after 18 h of contact. In this work, contact was limited to 18 h to minimize cross-migration, but longer durations are possible with other cell types (data not shown).
As a case study, we applied our dynamic platform to the study of cell–cell interactions between hepatocytes and stromal cells in coculture. As with many other cell types, interaction of epithelia with supportive stroma or “feeder layers” promotes tissue-specific gene expression in vitro. In the case of primary hepatocytes, cocultivation of hepatocytes with many different mesenchymal cell types (endothelia, fibroblasts, etc.) promotes retention of hepatocyte viability and liver-specific functions that are otherwise rapidly lost in vitro (2). This robust “coculture” phenomenon, although poorly understood, has wide-ranging applications in both therapeutic and diagnostic applications of engineered liver tissue (14–16). Using both conventional techniques and micropatterning approaches, we and others have previously found that the degree of interaction between the two cell types (“heterotypic interaction”) modulated the amount of liver-specific function retained in vitro (2, 17). These findings suggested an important role for proximity between the two cell types in the rescue of hepatocyte phenotype; however, the relative role of contact-mediated versus soluble signals, the dynamics of interaction, and the potential for reciprocal signaling had not been established.
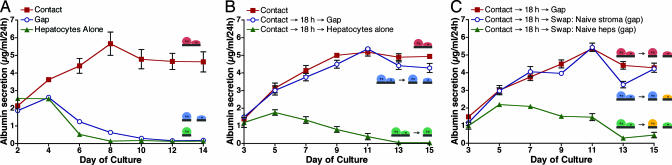
Hence, we set out to explore this system by using the μRC substrates, with primary rat hepatocytes and Swiss 3T3 murine fibroblasts cultured on opposing combs. Hepatocyte morphology and viability were assessed microscopically, and albumin production was measured as a quantitative marker of liver-specific function. Comparison of cultures in the contact, gap, and separated modes demonstrated that contact was necessary for maintenance of liver-specific function (Fig. 3A). Even in the gap mode, which corresponded to only an 80-μm separation between the two cell populations, hepatocyte function declined at a rate similar to that of hepatocytes cultured alone. Next, we conducted dynamic experiments in which cells were repositioned after 18 h of contact. Here, transient contact alone proved insufficient to rescue the hepatocyte phenotype, and liver-specific functions rapidly declined. In contrast, transient contact followed by sustained culture in the gap mode provided complete rescue of liver-specific function (Fig. 3B). These observations thus imply a necessary role for both heterotypic contact and soluble factors that diffuse across the gap.
Fig. 3.
Dynamic regulation of hepatocyte–stromal interactions reveals temporal dependencies in intercellular communication. (A) Contact between hepatocyte and fibroblast combs was required to maintain albumin secretion over a 2-wk period (red). In the gap mode (blue), function dropped almost as rapidly as with hepatocytes alone (green). (B) An 18-h period of transient initial contact followed by long-term culture in the gap mode (which allows diffusion of paracrine signals) resulted in sustained liver-specific function (blue) similar to that obtained with sustained contact (red). However, 18 h of initial contact followed by removal of adjacent stroma resulted in deterioration of function (green). (C) After 18 h of initial contact, stroma were removed and replaced by naïve stroma (in gap mode). Liver-specific function was maintained at similar levels (blue) to that obtained with no cell swapping (red). In a parallel experiment in which naïve hepatocytes were substituted, liver-specific function was not maintained (green).
Notably, it would appear that contact was required only initially, whereas soluble interactions were required for the duration of the experiment. This finding raised the possibility that reciprocal interactions, i.e., sustained alterations in fibroblast function as a result of hepatocyte contact, might play a role. To test this possibility, we exploited the “modular” nature of the μRC platform. Cocultures were conducted in contact mode for 18 h as before; however, the fibroblasts were then replaced with naïve fibroblasts (no exposure to heterotypic contact) in gap mode. Under these conditions, paracrine signals provided by naïve fibroblasts were still sufficient to sustain hepatic functions (Fig. 3C). Conversely, if naïve hepatocytes were substituted, hepatic function deteriorated. Hence, the data are consistent with constitutive expression of critical soluble factors by fibroblasts independent of hepatocyte interaction rather than supporting a role for reciprocal cell–cell interaction.
