Abstract
Ca2+-ATPase of skeletal muscle sarcoplasmic reticulum is an ATP-driven Ca2+ pump consisting of three cytoplasmic domains and 10 transmembrane helices. In the absence of Ca2+, the three cytoplasmic domains gather to form a compact headpiece, but the ATPase is unstable without an inhibitor. Here we describe the crystal structures of Ca2+-ATPase in the absence of Ca2+ stabilized with cyclopiazonic acid alone and in combination with other inhibitors. Cyclopiazonic acid is located in the transmembrane region of the protein near the cytoplasmic surface. The binding site partially overlaps with that of 2,5-di-tert-butyl-1,4-dihydroxybenzene but is separate from that of thapsigargin. The overall structure is significantly different from that stabilized with thapsigargin: The cytoplasmic headpiece is more upright, and the transmembrane helices M1–M4 are rearranged. Cyclopiazonic acid primarily alters the position of the M1′ helix and thereby M2 and M4 and then M5. Because M5 is integrated into the phosphorylation domain, the whole cytoplasmic headpiece moves. These structural changes show how an event in the transmembrane domain can be transmitted to the cytoplasmic domain despite flexible links between them. They also reveal that Ca2+-ATPase has considerable plasticity even when fixed by a transmembrane inhibitor, presumably to accommodate thermal fluctuations.
Keywords: Ca2+-ATPase, ion pump, membrane protein
Ca2+-ATPase of skeletal muscle sarcoplasmic reticulum (SERCA1a) is structurally and functionally the best studied member of the P-type ion-translocating ATPases (1, 2). Located in the sarcoplasmic reticulum membrane, SERCA1 pumps Ca2+ released into muscle cells during contraction and thereby causes relaxation. The reaction cycle consists of at least eight intermediates, and crystal structures have been determined for six of them (3–9). These crystal structures show that SERCA1 consists of three cytoplasmic domains (A, actuator; N, nucleotide binding; and P, phosphorylation) and 10 (M1–M10) transmembrane helices (3). They also demonstrate that ATPase undergoes large and global conformational changes during the reaction cycle. In conventional E1/E2 theory (10–12), active transport of Ca2+ is achieved by alternating the affinity and accessibility of the transmembrane Ca2+-binding sites. That is, in E1, the binding sites have high affinity and face the cytoplasm; in E2, the binding sites have low affinity to Ca2+ and face the lumen of the sarcoplasmic reticulum. Although H+ and water, instead of Ca2+, stabilize the binding sites in E2 (13), this form of the ATPase is unstable, particularly when solubilized (14). Therefore, so far all successful crystallization experiments in the absence of Ca2+ used inhibitors that stabilize the transmembrane domain.
Several inhibitors specific to SERCA1 are available (15–17). By far the best known and most potent inhibitor is thapsigargin (TG), a plant sesquiterpene lactone (18), with the dissociation constant in the subnanomolar range (15). Another commonly used inhibitor is 2,5-di-tert-butyl-1,4-dihydroxybenzene (BHQ), an antioxidant, which has a much simpler chemical structure and a lower affinity (19). We have determined the crystal structures of the ATPase with either one [E2(TG) or E2(BHQ)] or both [E2(TG+BHQ)] of them bound (4, 13). The crystal structures were essentially the same. Thus, only one conformation of the ATPase has been published so far for the E2 state.
Cyclopiazonic acid (CPA) is another well known and potent inhibitor of SERCA1 (16, 20). It binds to the ATPase in the E2 state and affects the enzymatic activity of Ca2+-ATPase in a way very similar to TG (21, 22). Because of some structural similarity to TG, which binds to the space surrounded by the M3, M5, and M7 transmembrane helices (4), it has been supposed that they share the binding sites (21). Site-directed mutagenesis and chimera results have been interpreted to support this idea, although the most influential residue on TG binding, that is, Phe-256 on the M3 helix, affected the CPA binding only slightly (21, 23). It has also been shown that a malarial Ca2+-ATPase (PfATP4) is sensitive to CPA but insensitive to TG (24).
Here we report crystal structures of SERCA1a stabilized with bound CPA alone [E2(CPA)] and in combination with other inhibitors. We describe the finding that the binding site of CPA is distinctly different from that of TG but partially overlaps with that of BHQ; yet, the interaction with the ATPase is different from BHQ and even the overall structure of the ATPase is different from that with E2(TG) or E2(TG+BHQ). Thus, these crystal structures unambiguously demonstrate that Ca2+-ATPase in the E2 state takes multiple conformations and show how changes that occur in transmembrane domains are transmitted to the cytoplasmic domains despite flexible links between them.
