Abstract
The molecular mechanism and significance of endocytic processes involved in directional axon elongation are not well understood. The Unc-51 family of serine/threonine kinases was shown to be important for axon growth and was also linked to endocytosis, providing an entry point to study this problem. We found that mouse Unc-51-like kinase 1/2 (Ulk1/2) proteins are localized to vesicular structures in growth cones of mouse spinal sensory neurons. RNAi-mediated knockdown of Ulk1 and/or Ulk2 resulted in impaired endocytosis of nerve growth factor (NGF), excessive axon arborization, and severely stunted axon elongation. The evidence also indicates that Ulk1/2 mediates a non-clathrin-coated endocytosis in sensory growth cones. Interestingly, NGF can induce the interaction of Ulk1 with TrkA receptor complexes through promoting K63-polyubiquitination of Ulk1 and binding of Ulk1 to the scaffolding protein p62. These results and additional studies suggest that Ulk1/2 proteins regulate filopodia extension and neurite branching during sensory axon outgrowth, probably through regulating TrkA receptor trafficking and signaling.
Keywords: axon growth, dorsal root ganglion neurons, endocytosis, p62
Endocytic processes play an active role during polarized axon elongation and growth cone navigation (1–7). However, the exact molecular mechanisms linking endocytosis to the signaling events downstream of growth factor-stimulated axon extensions are largely unknown. In the case of neurotrophin-induced axon outgrowth, many lines of evidence support the formation of signaling endosomes that are retrogradely transported back to cell bodies to promote neuronal survival (8, 9). However, little is known about whether (or how) endocytic processes play a role in local axon growth signaling in response to neurotrophins.
The Unc51/Ulk1 (for Unc51-like-kinase 1) family of serine/threonine kinases participate in a phylogenetically conserved pathway involving both axon growth and endocytosis (10–13). In Caenorhabditis elegans, mutation in the Unc51 gene resulted in stalled axon outgrowth, increased axon numbers (all short and stunted), and abnormal accumulation of intracellular membranous structures (10, 14). In mammals, Ulk1 was shown to be important for axon formation in cerebellar granule neurons (12, 13). Furthermore, a yeast two-hybrid screen identified SynGAP and syntenin as binding partners for Ulk1 and Ulk2 proteins in cerebellar granule neurons (13). Both molecules are modulators of the Rab5-mediated endocytic pathway, indicating a link between endocytosis and axon growth in these neurons. Interestingly, we previously found that Ulk1 can interact with p62, a molecule required for internalization of TrkA as well as for nerve growth factor (NGF)-induced neurite outgrowth in PC12 cells in an in vitro binding assay (15–17). These observations suggest the possible involvement of Ulk1/2-mediated endocytosis in regulating NGF-induced neurite outgrowth. We tested this hypothesis by studying the expression and localization of the Ulk1 and Ulk2 proteins in mouse embryonic sensory neurons, the phenotypes caused by the loss-of-function of Ulk1/2 in NGF-induced sensory axon outgrowth, and the possible mechanism by which p62 recruits Ulk1 to the NGF receptor TrkA to regulate TrkA/NGF signaling.
Results
Ulk1 and Ulk2 Proteins Are Expressed in Sensory Neurons and Are Present in Growth Cones.
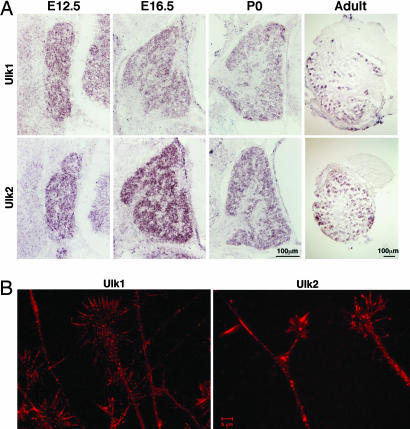
In situ hybridization experiments revealed that both mouse homologs of the Unc-51-like family gene, Ulk1 and Ulk2, are expressed in all dorsal root ganglion (DRG) neurons throughout development, and in a subset of neurons in adult DRG (Fig. 1A). Antibody staining on cultured embryonic DRG neurons showed that both Ulk1 and Ulk2 have punctuated, presumably vesicular distribution patterns along the axon shaft and within growth cones, especially in the center of growth cones and at branching points (Fig. 1B). In addition, both proteins are also present in filopodia with Ulk2 showing a more prominent signal. Ultracentrifugation fractionation experiments revealed that ≈50% of Ulk1 and Ulk2 proteins are associated with the membrane fraction (data not shown). These results suggest the possible involvement of Ulk1/2 in the growth and morphogenesis of sensory axons.