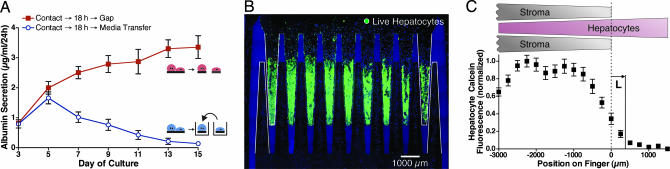
To investigate the importance of cell proximity, hepatocytes and fibroblasts were separated into different wells after 18 h of initial contact. Conditioned medium was then transferred from the fibroblast well to the hepatocyte well every 2 days. However, hepatic function was not maintained (Fig. 4A), underscoring the importance of close positioning in the gap configuration. Further, microscopic examination of cocultures yielded a striking observation: in cultures stabilized via transient contact followed by gap mode, hepatocytes toward the rear of each comb finger lost viability over the course of 2 weeks (Fig. 4B). Hepatocyte–fibroblast distance is greater in this region compared with the rest of the comb finger because of the geometry of the device in the gap configuration (Fig. 1A Inset). Quantifying viability with a fluorescent membrane integrity dye yielded a characteristic length of decay in viability of ≈325 μm (Fig. 4C). It was demonstrated through finite element modeling that diffusion of a rapidly decaying (≈hours) or rapidly consumed (comparable to rate of production) soluble factor could produce concentration profiles similar to the survival pattern of Fig. 4B (SI Appendix). These data suggest that the fibroblast-derived soluble signals critical for rescue of the hepatocyte phenotype and viability are effective over a very limited range, on the order of only 10 cell diameters.
Fig. 4.
Spatial reconfiguration reveals short-range soluble signaling. (A) After 18 h of initial contact, hepatocytes and stroma were separated into individual wells. Stromal conditioned medium was transferred every 2 days to the hepatocytes, but liver-specific function declined (blue). In contrast, transient contact followed by microscale separation (using the gap mode) resulted in sustained function (red). (B) Loss in liver-specific function progresses to loss in hepatocyte viability. Hepatocyte viability was probed by using a membrane integrity dye (calcein AM, green) with a nuclear counterstain for both cell types (blue). After initial contact, cultures were maintained for 2 weeks in the gap mode, resulting in a sharp gradient in hepatocyte viability dependent on proximity to stroma (n > 3; representative image shown). Selected comb fingers are outlined in white for clarity. (C) Quantified calcein fluorescence along the length of a comb finger (n = 9). L, the characteristic decay length of viability, is measured to be 325 μm by using an exponential fit over x > 0.
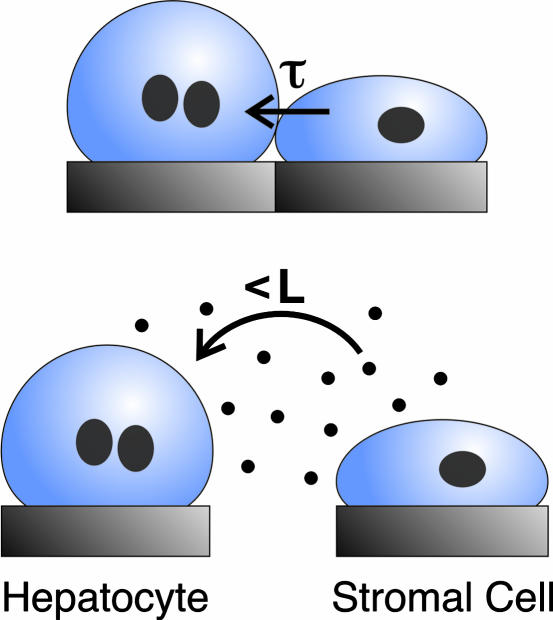
In summary, we hypothesize that preservation of hepatocyte viability and liver-specific functions in coculture depends on an initial contact-mediated signal followed by a sustained short-range soluble signal, from fibroblasts to hepatocytes (Fig. 5). In terms of the contact-mediated signal, it is not clear whether this is junctional in nature (hepatocytes and 3T3 fibroblasts do not express similar cadherin or connexin subtypes) or caused by cell-associated matrix molecules. It is also unknown why only transient contact is required. One possibility is that transient contact triggers an irreversible signaling pathway. Alternatively, the contaminating cells that remain after separation (Fig. 2C) may play a role in the response. But that seems unlikely because hepatic function could not be maintained in gap mode without initial contact, even when low numbers of fibroblasts were doped onto the hepatocyte fingers (data not shown). A third possibility is that fibroblasts secrete critical extracellular matrix components onto the hepatocyte fingers during the transient contact period that help to sustain function thereafter. Regardless, these data point to the possibility that hepatocytes could be preconditioned and subsequently sustained without supportive stromal cells, a finding with significant practical implications for the therapeutic and diagnostic applications of hepatocytes. Notably, only the peripheral hepatocytes can directly contact fibroblasts, yet the entire population is affected. This finding is consistent with previous reports (2) but the precise mechanism has not been established. Finally, the possible reasons that soluble signals are effective over very limited distances include: that the critical factors are highly labile, are active at relatively high local concentration, or are rapidly sequestered extracellularly via binding to extracellular matrix proteins.