Results
Structure Determination and the Overall Structure.
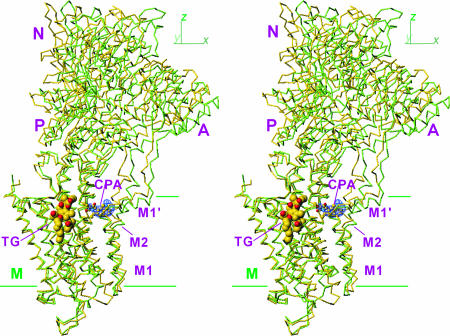
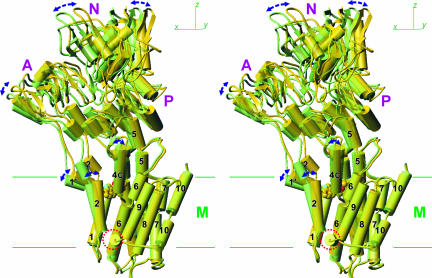
Unlike E2(TG), which crystallized in both P21 and P41212 space groups‡ (4), we could obtain crystals of E2(CPA) only in P21. Because the unit cell parameters were similar to P21 crystals of E2(TG) (E2(TG)P21), an atomic model was built by molecular replacement starting from that of E2(TG)P21 and refined to Rfree of 32.4% at 3.4 Å (Table 1). The overall structure of E2(CPA) was similar to that of E2(TG)P21 (rmsd = 1.2 Å for all Cα atoms) but more different from that of E2(TG+BHQ) and E2(TG) belonging to the P41212 space group (rmsd >1.9 Å). For the cytoplasmic region, the differences were mostly due to a movement of the whole cytoplasmic headpiece, which was more upright in E2(CPA) (Fig. 1). The differences were more complicated in the transmembrane domain.
Table 1.
Data and refinement statistics
| Data collection | ||||
| Crystal | E2(CPA) | E2(CPA+CC) | E2(CPA+TG) | E2(TG)P21 |
| No. of crystals | 2 | 2 | 2 | 2 |
| Resolution range, Å | 20.0–3.4 (3.52–3.40) | 20.0–2.8 (2.88–2.80) | 20.0–2.9 (2.97–2.90) | 20.0–3.1 (3.18–3.10) |
| Space group | P21 | P41212 | P21 | P21 |
| Cell dimensions | ||||
| a, Å | 63.04 | 71.65 | 62.90 | 62.85 |
| b, Å | 96.03 | 71.65 | 95.65 | 95.95 |
| c, Å | 155.42 | 586.25 | 155.10 | 154.50 |
| β, ° | 95.09 | 90.00 | 95.24 | 94.9 |
| Rmerge, % | 5.3 (30.1) | 14.1 (53.4) | 4.8 (36.1) | 13.4 (42.9) |
| I/σ(I) | 34.2 (3.4) | 32.0 (7.2) | 33.6 (2.0) | 27.6 (2.7) |
| Completeness, % | 99.9 (99.8) | 99.9(99.9) | 99.2(92.5) | 99.9 (99.4) |
| Redundancy | 7.3 (6.7) | 17.1 (14.5) | 7.0 (3.9) | 6.4 (5.6) |
| Refinement statistics | ||||
| Resolution range, Å | 15.0–3.4 | 15.0–2.8 | 15.0–2.9 | 15.0–3.1 |
| No. of reflections | 25,100 | 38,263 | 38,171 | 29,357 |
| Rwork/Rfree, % | 26.7/32.4 | 24.7/27.2 | 24.4/29.2 | 23.5/28.4 |
| No. of atoms | 7,695 | 7,753 | 7,741 | 7,717 |
| Overall B-factor, Å2 | 104.7 | 62.8 | 77.4 | 92.8 |
| rmsd bond, Å | 0.010 | 0.009 | 0.011 | 0.009 |
| rmsd angles, ° | 1.4 | 1.4 | 1.4 | 1.4 |
| Ramachandran,* % | 72.4/27.6/0 | 82.6/17.4/0 | 85.5/14.5/0 | 76.4/23.6 /0 |
Parentheses denote statistics in the highest-resolution shells.
*Fractions of residues in the most favored/additionally allowed/generously allowed regions of the Ramachandran plot according to ProCheck (32). No residue was found in the generously allowed and disallowed regions.