Fig. 1.
Expression and localization of Ulk1 and Ulk2 in mouse sensory neurons. (A) In situ hybridization experiments show the expression of Ulk1 (Upper) and Ulk2 (Lower) genes in mouse DRG neurons at four different stages: E12.5, E16.5, postpartum day 0 (P0), and adult. (B) Antibody staining reveals that Ulk1 and Ulk2 are present in sensory axons and growth cones and often localize to punctuated structures (presumably vesicles).
RNAi-Mediated Knockdown of Ulk1 and Ulk2 Reduces Axonal Length and Increases Axonal Branching in Embryonic Sensory Neurons.
To gain insight into the physiological functions of the Ulk1/2 proteins, we performed RNAi analyses in sensory neurons. We generated constructs based on the pSUPER vector system (18) to express shRNA directed against Ulk1 or Ulk2. All constructs were first tested in COS cells for efficiency and specificity in knockdown of their target proteins [supporting information (SI) Fig. 6]. This knockdown effect was then confirmed with antibody staining on RNAi-expressing DRG neurons (SI Fig. 7).
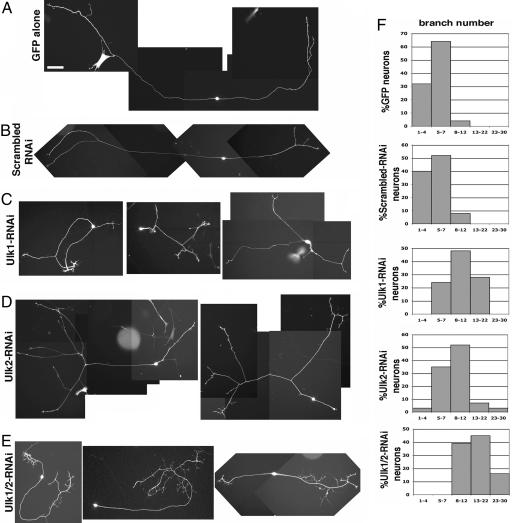
Embryonic DRG neurons were transfected with one of the following five plasmid combinations: (i) a plasmid expressing GFP (EGFP driven by the β-actin promoter), (ii) GFP plus a scrambled-RNAi plasmid, (iii) GFP plus an Ulk1-RNAi plasmid, (iv) GFP plus an Ulk2-RNAi plasmid, and (v) GFP plus both Ulk1-RNAi and Ulk2-RNAi plasmids. Representative images after 60 h in culture are shown in Fig. 2. Control neurons (GFP alone or GFP with scrambled-RNAi plasmid) mostly grew in a bipolar mode with two main axons that extend over a long distance. In contrast, reducing Ulk1 and/or Ulk2 in DRG neurons led to an increased number of axon branches and/or an exuberant number of long filopodia, and significantly shortened axon elongations (Fig. 2 and Table 1). Quantitative analyses of these phenotypes are described below and summarized in Table 1.
Fig. 2.
RNAi-mediated knockdown of Ulk1/2 caused exuberant axon branching in embryonic sensory neurons. Different pSUPER-based constructs were cotransfected with EGFP into E12.5 DRG neurons and cultured for 60 h. Representative images are shown here. All pictures are taken with the same magnification. (Scale bar: 100 μm.) (A) A neuron transfected with GFP alone shows bipolar morphology with two main long axons and two terminal branches. (B) A neuron transfected with scrambled-RNAi plus GFP plasmid shows bipolar morphology with two long axons and limited terminal branching. (C) Three Ulk1-RNAi plus GFP-transfected neurons all have multiple branches and shorter axons. (D) Two Ulk2-RNAi plus GFP-transfected neurons grow multiple branches. (E) Three neurons transfected with both Ulk1-RNAi and Ulk2-RNAi, as well as GFP, all have complex axon arborizations. (F) Percentage of neurons with different numbers of total axon branches. In each histogram, the y axis is the percentage of neurons; the x axis is the number of total branches grouped into five columns: 1–4, 5–7, 8–12, 13–22, and 23–30.
Table 1.
