Abstract
In all three domains of life, DNA replication begins at specialized loci termed replication origins. In bacteria, replication initiates from a single, clearly defined site. In contrast, eukaryotic organisms exploit a multitude of replication origins, dividing their genomes into an array of short contiguous units. Recently, the multiple replication origin paradigm has also been demonstrated within the archaeal domain of life, with the discovery that the hyperthermophilic archaeon Sulfolobus has three replication origins. However, the evolutionary mechanism driving the progression from single to multiple origin usage remains unclear. Here, we demonstrate that Aeropyrum pernix, a distant relative of Sulfolobus, has two origins. Comparison with the Sulfolobus origins provides evidence for evolution of replicon complexity by capture of extrachromosomal genetic elements. We additionally identify a previously unrecognized candidate archaeal initiator protein that is distantly related to eukaryotic Cdt1. Our data thus provide evidence that horizontal gene transfer, in addition to its well-established role in contributing to the information content of chromosomes, may fundamentally alter the manner in which the host chromosome is replicated.
Keywords: Archaea, DNA replication, viral integration
It is well established that Archaea and Eukarya possess orthologous machineries for DNA replication (1, 2). Despite these similarities in the protein machineries, initial studies suggested that there may be a fundamental difference in the modes that archaea and eukaryotes employ to ensure genome duplication. More specifically, studies of Pyrococcus abyssi and Halobacterium NRC-1 revealed that these species appear to use a single origin of DNA replication per chromosome (3, 4). This contrasts markedly with the situation in eukaryotes where many origins are present per chromosome (5, 6). However, multiple replication origins have now been discovered in the chromosomes of the crenarchaeal hyperthermophiles Sulfolobus solfataricus and Sulfolobus acidocaldarius (7–9). It is currently unclear whether other archaeal species also utilize multiple replication origins. Furthermore, is not yet understood how Sulfolobus acquired these multiple initiation sites during the evolution of its genome. In general, eukaryal DNA replication represents a more complicated version of that in archaea, and it is clear that multiple gene duplication events have given rise to some of this complexity. For example, the archaeal MCM (minichromosome maintenance) complex is typically a homomultimer. Contrastingly, all eukaryotes have at least six related MCMs that form a heterohexamer (10). Although gene duplication events can explain the evolution of heteromultimeric protein assemblies, they do not readily account for the development of multiple origin systems. This is particularly evident when the sequences of the three Sulfolobus replication origins (termed oriC1, oriC2, and oriC3) are compared. Although all three are bound by the candidate initiator proteins, the sequence motifs used at the three are strikingly diverse, hinting at independent derivations of the three origins (7, 9). In archaea, the candidate initiator proteins are homologous to the eukaryotic initiator proteins Cdc6 and Orc1. In eukaryotes, these proteins together with Orc2–6 act to recruit MCM to origins of replication in a reaction that absolutely requires an additional factor, Cdt1 (6). Although archaea possess orthologs of Orc1, Cdc6, and MCM, no archaeal homolog of Cdt1 has yet been identified.
In the current work, we reveal that Aeropyrum pernix has at least two replication origins, indicating that the multiple replication origin paradigm is not restricted to the Sulfolobus genus. Comparison of the A. pernix and Sulfolobus origins reveals a clear relationship between these loci. Further, analyses of the gene order and identity in the environment of the origins provides evidence for the evolution of replicon complexity by capture of extrachromosomal elements. Additionally, we identify a conserved ORF adjacent to one of the origins in Sulfolobus and Aeropyrum that has sequence similarity to the essential eukaryal replication factor, Cdt1. This archaeal factor is predicted to have domain organization reminiscent of bacterial plasmid replication initiator proteins, hinting at the evolutionary derivation of Cdt1. Finally, we reveal that this factor binds sequence specifically to replication origins.