Fig. 1.
Superimposition of E2(CPA) and E2(TG+BHQ), which represent the two most distant structures in the five E2 crystals. Cα traces are viewed in stereo along the plane of the lipid bilayer (i.e., xy-plane). Two structures are fitted with the M7–M10 transmembrane helices. Yellow, E2(CPA); light green, E2(TG+BHQ). TG is shown in space fill, and CPA is in ball-and-stick. The blue net shows an ∣Fobs∣ − ∣Fcalc∣ electron density map (contoured at 5σ) before introduction of CPA in the atomic model. Three cytoplasmic (A, N, and P) and transmembrane (M) domains are marked.
P41212 crystals of E2(TG) [Protein Data Bank (PDB) ID 1IWO] and E2(TG+BHQ) (PDB ID 2AGV) diffracted to 2.5-Å resolution (13), whereas E2(TG)P21 crystals diffracted to only 3.3 Å (4). The overall structure was certainly affected by crystal symmetry, given that the cytoplasmic domain of E2(TG)P21 was more upright than E2(TG) of P41212 symmetry. To enable comparison at a higher resolution, therefore, we needed P41212 crystals with bound CPA. Such crystals were obtained by combining CPA with curcumin (CC). With 60 μM CC in the dialysis buffer for crystallization, this combination of inhibitors yielded crystals [E2(CPA+CC)] that diffracted to 2.8-Å resolution with a slightly shorter unit cell dimension along the c axis than those of E2(TG) and E2(TG+BHQ) crystals (Table 1). An atomic model was built by molecular replacement starting from that of E2(TG+BHQ) and refined to Rfree of 27.2% (Table 1). Although the crystal symmetry was altered by the addition of CC, there was no hint of CC in the electron density map. Consistent with what was observed with the P21 crystals, the cytoplasmic headpiece in E2(CPA+CC) was more upright than that in E2(TG) [and E2(TG+BHQ)].
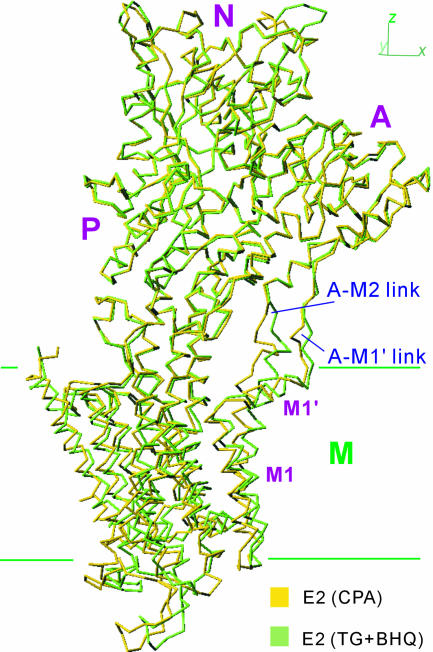
Despite the overall structures and dispositions of several transmembrane helices differing between E2(TG) and E2(CPA), crystallization of E2(CPA+TG) was successful. The space group of E2(CPA+TG) crystals was always P21, and the resultant structure was intermediate between E2(TG) and E2(CPA). When all five structures [i.e., E2(CPA), E2(CPA+CC), E2(CPA+TG), E2(TG)P21, and E2(TG+BHQ)] were compared, the two most distant ones were E2(TG+BHQ) and E2(CPA). Thus, the two structures in Fig. 1 show the maximum extent of variation within E2 crystal structures. Differences in the headpiece are very small (rmsd < 0.7 Å), and the two structures superimposed well with the cytoplasmic headpiece. However, the links between the A-domain and the transmembrane helices did not superimpose (Fig. 2), indicating that these links are flexible.
Fig. 2.
Superimposition using the P-domain showing that the headpiece moves together and that the links between the A-domain and transmembrane helices M1–M2 are flexible. The structures shown here are viewed in the direction shown in Fig. 1.
CPA Binding Site.
A Fourier difference (∣Fobs∣ − ∣Fcalc∣) map for E2(CPA), before introduction of CPA in the atomic model, clearly located CPA near the cytoplasmic surface of the transmembrane domain (Fig. 1), as the difference map contoured at 5σ showed only one peak. The map was, however, insufficient for determining the orientation and conformation of CPA, because the resolution was only 3.4 Å. For this purpose, E2(CPA+TG) and E2(CPA+CC) crystals were most useful. The difference map (Fig. 3) showed clear electron density at the same place as in E2(CPA), with two lobes of different sizes. These features were expected from the structure of CPA. One conformation of CPA registered in the Cambridge Database [entry: VOMTIA (25)] fitted nicely to the electron density but only after inversion (Fig. 3).