Quantitative analyses of axon outgrowth and branching phenotype
| Transfection | N* | Average length of the longest axon, μm | Average no. of total axon branches per neuron† | % neurons with quaternary arbors |
|---|---|---|---|---|
| GFP | 25 | 1,513 ± 69 | 5.1 ± 0.3 | 0 |
| Scrambled-RNAi | 25 | 1,435 ± 81 (P > 0.27) | 5.2 ± 0.4 (P > 0.93) | 0 |
| Ulk1-RNAi | 29 | 726 ± 45 (P < 0.001) | 10.9 ± 0.8 (P < 0.001) | 34 |
| Ulk2-RNAi | 29 | 906 ± 41 (P < 0.001) | 8.9 ± 0.8 (P < 0.001) | 31 |
| Ulk1/Ulk2-RNAi | 31 | 658 ± 35 (P < 0.001) | 15.5 ± 0.9 (P < 0.001) | 93 |
*N, number of neurons analyzed.
†Fibers longer than 20 μm are counted as axon branches. Primary axon arbor is defined as the main axon growing out from the cell body, secondary arbors are those sprouted from the primary axons, and so on. Thus, quaternary arbors are those that grew out from tertiary arbors.
Axon Branch Numbers.
Fibers longer than 20 μm were all counted as branches. Compared with control neurons, reducing either Ulk1 or Ulk2 nearly doubled (2×) the average branch number per neuron, whereas knockdown of both proteins tripled (3×) the number of axon branches (Table 1). The histogram of the branch number distribution for each transfection category is shown in Fig. 2F. More than 90% of control neurons had <7 axon branches total. In comparison, all of the Ulk1/2 double RNAi-expressing neurons had 8 or more branches, and >60% had 13 or more branches. In the individual RNAi-expressing neurons, there was also an increase in the percentage of neurons with >8 axon branches.
Complexity of Branching Patterns.
GFP or control-RNAi-transfected neurons had no quaternary arbors (the primary branches are defined as the axons coming directly from the cell body), whereas approximately one-third of Ulk1-RNAi or Ulk2-RNAi-expressing neurons, and almost all Ulk1/2 double-RNAi-expressing neurons possessed complex branching structures with quaternary or even higher order arbors (Fig. 2E and Table 1). This excessive arborization indicates that Ulk1 and Ulk2 play synergistic roles in preventing the formation and/or stabilization of higher order filopodia (Table 1).
Axon Length.
We also measured the average length of the longest axon for each of the transfection categories. The longest axons in Ulk1/2-double-RNAi-expressing neurons were less than half the length of those of the controls (Table 1, third column). Taken together, our data suggest that loss of function of Ulk1/2 leads to shortened axonal elongation and increased branching in cultured sensory neurons.
The Effect of Reducing Ulk1/2 on NGF Internalization into the Growth Cones.
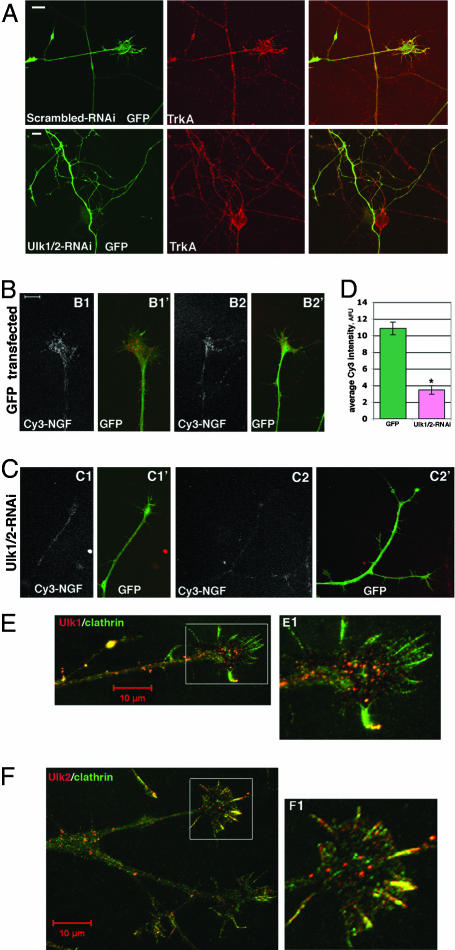
We next explored the possible mechanisms underlying the axon morphology changes that resulted from suppressing Ulk1/2 activity. Based on previous findings demonstrating that Ulk1/2 affects endocytic processes (11, 13) and that NGF triggers the endocytosis of TrkA receptor (8), it is possible that Ulk1/2 may participate in TrkA receptor-mediated NGF endocytosis in sensory neurons. To test this possibility, we used a Cy3-conjugated NGF-based endocytosis assay as described by Tani et al. (19). Neurons transfected with GFP alone showed clear Cy3-fluorescent signals within their growth cones, indicating efficient internalization of NGF (Fig. 3B). However, neurons expressing Ulk1/2-RNAi constructs had a faint Cy3-NGF signal (Fig. 3C). Quantitative analyses revealed that the average Cy3-intensity in growth cones of GFP-transfected neurons was 10.9 ± 0.8 arbitrary fluorescence units (AFU; n = 11), whereas that for Ulk1/2-double-RNAi-transfected neurons was 3.5 ± 0.5 AFU (n = 11, Fig. 3D). Thus reducing the expression of Ulk1/2 reduced the endocytosis of NGF (and by implication, TrkA) in sensory growth cones. This reduction is not due to the lack of cell surface expression of TrkA. Ulk1/2-RNAi-expressing neurons had similar levels of surface TrkA staining as controls, even on surfaces associated with highly branched termini (Fig. 3A and data not shown). Therefore, all neurons were equally capable of binding to NGF.