Results and Discussion
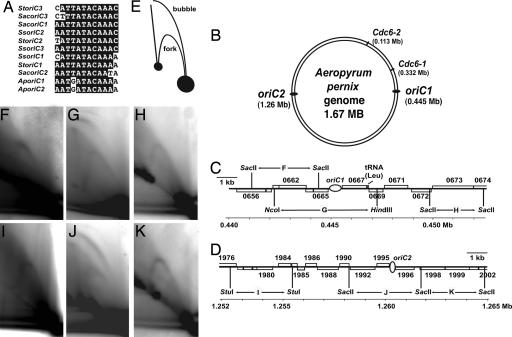
Previously we have demonstrated that the highly conserved Sulfolobus Cdc6-1 protein binds sequence specifically to a consensus motif, the ORB element, that is conserved at many of the predicted origins in a variety of archaeal species. We had formerly identified several ORB elements within a 700-bp noncoding region in the hyperthermophilic crenarcheote A. pernix (7). Recently, biochemical analysis has confirmed origin activity at this site (11). Comparison of the nucleotide sequence of the three Sulfolobus origins with this A. pernix origin (AporiC1) revealed a previously uncharacterized motif (UCM) in the center of all four sites (Fig. 1). A second copy of the UCM was found in the Aeropyrum genome, within a 270-bp noncoding region, on the opposite side of the circular chromosome from AporiC1 (12). Although we could not detect any ORB motifs at this second UCM-containing locus, we note that both UCM-containing loci sites coincide with two GC skew disparity minima, predicted by a bioinformatic Z-curve analysis of the A. pernix genome (13). We hypothesized that the A. pernix genome harbors at least two initiation sites, centered on these UCMs. The activity of these replication origins was subsequently confirmed in vivo, by two-dimensional agarose gel electrophoresis (Fig. 1). Arcs corresponding to active replication initiation sites were detectable at both putative origin sites; flanking restriction sites only show evidence of replication forks. Thus, A. pernix replicates its genome from at least two initiation sites, revealing that multiple origin usage is not exclusive to the Sulfolobus genus. The UCM appears to be an important signature motif in the center of origins in Sulfolobus and Aeropyrum. We speculated that this motif might also be found in origins of replication in other relatives of Sulfolobus. Indeed, examination of the Hyperthermus butylicus (14) and Metallosphaera sedula genomes (www.ebi.ac.uk/genomes/wgs.html) reveals the presence of UCMs in these species. We observed that three out of four of the UCMs in the H. butylicus and M. sedula genomes are located adjacent to ORB elements, whilst the fourth UCM lies in a locus that is similar to S. solfataricus oriC3. [For details, see supporting information (SI) Fig. 6.]
Fig. 1.
Characterization of the Aeropyrum pernix replication origins by two-dimensional (2D) gel electrophoresis. (A) Alignment of the UCM sequences located centrally in the two A. pernix and three Sulfolobus origins (St, Sulfolobus tokodaii; Sac, S. acidocaldarius; Ss, S. solfataricus). (B) Cartoon representing the A. pernix genome, indicating the position of the two origins relative to the initiator genes Cdc6-1 and Cdc6-2. (C and D) Representation of the two origin loci. Ovals denote the origin locations. Gene ID numbers are indicated; ORFs above the midline are transcribed left to right, and those below the line are transcribed from right to left. (E) Illustration of the species of replication intermediates detectable at the origins. (F–K) 2D gel examination of the A. pernix origin loci. DNA isolated from asynchronous replicating A. pernix cells was digested with restriction enzymes to produce the fragments indicated in C and D and subjected to the 2D procedure. G and J display bubble arc intermediates that are indicative of an active bidirectional origin of replication within the central third of the restriction fragment.