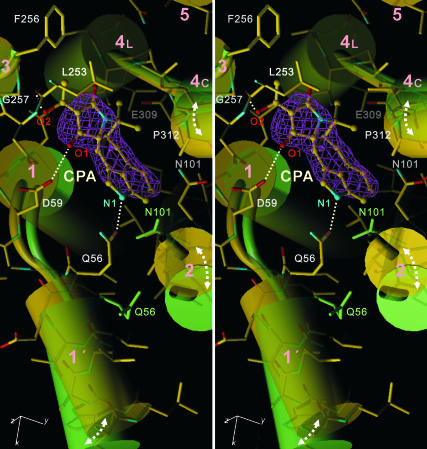
Fig. 3.
The CPA binding site in E2(CPA+CC) superimposed with that of E2(TG+BHQ) viewed in stereo approximately perpendicular to the membrane. Yellow, E2(CPA+CC); light green, E2(TG+BHQ). Cylinders represent transmembrane helices (M1–M5). The pink net represents an ∣Fobs∣ − ∣Fcalc∣ electron density map at 2.8-Å resolution contoured at 4σ before CPA (shown in ball-and-stick) was introduced into the atomic model. Dotted lines show likely hydrogen bonds. The atomic model is shown in stick representation for E2(CPA+CC). Side chains of Asn-101 and Gln-56 also are shown for E2(TG+BHQ).
CPA binding rearranges transmembrane helices M1–M5. The binding site, surrounded by M1–M4, is more confined in E2(CPA) compared with E2(TG) and E2(TG+BHQ) by the movement of M1′ and M2 (Fig. 3). N1 of CPA apparently pulls the short amphipathic helix M1′ to form a hydrogen bond with Gln-56 located at the C-terminal end of M1′ (Fig. 3). A hydroxyl group of CPA appears to form a hydrogen bond with Leu-253 carbonyl (M3); one carbonyl oxygen (O1) is located within the hydrogen bond distance from Asp-59 carboxyl (M1). As a result of the rearrangement of the M1–M4 helices, a highly complementary surface is formed. There are at least 10 van der Waals contacts between CPA and the ATPase, with a distance ≤4.0 Å (Fig. 3). They include those with main chain atoms of Leu-93 (O, M2), Asn-101 (Cα, M2), Gly-257 (Cα, M3), and Glu-309 (N, M4) and Pro-312 (Cδ, M4). M2 moves toward M4, pushed by CPA, with a large conformational change around Asn-101 (M2). In fact, CPA would not be able to bind to E2(TG) unless Asn-101 is moved nor to E1·2Ca2+ because the binding pocket is occupied by the M1 helix (Fig. 4). Thus, the inhibitory action of CPA and a large impairment by the Gly257Ile mutation (21) can be readily explained. These features are consistent with all of the CPA-bound E2 crystals.
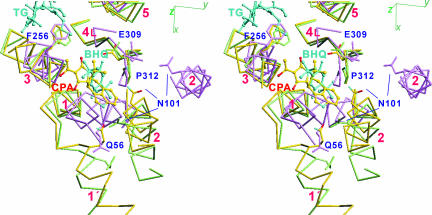
Fig. 4.
Three transmembrane inhibitors (CPA, TG, and BHQ) superimposed with Cα traces of the three crystal structures, E1·2Ca2+ (violet), E2(CPA+CC) (yellow), and E2(TG+BHQ) (light green), viewed in stereo. The side chains of several key residues are shown. The transmembrane helices (M1–M6) are numbered.
The CPA binding site partially overlaps with that of BHQ (Fig. 4). Even the likely hydrogen bond with Asp-59 carboxyl (Fig. 3) is the same. However, the M2 side of CPA is bulkier than that of BHQ and makes much more extensive hydrophobic contacts with the ATPase (Fig. 4). These features readily explain the higher affinity of CPA [Kd = 7 nM (26)] compared with that of BHQ [Kd = 0.2 μM (17)]. Also, the more superficial binding site of CPA, with an obvious entry/exit route, explains the rather fast kinetics of dissociation of CPA compared with TG (26), which is deeply inserted into the membrane (Fig. 1).