Fig. 5.
Proposed model for intercellular communication. Maintenance of liver-specific function in hepatocytes requires an initial short-term (τ ≈ 18 h) contact-mediated signal from stromal cells (Upper), followed by sustained short-range (L ≈ 325 μm) soluble signaling from the stroma (Lower).
Through this case study, we have used μRC to execute a number of previously inaccessible experiments. It was possible to decouple contact-mediated and soluble signals, dynamically modulate both contact-mediated and soluble cell–cell signaling, examine the reversibility of a pathway upon removal of the triggering signal, test for the presence of reciprocal cell–cell signaling, and measure the effective range of soluble signals. We propose that micromechanical culture substrates are a robust and generalizable tool. Because our device surface is comparable to tissue culture plastic, it should be readily adapted to a variety of cell types and molecular techniques. For example, we have demonstrated compatibility with liver progenitors, sinusoidal endothelial cells, and bone marrow stromal cells, as well as transfection of siRNA into individual cell populations (SI Fig. 6). We expect this methodology to find utility in the investigation of cellular niches (18), the dissection of developmental processes (19), and the study of disease progression, in particular in tissues where stromal interactions are thought to play a role (e.g., tumorigenesis) (20). Future directions in device engineering could include embedded microfluidics and sensors for local delivery of soluble factors and in situ monitoring (21) and integrated actuation for heterogeneous mechanical control of array elements.
Materials and Methods
Materials.
Collagen-I was purified from rat tails as described (22). Briefly, rat-tail tendons were denatured in acetic acid, salt-precipitated, dialyzed against HCl, and sterilized with chloroform. Because the silicon substrates are opaque, a reflecting noninverted microscope is required to inspect cells during culture. To examine cultures without compromising sterility, a microscopy system was required with an optical working distance greater than the thickness of a covered culture plate. We used a ×5 objective with a 36-mm working distance and a ×10 objective with a 38-mm working distance (Optical Product Development, Lexington, MA) mounted on a Meiji MA655/05 head (Microscope World, Encinitas, CA).
Device Fabrication.
Fabrication was performed at the University of California at Berkeley Microfabrication Laboratory and the Massachusetts Institute of Technology Microsystems Technology Laboratories, using a similar process at both locations. Device parts were fabricated by using well established microelectromechanical systems fabrication methods. Good references include papers by Ayon et al. (12) and Knobloch et al. (13). Briefly, a double-side-polished silicon wafer (4 inches, 500 μm; University Wafer, South Boston, MA) was oxidized (1,000°C, O2/H2O) to grow a 1-μm layer of silicon dioxide. A layer of thick photoresist (Megaposit SPR220; Rohm and Hass, Philadelphia, PA) was spin-coated, patterned by using a chrome mask and contact alignment (Karl Suss MA6; SUSS MicroTec, Waterbury Center, VT), and developed (LDD-26W; Shipley, Marlborough, MA). The patterned wafer, or device wafer, was then attached to a handle wafer by using a photoresist bond. After etching through the oxide layer (He/CHF3/CF4 plasma), deep reactive ion etching (ICP-ASE; Surface Technology Systems, Newport, UK) was used to etch through the entire device wafer as described (13). The parts were then released in acetone and cleaned in Piranha solution (4:1 H2SO4/H2O2, 120°C, 10 min). Finally, the silicon surface was functionalized for cell culture by spin-coating with polystyrene (100 mg/ml in toluene, 2,400 rpm, 1 min) (1-EC101D-R485; Headway Research, Garland, TX) followed by plasma treatment (O2, 200 mT, 200 W, 1 min), resulting in a surface comparable to tissue culture plastic. Devices were reused multiple times (>20). Between experiments, the parts were cleaned in toluene followed by Piranha solution, and polystyrene was reapplied.