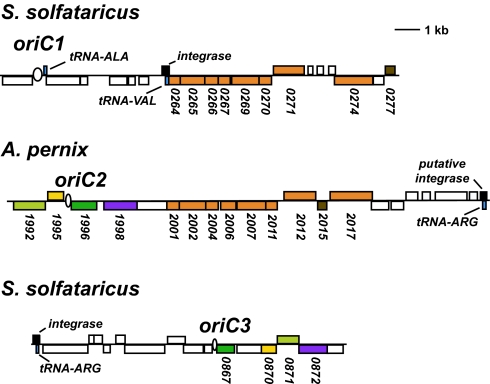
Comparisons of the origin loci of Aeropyrum and the Sulfolobus species revealed an intriguing mosaic-like nature to the genes at A. pernix oriC2 (AporiC2), with some homologues found at S. solfataricus oriC1 (SsoriC1) and others at SsoriC3; we could not detect any homologies with genes in the vicinity of SsoriC2. There was a marked transition point within this hybrid arrangement, separating the SsoriC1- and SsoriC3-locus homologues (Fig. 2). This abrupt change in homology was bordered by an ORF encoding a viral integrase element (ORF Sso0262) at SsoriC1 (Fig. 2), adjacent to a tRNA gene, a feature typical of many prokaryotic viral integration sites (15–17). Additionally, at the AporiC2 locus, a tRNA-Arg gene was found at the opposite end of the region of SsoriC1 homology (Fig. 2). Sequence analysis of the environment around this tRNA revealed homology to integrase elements. We also observed that tRNA genes and viral integration genes appear to be associated with SsoriC3 and AporiC1. Although it is possible that this phenomenon could simply indicate a preference for extrachromosomal element integration at origins of replication, we believe that these elements could play a more fundamental role in shaping the architecture of the origin-containing loci, for the reasons detailed below.
Fig. 2.
A hybrid arrangement of Sulfolobus origin associated gene homologues is observed at the A. pernix oriC2 locus. Genes located at two S. solfataricus initiation sites, oriC1 and oriC3, are positioned together at the A. pernix oriC2. Homologous genes are denoted by color, and the gene ID numbers are indicated. Transfer-RNA genes (blue) and viral integration sites (black) are also highlighted.
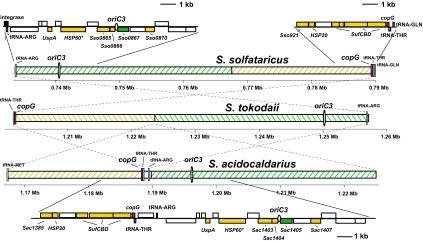
Examination of the genomic environment at the oriC3 loci of S. solfataricus and S. tokodaii (18) revealed a large inversion of ≈58 kb between the two species (Fig. 3 and Table 1). Significantly, the origin was contained within the inversion, and the gene order surrounding the initiation sites was completely conserved. In addition, the region was flanked by tRNA genes. An almost identical pattern of gene distribution was also observed at the S. acidocaldarius oriC3, although there was one clear intrafragment translocation of 21.5 kb (Fig. 3). The absence of insertion sequences and MITEs (miniature inverted-repeats transposable elements) in the S. acidocaldarius genome has led to the proposal that the gene order in S. acidocaldarius most closely resembles that of the last common ancestor of modern Sulfolobus species (19, 20). A double inversion of the 21.5-kb and 36.5-kb fragments in S. acidocaldarius could have produced the gene order displayed in the 58-kb S. tokodaii fragment. Notably, an S. acidocaldarius ORF (Sac1391), residing within the conserved region only 8.5 kb from oriC3, displayed strong homology to the Sulfolobus plasmid copy number control protein, copG. This gene is likely to have been introduced into the genome as a result of the integration of a hyperthermophilic plasmid or virus. Homologues of the copG gene were also observed in both the S. solfataricus and S. tokodaii conserved fragments (Sso6805 and ST3657, respectively).
Fig. 3.
Organization and orientation of the oriC3 loci in three Sulfolobus species. Colored hatching represents homologous regions of the genome in the three species. The direction of the hatching indicates the orientation of the homologous regions relative to the rest of the genome. A 21.5-kb intrafragment translocation is indicated by the yellow hatching. Origins are represented by ovals. The magnified segment of the S. acidocaldarius region illustrates the oriC3 proximal genes. S. solfataricus gene homologues are also displayed. ORF colors denote gene function: red, copG; green, WhiP; and yellow, probable stress response-related genes. HSP60*, thermosome α subunit belonging to the HSP60 family.
Table 1.