Global Structural Changes Caused by CPA Binding.
To examine the structural changes caused by CPA binding more quantitatively, we compared E2(CPA+CC) and E2(TG+BHQ). They both have relatively high resolution [2.8 Å for E2(CPA+CC) and 2.4 Å for E2(TG+BHQ)] and the same crystal symmetry (P41212), making it unnecessary to consider the effects of crystal packing.
Around CPA, two structural changes are evident. One is a 18° rotation of the M1′ helix toward M2 around an axis approximately perpendicular to the membrane (Figs. 3 and 5). This position of M1′ is apparently stabilized by a hydrogen bond between CPA and Gln-56. In E2(TG+BHQ), Leu-52, Gln-56, and Phe-57 on M1′ are in van der Waals contacts with Val-106, Ala-102, and Leu-98 on M2, respectively. As a result of the van der Waals contacts, M1′ pushes M2 near the cytoplasmic surface, causing a 12° change in inclination of the M2 helix, as the luminal end of M2 is fixed by contacts with a short amphipathic helix between M9 and M10 (4) (Fig. 5). This movement of M2 is then transmitted to the cytoplasmic half of M4 (M4C), mainly through Asn-101–Pro-312 (Fig. 4), as a result of a molecular domino effect. Then, in turn, M4C moves M5 (Fig. 5), as their cytoplasmic ends are clamped together by short β-strands (4). Finally, the P-domain and with it the whole cytoplasmic headpiece take a more upright position, because the cytoplasmic end of M5 is integrated into the P-domain as a part of the Rossmann fold (3).
Fig. 5.
Global structural changes induced by the binding of CPA. E2(CPA+CC) (yellow) and E2(TG+BHQ) (light green) crystal structures are superimposed with the M7–M10 helices. The structures are shown from the side opposite to that in Fig. 1. Arrows indicate the differences between the two structures. The circle indicates the amphipathic helix between M8 and M9. CPA is shown in space fill. M2, M5, and M7 are represented with two to three cylinders, although they are continuous helices.
The movement of M2 inevitably affects the positions of other transmembrane helices, in particular, M1 and the luminal half of M4 (M4L). The rotation of M1′ is not transmitted to M1 directly. The top part of M1 does not follow the rotation of M1′ (Fig. 4); it is fixed by van der Waals contacts with M3 and M4 (Fig. 3), indicating that the link between M1′ and M1 is flexible. Instead, M1 follows M2. This movement occurs because M1 and M2 form a stable V-shaped structure and move together throughout the entire reaction cycle except for E1·2Ca2+ (6), keeping tight van der Waals contacts between them. Although the link between M1 and M1′ is flexible, M1′ makes van der Waals contacts with M2 from E1·ATP to E2 and can move M1 through M2.
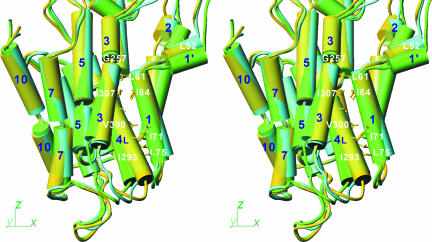
Similar van der Waals contacts are also maintained between the M1 and M4L helices at different levels along them throughout E1·ATP to E2. They include Leu-61, Leu-65, Ile-71, and Leu-75 on M1 and Ile-293, Phe-296, Val-300, and Ile-307 on M4 (Fig. 6). If the movement of the A-domain is transmitted to M1, it is transmitted to M4L at the same time. This appears to be the normal way of signal transmission in the forward direction of the reaction cycle [particularly in E1P→E2P and E2P→E2 (6)]. Here, the origin of the signal is different (i.e., binding of CPA), but the flow of movements and the usage of helix–helix contact in transmitting the movements appear to be the same: M1, guided by M2, moves M4L to take a more horizontal position than in E2(TG+BHQ) (Fig. 6).
Fig. 6.
Differences in orientation of the transmembrane helices in three crystal structures. E2(CPA) (yellow), E2(TG+BHQ) (light green), and E2·MgF42− (cyan) are superimposed with the M7–M10 helices and are shown in stereo in the same direction as in Fig. 1. M2, M3, M5, and M7 are represented with two to three cylinders, although they are continuous helices. Some of the residues used in helix–helix contacts are shown.
Effect of TG.