Cell Culture.
Primary hepatocytes were isolated from 2- to 3-month-old adult female Lewis rats (Charles River Laboratories, Wilmington, MA) weighing 180–200 g, following a modified procedure of Seglen (23). Detailed procedures for hepatocyte isolation and purification have been described (22). Hepatocyte culture medium consisted of DMEM with high glucose, 10% (vol/vol) FBS, 0.5 units/ml insulin, 7 ng/ml glucagon, 7.5 g/ml hydrocortisone, and 1% (vol/vol) penicillin-streptomycin. Swiss 3T3 fibroblasts were purchased from ATCC (Manassas, VA). J2–3T3 fibroblasts were a gift from Howard Green (Harvard Medical School, Cambridge, MA) (24). Fibroblast culture medium consisted of DMEM with high glucose, 10% bovine calf serum, and 1% penicillin-streptomycin.
Device Actuation.
Actuation was performed within a biosafety cabinet by using stainless-steel tweezers (2-mm round tips), sterilized in 70% ethanol before use. Substrates were pushed or picked up by using the round hole at the rear of each part. It is possible for the parts to be misaligned vertically when they are locked together. Therefore, after configuring substrates in the intended state, plates were covered and examined under the reflecting microscope to verify that interlocked fingers were in-plane. Typically, ≈5% of interlocked parts were misaligned. To fix alignment, parts were simply separated and locked back together.
Seeding of Cells onto Micromechanical Substrates.
Polystyrene-coated silicon substrates were placed into individual wells on standard 12-well culture plates. Substrates intended to support hepatocytes were incubated in collagen solution (400 μg/ml in water) at 37°C for at least 45 min. To provide a flat, uniform surface for seeding, substrates were each locked together with a complementary part, in the contact mode. These complementary parts were used only during cell seeding and were set aside afterward. Substrates were sterilized by soaking in 70% ethanol for 1 h and then washed twice in distilled water. Primary hepatocytes were typically seeded onto the male parts (no arms), whereas fibroblasts (Swiss 3T3 or J2–3T3) were seeded onto the female parts (with arms) (Fig. 1A). Cells were seeded at 500,000 cells per ml, with 1 ml per well, in the appropriate culture medium and incubated for 60 min at 37°C. Plates were shaken every 20 min to resuspend unattached cells. After 60 min, unattached cells were aspirated, the substrate was washed with culture medium, and seeding was repeated with a fresh cell suspension. This process was repeated until the substrate surface was fully coated, usually requiring two to four seeding cycles for hepatocytes and two seeding cycles for fibroblasts. Within 6 h of completing cell seeding, the complementary parts were removed from each substrate. Cell-coated substrates were then transferred to fresh wells and incubated overnight in the appropriate medium. The next day, a cell scraper (Fisher Scientific, Pittsburgh, PA) was used to remove hepatocytes from the rear half of the substrates, to leave only the cells attached directly on the comb fingers (plus a border of ≈1 mm caused by imprecise manual scraping) (Fig. 1A Inset). Hepatocyte- and fibroblast-coated substrates were then assembled into their initial configurations for a particular experiment.
Fluorescent Labels.
Hepatocytes were labeled with calcein AM (Molecular Probes, Eugene, OR) at 5 μg/ml in hepatoctye medium. Swiss 3T3 fibroblasts were labeled with CellTracker Orange CMTMR (Molecular Probes) at 0.5 μM in serum-free fibroblast medium. J2–3T3 fibroblasts were labeled with CellTracker Blue CMAC (Molecular Probes) at 2.5 μM in serum-free fibroblast medium. For high-magnification images, hepatocyte membranes were labeled with PHK67 (Sigma–Aldrich, St. Louis, MO) at 1:1,000 in Diluent C (Sigma). Fibroblast membranes were labeled with Vybrant DiI (Molecular Probes) at 5 μl/ml in serum-free fibroblast medium. Cell nuclei were labeled with Hoechst 33258 (Molecular Probes) at 0.001% in hepatocyte medium.
Functional Assays.
Albumin content was measured by using enzyme-linked immunosorbent assays (MP Biomedicals, Irvine, CA) with horseradish peroxidase detection and 3,3′,5,5′ tetramethylbenzidine (Pierce Biotechnology, Rockford, IL) as a substrate (22). All experiments were performed at least twice, with triplicate samples for each condition. One representative outcome is presented for each experiment, with similar trends observed in multiple trials. Fluorescence quantification was performed with MetaVue 6.2r0 software (Universal Imaging, Downingtown, PA).