Stress-related proteins associated with the oriC3 locus
| Gene ID | Known or predicted motif/function |
|---|---|
| SAC1385 SSO0921 ST1204 | Partial homology to HSP60 |
| SAC1386 SSO0922 ST1203 | Hsp20 |
| SAC1388 SSO0925 ST1201 | FeS assembly ATPase SufC |
| SAC1389 SSO0927 ST1200 | FeS assembly protein SufB |
| SAC1390 SSO0928 ST1199 | FeS assembly protein SufD |
| SAC1399 SSO0859 ST1256 | Homology to bacterial universal stress protein (USP) |
| SAC1401 SSO0862 ST1253 | Thermosome α subunit |
| SAC1403 SSO0865 ST1251 | CxxC thioredoxin motifs |
| SAC1404 SSO0866 ST1250 | Homology to HSP70 C terminus |
| SAC1407 SSO0870 ST1246 | Homology to heat-inducible transcription repressor (hrcA) |
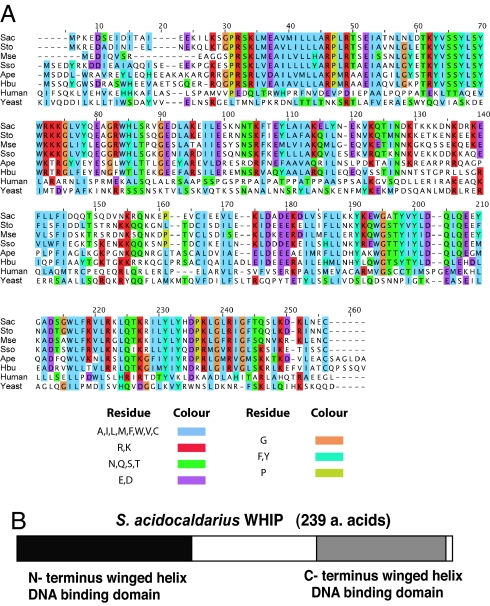
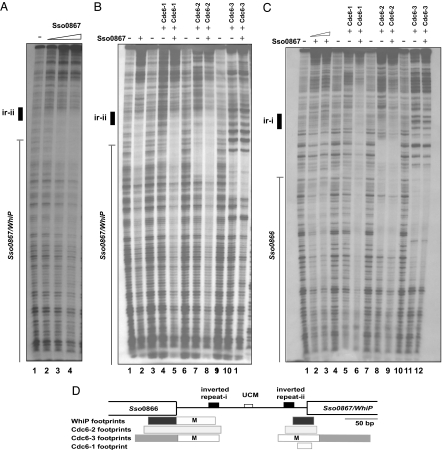
We wished to ascertain whether additional genes around Sulfolobus oriC3 could have originated from an extrachromosomal element. In all three Sulfolobus species the ORF Sso0867 (ST1249; Sac1405) was located beside oriC3. The A. pernix homologue of Sso0867 (Ape1996) was also adjacent to AporiC2. Although we could not detect any other homologues of the Sso0867 protein in the National Center for Biotechnology Information database by conventional BLAST searching, we note that this protein displays limited homology (34% similarity, 21% identity) to the C-terminal region of Saccharomyces cerevisiae replication initiator protein, Cdt1 (Fig. 4A). Although this level of homology is very low, the analogous regions of Cdt1 from budding and fission yeasts only have 32% similarity and 18% identity. With this modest homology in mind, and by analogy with the association of Sulfolobus oriC1 and oriC2 with Orc1/Cdc6 homologues, we speculated that the proximity of the Sso0867 homologues to the Sulfolobus and Aeropyrum origins might implicate this gene in origin function (see below). It should be noted that, in S. solfataricus, Sso0867 is located ≈85 kb away from the nearest Orc1/Cdc6 homolog (Cdc6-2). Further bioinformatics analysis of Sso0867 allowed us to identify potential functional domains. Alignment of the four archaeal Sso0867 homologues reveals two highly conserved domains in the N- and C-halves of the protein, separated by a less conserved central region (Fig. 4B and Table 2). Similarity searches demonstrated that the N-terminal region consisted of a winged helix–turn–helix (wHTH) DNA binding domain. In addition, the C-terminal domain also showed weak similarity to a wHTH (Fig. 4B). This arrangement of two wHTH domains is reminiscent of the RepA plasmid initiator protein from the bacterium Pseudomonas. Significantly, structural similarities have previously been observed between bacterial RepA and archaeal Cdc6 initiator proteins (21). This RepA family of proteins is closely related to the Escherichia coli RepA bacteriophage plasmid initiator and also the RepE initiator of the E. coli miniF plasmid (22). It is notable that the RepE initiator protein also contains N- and C-terminal DNA binding domains separated by a central region. Although only the RepE N-terminal domain primary sequence displays a clear wHTH motif, resolution of the crystal structure of this protein has revealed that the N- and C-domains have similar arrangements (22). Interestingly, eukaryal Cdt1 also contains a wHTH domain, and recent analyses by Iyer and Aravind (23) have suggested that eukaryal Cdt1 may be derived from an archaeal wHTH containing protein. For these reasons, we wished to test whether the Sso0867 gene encoded an initiator protein. We purified recombinant Sso0867, and examined the interaction of this protein with SsoriC3 by DNaseI footprinting. As can be seen in Fig. 5, Sso0867 binds the origin region producing a specific DNaseI footprint (Fig. 5). We also observed that the incubation of Sso0867 with Cdc6-1 or Cdc6-2 (but not Cdc6-3) seemed to enhance this interaction (Fig. 5). Furthermore, Sso0867 also binds to SsoriC1 and SsoriC2 (data not shown). In light of the modest homology with Cdt1 and likely structural homology to bacterial plasmid initiator protein, we propose the name WhiP (for Winged-Helix Initiator Protein).