E2(CPA) is the first E2 structure that is significantly different from E2(TG). Yet we could obtain E2(CPA+TG) crystals, and the structure was intermediate between E2(TG) and E2(CPA). Because the binding site of TG is the cavity surrounded by M3, M5, and M7 (Figs. 1 and 4), it is of interest to examine differences in these helices, in particular, M3. In fact, the luminal half of M3 is more detached from M5 (Fig. 6), conforming with a more horizontal orientation of M4L. However, this disposition is probably influenced by CPA, because, as mentioned before, the hydroxyl group at one end of CPA is likely to form a hydrogen bond with the carbonyl of Leu-253 and fix the M3 helix at that point (Fig. 3).
Discussion
As described, the structure of E2(CPA) is different from that of E2(TG). Although both CPA and TG bind to the transmembrane domain, the most prominent difference is in the orientation of the headpiece, primarily as a result of a rearrangement of the M1′, M2, M4, and M5 helices. Even though the crystal structures of E2(CPA) and E2(TG) are different, it was possible to generate a E2(CPA+TG) crystal. These observations mean that Ca2+-ATPase in E2 possesses structural plasticity and exhibits substantial thermal movements even when stabilized with potent inhibitors. They also point to flexible regions in the ATPase that allow such movements. This finding is in line with the results of limited proteolysis, which demonstrate that the A-domain–M3 and the A-domain–M2 loops are flexible and susceptible to proteinase K attack in E2. If the M3 helix is stabilized with TG, the A-domain–M3 loop is largely protected from proteinase K attack, making the A-domain–M2 link the predominant target (27). Our present results show that the A-domain–M1′ link is also flexible as well as the M1–M1′ link (Figs. 2 and 3).
CPA interacts primarily with M1, M1′, and M2, all of which have flexible links with the A-domain. Yet the binding of CPA changes the orientation of the cytoplasmic headpiece. How, then, can a binding change or signal be transmitted from the transmembrane region to move the cytoplasmic domain despite the flexible links? In fact, why are flexible links necessary?
Change in Orientation of M4L.
Another interesting observation here is that M4L, the luminal half of the M4 helix, has a fairly large change (≈10°) in orientation among different E2 structures (Fig. 6). This means that M4L can change its orientation at least within this range by thermal movements in E2. Our molecular dynamics simulation of Ca2+-ATPase in E2 (Y. Sugita, M. Ikeguchi, and C. Toyoshima, unpublished data) certainly shows this feature. Such a large movement is remarkable, because the change in inclination of M4L is likely to be the mechanism for opening and closing the luminal gate (13). M4L in the E2·MgF42− complex (PDB ID 1WPG), regarded as an analogue of the product state of the hydrolysis of phosphate (E2·Pi), is certainly more horizontal than in E2(TG) but not enough for Ca2+ to pass through. In fact, M4L in E2(CPA) is slightly more horizontal than that in E2·MgF42− (Fig. 6). Intrinsic Trp fluorescence levels substantiate this finding (28). The fluorescence of E2·MgF42− is low and similar to that of E2(TG), and the difference between E2 and E2P is likely to reflect the environment of Trp-288 at the bottom of M4, because this is the most prominent change among the transmembrane helices between these two structures.
Because the A-domain is located at one end of the cytoplasmic headpiece, a small change in the orientation of M5, the central pillar helix, will be amplified and transmitted to the transmembrane domain. If the links between the A-domain and M1–M3 helices were rigid, therefore, thermal fluctuation would open the gate stochastically like ion channels. This needs to be avoided in pumps and is one of the reasons why the links between the A-domain and M1–M3 helices have to be flexible.
Modes of Communication Between Domains Connected by Flexible Links.
Previous crystallographic studies on Ca2+-ATPase have provided examples of interdomain communication. In E1·2Ca2+ → E1·ATP or E1·AlFx·ADP, tilting of the A-domain by Mg2+ and γ-phosphate (analogue) pulls the A-domain–M2 link and hence the M1 and M2 helices toward the cytoplasm (5, 7). At the same time, the A-domain–M3 link is strained (5). This appears to be the driving force for a 110° rotation of the A-domain in E1P → E2P, which pulls the V-shaped M1–M2 assembly toward M3 (6). This pulling is transmitted to M4L through M1 by the van der Waals contacts between them. In E2P→E2, release of the A-domain relaxes the A-domain–M1′ link and so too the whole M1–M2 assembly, because M1′ makes van der Waals contacts with M2. This means that the strain exerted on M4L by M1 is relieved and the helix assumes a more relaxed position to close the luminal gate (6).