Fig. 3.
Ulk1/2 may mediate a non-clathrin-coated vesicle endocytosis to regulate NGF internalization. (A) DRG were transfected with constructs as indicated on the figures and cultured for 60 h. Cells were fixed and stained with anti-TrkA under nonpermeabilizing (surface TrkA) conditions. TrkA was equally expressed on the surface of axons and growth cones in both control and Ulk1/2-RNAi-transfected neurons and on the numerous arbors of the Ulk1/2-RNAi-expressing neurons. (Scale bar: 10 μm.) (B) Representative images of Cy3-NGF internalization into the growth cones of GFP-expressing neurons. B1 and B2 show Cy3-images alone; B1′ and B2′ are merged images of Cy3 and GFP. (C) Representative images of Cy3-NGF internalization into the growth cones of Ulk1/2-RNAi-expressing neurons. C1 and C2 show Cy3-images alone; C1′ and C2′ are merged images of Cy3 and GFP. Pictures in B and C are at the same magnification. (Scale bar: 10 μm.) (D) Quantitative analyses of Cy3-NGF internalization into growth cones. The y axis is the average Cy3 intensity using AFU. The signal in GFP-transfected neurons was 10.9 ± 0.8 AFU (n = 11); in Ulk1/2-RNAi-expressing neurons, it was 3.5 ± 0.5 AFU (n = 11, P < 0.001). (E) Two-color immunofluorescence staining of anti-Ulk1 (red) and anti-clathrin (green) heavy chain on a DRG growth cone. (E1) Boxed area is shown enlarged. Most green staining does not colocalize with red staining. (F) Two-color immunofluorescence staining of anti-Ulk2 (red) and anti-clathrin (green) heavy chain. A DRG growth cone is shown. (F1) Boxed area is shown enlarged. Most green staining does not colocalize with red staining, although several filopodia seem to be “yellow” (indicating partial colocalization of Ulk2 and clathrin).
Ulk1/2 May Mediate a Non-Clathrin-Coated Endocytosis.
To further characterize Ulk1/2-mediated endocytosis processes in sensory neurons, we performed two-color immunofluorescence staining of both Ulk1/2 and the clathrin heavy chain. Interestingly, the puncta of clathrin do not colocalize with the puncta of Ulk1 or Ulk2 proteins in the sensory growth cones (Fig. 3 E and F), although some colocalization is observed in filopodia. This result suggests that Ulk1/2 proteins mediate a non-clathrin-coated endocytic process in sensory neurons. To directly test the possibility that the clathrin-coated vesicle (CVV)-mediated endocytic process in Ulk1/2 knockdown neurons is not affected, we assayed the ability of neurons to take up fluorescent-labeled transferrin, which is known to enter the cell specifically through CVV. Internalization of transferrin can be clearly seen in both control and in Ulk1/2 knockdown neurons, and statistical analyses of the average fluorescence intensity revealed no significant differences (SI Fig. 8). These data support the idea that CVV endocytosis is largely intact in the absence of Ulk1/2 proteins, and is also consistent with our finding that total endocytosis of NGF was reduced but not abolished in Ulk1/2 knockdown neurons (Fig. 3D).
NGF Controls Polyubiquitination of Ulk1 and Interaction of Ulk1 with the NGF Signaling Complex.