Fig. 4.
Organization of WhiP and its relationship to eukaryal Cdt1. (A) Sequence alignment of archaeal WhiP proteins [Sac, S. acidocaldarius (GenBank accession no. YP_256028); Sto, S. tokodaii (GenBank accession no. NP_377180); Mse, M. sedula (GenBank accession no. ZP_01600793); Sso, S. solfataricus (GenBank accession no. NP_342366); Ape, A. pernix (GenBank accession no. NP_148313); Hbu, H. butylicus (GenBank accession no. YP_001013395)] with human Cdt1 residues 296–546 (GenBank accession no. EAW66763) and yeast Saccharomyces cerevisiae Cdt1 residues 354–604 (GenBank accession no. NP_012580). The alignment was generated by using ClustalW and colored in Jalview (30). (B) Cartoon depicting the two conserved DNA-binding domains in the S. acidocaldarius WhiP ORF. These domains, of the “winged-helix” DNA-binding domain superfamily (SSF46785), were identified by using InterProScan (version 14.0; www.ebi.ac.uk/InterProScan).
Table 2.
Probabilities of the conserved domains in three Sulfolobus species and A. pernix
| Species | N residues | P | C residues | P |
|---|---|---|---|---|
| S. acidocaldarius | 1–95 | 9.9 × 10−6 | 166–233 | 1.1 × 10−2 |
| S. tokodail | 33–101 | 9.4 × 10−6 | 179–246 | 2.4 × 10−3 |
| S. solfataricus | 21–91 | 1.2 × 10−5 | 159–226 | 1.1 × 10−2 |
| A. pernix | 54–172 | 1.9 × 10−7 | 225–292 | 3.5 × 10−3 |
Fig. 5.
The Winged-Helix Initiator Protein (WhiP) binds S. solfataricus oriC3. DNaseI footprinting analysis of WhiP, Cdc6-1, Cdc6-2, and Cdc6-3 interactions with oriC3 is shown. The position of previously described 12-bp inverted repeats (ir) and Sso0866 and Sso0867 ORFs are indicated at the side of the panel. (A) 0, 85, 100, and 130 nM WhiP on the oriC3 upper strand. (B and C) Cooperative effects of WhiP on Cdc6 binding on the oriC3 upper and lower strands, respectively. (B) Lanes 1, 3, 6, and 9: no protein; lanes 2, 4, 7, and 10: 130 nM WhiP, 500 nM Cdc6-1, 250 nM Cdc6-2, and 100 nM Cdc6-3, respectively; lanes 5, 8, and 11: 130 nM WhiP plus 500 nM Cdc6-1, 250 nM Cdc6-2, or 100 nM Cdc6-3, respectively. (C) Lanes 1, 4, 7, and 10: no protein; lanes 2, 3, 5, 8, and 11: 125 nM WhiP, 150 nM WhiP, 1,000 nM Cdc6-1, 200 nM Cdc6-2, and 75 nM Cdc6-3, respectively; lanes 6, 9, and 12: 150 nM WhiP plus 1,000 nM Cdc6-1, 200 nM Cdc6-2, or 75 nM Cdc6-3, respectively. (D) Cartoon summarizing the footprinting at oriC3. The open boxes labeled M denote regions of modification rather than discrete footprints.