Thus, we see three modes of signal transmission here. Pulling on flexible links and straining them appear to be primary. Then removing strain can guide the system to another state. In this way, a seemingly unfavorable state is made favorable, and direction is provided to the reaction cycle. For instance, the A-domain will not be raised by pushing on slacked links.
Flexible links between rigid segments may also be needed to generate complex domain movements, because more combinations between the segments become possible and because movements of one particular segment can be guided by other, fixed segments. We saw that the binding of CPA to the transmembrane domain moves the whole cytoplasmic headpiece through a complex rearrangement of transmembrane helices. In this way, different steps of a reaction cycle could be converted to distinct movements required to fulfill a complex task, such as ion pumping across membranes.
Materials and Methods
Crystallization.
Crystallization conditions were essentially the same as those reported previously for E2(TG) (4). For the crystals with CPA, affinity-purified enzyme (20 μM) in octaethyleneglycol mono-n-dodecylether (C12E8) and 1 mM Ca2+ were mixed with 0.1 mM CPA and 3 mM EGTA and then dialyzed against a buffer consisting of 2.75 M glycerol, 4% PEG 400, 3 mM MgCl2, 0.05 mM CPA, 2.5 mM NaN3, 2 μg/ml butylhydroxytoluene, 0.2 mM DTT, 0.1 mM EGTA, and 20 mM MES, pH 6.1, for ≈1 month. For E2(CPA+TG) crystals, 10 μM TG was added to the specimen before dialysis. For E2(CPA+CC) crystals, initial specimen and dialysis buffer contained 0.12 and 0.06 mM CC, respectively. Crystals were allowed to grow for 1 month and were flash-frozen in cold nitrogen gas.
Data Collection.
Diffraction data were collected at BL41XU of SPring-8 by using Mar165 [for E2(TG)P21 only] and ADSC Q315 CCD detectors. For each structure, diffraction intensities from the two best crystals were merged by using Denzo and Scalepack (29). Unit cell parameters and data statistics are listed in Table 1.
Modeling.
The atomic models of E2(TG)P21 and E2(CPA+CC) were made by molecular replacement starting from the model for E2(TG+BHQ) (PDB ID 2AGV). Those for E2(CPA) and E2(CPA+TG) were made similarly but from the model for E2(TG)P21. The atomic models were refined with CNS (30); TLS refinement with REFMAC (31) was further applied only to E2(CPA+TG). The geometry of the models was examined with ProCheck (32). Refinement statistics are summarized in Table 1.
Acknowledgments
We thank N. Shimizu, M. Kawamoto, and T. Tsuda for data collection at BL41XU of SPring-8; H. Nomura and J. Tsueda for expert technical assistance; and D. B. McIntosh for help in improving the manuscript. This work was supported in part by a Creative Science Project Grant from the Ministry of Education, Culture, Sports, Science and Technology and the Japan New Energy and Industry Technology Development Organization.
Abbreviations
- SERCA1
sarco(endo)plasmic reticulum calcium ATPase 1
- TG
thapsigargin
- BHQ
2,5-di-tert-butyl-1,4-dihydroxybenzene
- CC
curcumin
- CPA
cyclopiazonic acid
- PDB
Protein Data Bank.
Note.
During the preparation of this article, a report on the crystal structure of Ca2+-ATPase with bound CPA appeared (33). The report placed CPA at the same position but in the orientation opposite to that described here.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates for the E2(TG)P21, E2(CPA), E2(CPA+TG), and E2(CPA+CC) forms of Ca2+-ATPase have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2EAR, 2EAS, 2EAT, and 2EAU, respectively).