Supplementary Material
Acknowledgments
We thank Salman Khetani, Jared Allen, Chris Flaim, and Austin Derfus for insightful discussions and helpful suggestions; Alice Chen, Gregory Underhill, and Sandra March for technical assistance; and Mary Weiss and Helene Strick-Marchand (Institute Pasteur, Paris, France) for the BMEL 9A1 cell line. This work was supported by the National Science Foundation Faculty Early Career Development Program, National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases, and the David and Lucile Packard Foundation. E.E.H. was supported by a Ruth L. Kirschstein National Research Service Award.
Abbreviation
- μRC
micromechanical reconfigurable culture.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608660104/DC1.
References
- 1.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia SN, Balis UJ, Yarmush ML, Toner M. FASEB J. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 3.Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Proc Natl Acad Sci USA. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu WF, Nelson CM, Pirone DM, Chen CS. J Cell Biol. 2006;173:431–441. doi: 10.1083/jcb.200510087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okano T, Yamada N, Okuhara M, Sakai H, Sakurai Y. Biomaterials. 1995;16:297–303. doi: 10.1016/0142-9612(95)93257-e. [DOI] [PubMed] [Google Scholar]
- 6.Lahann J, Mitragotri S, Tran TN, Kaido H, Sundaram J, Choi IS, Hoffer S, Somorjai GA, Langer R. Science. 2003;299:371–374. doi: 10.1126/science.1078933. [DOI] [PubMed] [Google Scholar]
- 7.Jiang X, Ferrigno R, Mrksich M, Whitesides GM. J Am Chem Soc. 2003;125:2366–2367. doi: 10.1021/ja029485c. [DOI] [PubMed] [Google Scholar]
- 8.Cheng XH, Wang YB, Hanein Y, Bohringer KF, Ratner BD. J Biomed Mater Res A. 2004;70:159–168. doi: 10.1002/jbm.a.30053. [DOI] [PubMed] [Google Scholar]
- 9.Yeo WS, Yousaf MN, Mrksich M. J Am Chem Soc. 2003;125:14994–14995. doi: 10.1021/ja038265b. [DOI] [PubMed] [Google Scholar]
- 10.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 11.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 12.Ayon AA, Braff R, Lin CC, Sawin HH, Schmidt MA. J Electrochem Soc. 1999;146:339–349. [Google Scholar]
- 13.Knobloch AJ, Wasilik M, Fernandez-Pello C, Pisano AP. In: 2003 ASME International Mechanical Engineering Congress. Turner KL, editor. Vol 5. New York: Am Soc Mechanical Engineers; 2003. pp. 115–123. [Google Scholar]
- 14.Tilles AW, Baskaran H, Roy P, Yarmush ML, Toner M. Biotechnol Bioeng. 2001;73:379–389. doi: 10.1002/bit.1071. [DOI] [PubMed] [Google Scholar]
- 15.Allen JW, Khetani SR, Bhatia SN. Toxicol Sci. 2005;84:110–119. doi: 10.1093/toxsci/kfi052. [DOI] [PubMed] [Google Scholar]
- 16.Guillouzo A. Environ Health Perspect. 1998;106(Suppl 2):511–532. doi: 10.1289/ehp.98106511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guguen-Guillouzo C, Guillouzo A. Mol Cell Biochem. 1983;53/54:35–56. doi: 10.1007/BF00225245. [DOI] [PubMed] [Google Scholar]
- 18.Moore KA, Lemischka IR. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 19.Lemaigre F, Zaret KS. Curr Opin Genet Dev. 2004;14:582–590. doi: 10.1016/j.gde.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Zigrino P, Loffek S, Mauch C. Biochimie. 2005;87:321–328. doi: 10.1016/j.biochi.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 21.Papageorgiou DP, Shore SE, Bledsoe SC, Jr, Wise KD. J Microelectromech Syst. 2006;15:1025–1033. [Google Scholar]
- 22.Dunn JC, Tompkins RG, Yarmush ML. Biotechnol Prog. 1991;7:237–245. doi: 10.1021/bp00009a007. [DOI] [PubMed] [Google Scholar]
- 23.Seglen PO. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 24.Rheinwald JG, Green H. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.