Thus, we have revealed that a copG plasmid copy number control homologue is closely associated with oriC3, and in addition we have identified an archaeal origin associated protein, WhiP. This previously unrecognized protein binds replication origins and is reminiscent of the bacterial plasmid initiator, RepA. Furthermore, a number of genes, which may play roles in stress responses, lie in the vicinity of these two plasmid-derived genes (Fig. 3 and Table 1). In this regard, it may be significant that the megaplasmid of the hyperthermophilic bacterium Thermus thermophilus possesses a number of genes that are thought to confer growth advantages in thermogenic environments (24). We therefore suggest that this portion of the Sulfolobus genome, including the copG, whiP, stress-related genes, and also the origin itself, was introduced by an extrachromosomal element. Such an event could have conveyed a selective advantage for cellular proliferation in adverse environments by both supplying additional stress response genes and reducing the time taken to replicate the host chromosome.
It has been theorized that, in all three domains of life, some DNA informational proteins (those involved in replication, recombination, repair and transcription) may have originated from a plasmid or virus. Indeed, it has been suggested that functional analogues of DNA informational proteins derived from extrachromosomal elements could be responsible for the numerous nonorthologous gene displacements observed in a variety of organisms (25). It has even been proposed that the cellular DNA genomes of the eukaryal, archaeal, and bacterial progenitors themselves may have originated from a viral source (26, 27). We have provided evidence that the genomic region encompassing Sulfolobus oriC3 has been captured from a virus or a viral/plasmid hybrid. Could extrachromosomal element capture also have played a role in the evolution of multiple initiation sites in the eukaryotic domain? Certainly, horizontal gene transfer has been confirmed as a significant evolutionary mechanism in unicellular eukaryotes (28). Further evidence for horizontal gene transfer in eukaryotic cells is provided by the yeast plasmid 2μ circle, a naturally occurring element present in most Saccharomyces strains. The 2μ initiation site is structurally reminiscent of the phage λ origin of replication, but also encompasses the ARS motifs conserved at all S. cerevisiae origins (29). It therefore seems likely that this plasmid was derived following the interaction of a viral element with the yeast genome. Perhaps similar interactions between the genomes of ancient eukaryotes and extrachromosomal elements may have contributed to the development of the multiplicity of replication origins that we observe today.
Materials and Methods
Neutral/Neutral 2D Agarose Gel Electrophoresis.
Asynchronous A. pernix cultures were grown under aerobic conditions at 90°C on a magnetically stirred heating block (Stuart CB302) in Difco Marine Broth 2216 (BD Biosciences, San Jose, CA), supplemented with 4 mM sodium thiosulfate. Cells at early/mid-log phase (OD600 of 0.3), with a doubling time of 4 h, were harvested by centrifugation (7,000 × g for 10 min at 4°C), and washed and suspended in chilled 0.5 M NaCl/50 mM Tris (pH 7.4) to an OD600 of 600. Genomic plugs were made by immobilizing the cells in an equal volume of 0.8% low melting point agarose (Biogene, Cambridge, MA) and poured into plug molds (Bio-Rad, Hercules, CA). The treatment of the genomic DNA within agarose plugs and 2D gel analysis was performed as described (7).
DNaseI Footprinting and Purification of the WhiP and Cdc6 Proteins.
DNaseI footprinting assays were performed as described (7). The ORF of WhiP was amplified by PCR with primers that introduced restriction sites for NdeI and XhoI at the start and stop codons, respectively. The gene was cloned into the pET30a expression vector (Novagen, Madison, WI). The resultant plasmid encoded the WhiP protein fused to a hexahistidine tag. The WhiP and Cdc6 proteins were purified as described (7).
Supplementary Material
Abbreviations
- UCM
uncharacterized motif
- wHTH
winged helix–turn–helix.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700206104/DC1.