References
- 1.Møller JV, Juul B, le Maire M. Biochim Biophys Acta. 1996;1286:1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 2.Toyoshima C, Inesi G. Annu Rev Biochem. 2004;73:269–292. doi: 10.1146/annurev.biochem.73.011303.073700. [DOI] [PubMed] [Google Scholar]
- 3.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 4.Toyoshima C, Nomura H. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 5.Toyoshima C, Mizutani T. Nature. 2004;430:529–535. doi: 10.1038/nature02680. [DOI] [PubMed] [Google Scholar]
- 6.Toyoshima C, Nomura H, Tsuda T. Nature. 2004;432:361–368. doi: 10.1038/nature02981. [DOI] [PubMed] [Google Scholar]
- 7.Sørensen TL, Møller JV, Nissen P. Science. 2004;304:1672–1675. doi: 10.1126/science.1099366. [DOI] [PubMed] [Google Scholar]
- 8.Olesen C, Sørensen TL, Nielsen RC, Møller JV, Nissen P. Science. 2004;306:2251–2255. doi: 10.1126/science.1106289. [DOI] [PubMed] [Google Scholar]
- 9.Jensen AM, Sørensen TL, Olesen C, Møller JV, Nissen P. EMBO J. 2006;25:2305–2314. doi: 10.1038/sj.emboj.7601135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albers RW. Annu Rev Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 11.Post RL, Hegyvary C, Kume S. J Biol Chem. 1972;247:6530–6540. [PubMed] [Google Scholar]
- 12.de Meis L, Vianna AL. Annu Rev Biochem. 1979;48:275–292. doi: 10.1146/annurev.bi.48.070179.001423. [DOI] [PubMed] [Google Scholar]
- 13.Obara K, Miyashita N, Xu C, Toyoshima I, Sugita Y, Inesi G, Toyoshima C. Proc Natl Acad Sci USA. 2005;102:14489–14496. doi: 10.1073/pnas.0506222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamasaki K, Daiho T, Suzuki H. J Biol Chem. 2002;277:13615–13619. doi: 10.1074/jbc.M200625200. [DOI] [PubMed] [Google Scholar]
- 15.Sagara Y, Inesi G. J Biol Chem. 1991;266:13503–13506. [PubMed] [Google Scholar]
- 16.Goeger DE, Riley RT, Dorner JW, Cole RJ. Biochem Pharmacol. 1988;37:978–981. doi: 10.1016/0006-2952(88)90195-5. [DOI] [PubMed] [Google Scholar]
- 17.Logan-Smith MJ, Lockyer PJ, East JM, Lee AG. J Biol Chem. 2001;276:46905–46911. doi: 10.1074/jbc.M108778200. [DOI] [PubMed] [Google Scholar]
- 18.Christensen SB, Andersen A, Smitt UW. Fortschr Chem Org Naturst. 1997;71:129–167. doi: 10.1007/978-3-7091-6529-4_2. [DOI] [PubMed] [Google Scholar]
- 19.Khan YM, Wictome M, East JM, Lee AG. Biochemistry. 1995;34:14385–14393. doi: 10.1021/bi00044a015. [DOI] [PubMed] [Google Scholar]
- 20.Seidler NW, Jona I, Vegh M, Martonosi A. J Biol Chem. 1989;264:17816–17823. [PubMed] [Google Scholar]
- 21.Ma H, Zhong L, Inesi G, Fortea I, Soler F, Fernandez-Belda F. Biochemistry. 1999;38:15522–15527. doi: 10.1021/bi991523q. [DOI] [PubMed] [Google Scholar]
- 22.Plenge-Tellechea F, Soler F, Fernandez-Belda F. J Biol Chem. 1997;272:2794–2800. doi: 10.1074/jbc.272.5.2794. [DOI] [PubMed] [Google Scholar]
- 23.Wootton LL, Michelangeli F. J Biol Chem. 2006;281:6970–6976. doi: 10.1074/jbc.M510978200. [DOI] [PubMed] [Google Scholar]
- 24.Krishna S, Woodrow C, Webb R, Penny J, Takeyasu K, Kimura M, East JM. J Biol Chem. 2001;276:10782–10787. doi: 10.1074/jbc.M010554200. [DOI] [PubMed] [Google Scholar]
- 25.van Rooyen PH. Acta Crystallogr C. 1992;48:551–552. [Google Scholar]
- 26.Soler F, Plenge-Tellechea F, Fortea I, Fernandez-Belda F. Biochemistry. 1998;37:4266–4274. doi: 10.1021/bi971455c. [DOI] [PubMed] [Google Scholar]
- 27.Xu C, Ma H, Inesi G, Al Shawi MK, Toyoshima C. J Biol Chem. 2004;279:17973–17979. doi: 10.1074/jbc.M313263200. [DOI] [PubMed] [Google Scholar]
- 28.Danko S, Yamasaki K, Daiho T, Suzuki H. J Biol Chem. 2004;279:14991–14998. doi: 10.1074/jbc.M313363200. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Winn MD, Isupov MN, Murshudov GN. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 32.Collaborative Computational Project N. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 33.Moncoq K, Trieber CA, Young HS. J Biol Chem. 2007 doi: 10.1074/jbc.M611653200. in press. [DOI] [PubMed] [Google Scholar]