References
- 1.Kelman Z, White MF. Curr Opin Microbiol. 2005;8:669–676. doi: 10.1016/j.mib.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Barry ER, Bell SD. Microbiol Mol Biol Rev. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myllykallio H, Lopez P, Lopez-Garcia P, Heilig R, Saurin W, Zivanovic Y, Philippe H, Forterre P. Science. 2000;288:2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- 4.Berquist BR, DasSarma S. J Bacteriol. 2003;185:5959–5966. doi: 10.1128/JB.185.20.5959-5966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson NP, Bell SD. FEBS J. 2005;272:3757–3766. doi: 10.1111/j.1742-4658.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- 6.Bell SP, Dutta A. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 7.Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 8.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. Proc Natl Acad Sci USA. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson NP, Blood KA, McCallum SA, Edwards PAW, Bell SD. EMBO J. 2007 doi: 10.1038/sj.emboj.7601529. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tye BK. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 11.Grainge I, Gaudier M, Schuwirth BS, Westcott SL, Sandall J, Atanassova N, Wigley DB. J Mol Biol. 2006;363:355–369. doi: 10.1016/j.jmb.2006.07.076. [DOI] [PubMed] [Google Scholar]
- 12.Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Ankai A, Kosugi H, et al. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- 13.Zhang R, Zhang CT. Archaea. 2005;1:335–346. doi: 10.1155/2005/509646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brugger K, Chen L, Stark M, Zibet A, Redder P, Ruepp A, Awayez MJ, She Q, Garrett RA, Klenk H-P. Archaea. 2007;2:127–135. doi: 10.1155/2007/745987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiter WD, Palm P, Yeats S. Nucleic Acids Res. 1989;17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng X, Holz I, Zillig W, Garrett RA, She Q. J Mol Biol. 2000;303:449–454. doi: 10.1006/jmbi.2000.4160. [DOI] [PubMed] [Google Scholar]
- 17.She Q, Peng X, Zillig W, Garrett RA. Nature. 2001;409:478. doi: 10.1038/35054138. [DOI] [PubMed] [Google Scholar]
- 18.Kawarabayasi Y, Hino Y, Horikawa H, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, Kosugi H, Hosoyama A, et al. DNA Res. 2001;8:123–140. doi: 10.1093/dnares/8.4.123. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Brugger K, Skovgaard M, Redder P, She Q, Torarinsson E, Greve B, Awayez M, Zibat A, Klenk H-P, Garrett RA. J Bacteriol. 2005;187:4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brugger K, Torarinsson E, Redder P, Chen L, Garrett RA. Biochem Soc Trans. 2004;32:179–183. doi: 10.1042/bst0320179. [DOI] [PubMed] [Google Scholar]
- 21.Giraldo R, Diaz-Orejas R. Proc Natl Acad Sci USA. 2001;98:4938–4943. doi: 10.1073/pnas.081079298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komori H, Matsunaga F, Higuchi Y, Ishiai M, Wada C, Miki K. EMBO J. 1999;18:4597–4607. doi: 10.1093/emboj/18.17.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer LM, Aravind L. In: DNA Replication and Human Disease. DePamphilis ML, editor. New York: CSH Press; 2006. pp. 751–757. [Google Scholar]
- 24.Bruggemann H, Chen C. J Biotechnol. 2006;124:654–661. doi: 10.1016/j.jbiotec.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 25.Forterre P. Mol Microbiol. 1999;33:457–465. doi: 10.1046/j.1365-2958.1999.01497.x. [DOI] [PubMed] [Google Scholar]
- 26.Forterre P. Proc Natl Acad Sci USA. 2006;103:3669–3674. doi: 10.1073/pnas.0510333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmer C. Science. 2006;312:870–872. doi: 10.1126/science.312.5775.870. [DOI] [PubMed] [Google Scholar]
- 28.Andersson JO, Sjogren AM, Davis LA, Embley TM, Roger AJ. Curr Biol. 2003;13:94–104. doi: 10.1016/s0960-9822(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 29.Broach JR. Cell. 1982;28:203–204. doi: 10.1016/0092-8674(82)90337-3. [DOI] [PubMed] [Google Scholar]
- 30.Clamp N, Cuff JA, Searle SM, Barton GJ. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.