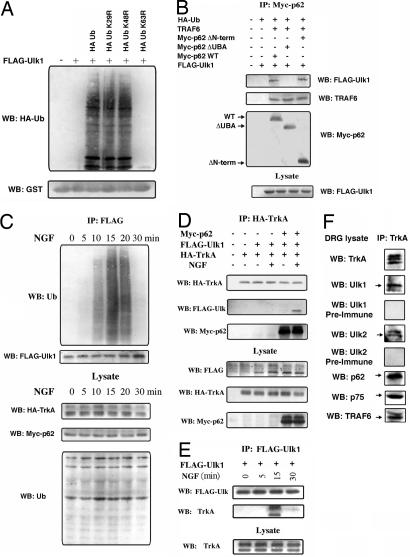
We next examined the possible biochemical basis of Ulk1/2 cross-talk with the NGF/TrkA signaling pathway. It is known that NGF recruits p62, which is required for the internalization of NGF/TrkA signaling complexes (15, 16). We previously demonstrated a possible interaction between the UBA domain (ubiquitin association domain) of p62 (p62-UBA) and polyubiquitinated-Ulk1 (20). Thus, p62 can potentially recruit Ulk1/2 to the TrkA complex. Using a GST pull-down assay, we confirmed that GST-p62UBA can directly pull down polyUb-Ulk1 (Fig. 4A). It is known that the UBA domain of p62 selectively interacts with K63-chains of polyubiquitin. Thus expressing the mutant ubiquitin, K63R-Ub, eliminated the ability of Ulk1 to interact with the p62-UBA domain (Fig. 4A). The identities of the endogenous E3 ligases for catalyzing the K63-polyubiquitination of Ulk1 are not completely known, although we found that TRAF6 protein can catalyze this reaction in vitro (SI Fig. 9). We also tested the interaction between K63-polyubiquitinated Ulk1 and p62-UBA in HEK cells by cotransfecting Ulk1, TRAF6 (to increase ubiquitination), and various forms of p62, and performed coimmunoprecipitation experiments from the cell lysate. Ulk1 protein interacts with WT or N-terminal-deleted p62 (lanes 3 and 5 in Fig. 4B), but not with p62 where the UBA-domain is deleted (lane 4 in Fig. 4B). Consistent with previous findings, TRAF6 itself interacts with p62 in a manner that is independent of the p62-UBA domain (21).
Fig. 4.
NGF stimulates ubiquitination of Ulk1 and recruitment of Ulk1 to the TrkA receptor complex. (A) K63-ubiquitinated Ulk1 interacts with p62-UBA domain in GST pull-down assay. HEK cells coexpressing FLAG-Ulk1 along with ubiquitin or its mutants were used for a p62 UBA-domain pull-down assay. The interactions were analyzed by immunoblotting with anti-HA (Ub) and GST tags. (B) Ulk1 interacts with the UBA domain of p62. Subconfluent cultures of HEK 293 cells were cotransfected as indicated. Cell lysates were immunoprecipitated with anti-myc followed by Western blotting (WB) with antibody to FLAG epitope, TRAF6, and myc to detect interaction. The lysates were Western blotted (WB) with anti-FLAG to verify the expression levels of Ulk1. (C) Time course of NGF-stimulated polyubiquitination of Ulk1. HEK cells were transfected with FLAG-tagged-Ulk1, HA-TrkA, myc-p62, and HA-Ub. Forty-eight hours after transfection, the cells were stimulated with 50 ng/ml NGF for different times (0, 5, 10, 15, 20, and 30 min) and lysed, and ubiquitination was determined by immunoprecipitation of the cell lysates with anti-FLAG antibody followed by Western blot (WB) analysis with anti-ubiquitin and FLAG. As a control, a fraction of the lysate was blotted with anti-HA and myc to check for the expression of TrkA, p62, and Ub. (D) Interaction of Ulk1 with TrkA through p62. HEK cells were transfected as indicated, stimulated with or without NGF (50 ng/ml) for 15 min followed by immunoprecipitation of HA-TrkA and Western blot (WB) with HA (TrkA), FLAG (Ulk1), and myc (p62) antibodies. The lysates were also blotted with antibody to FLAG (Ulk1), HA (TrkA), and myc (p62). Ulk1 was coimmunoprecipitated with TrkA only when p62 was present and also required NGF stimulation. (E) TrkA coimmunoprecipitated with Ulk1 in an NGF-stimulated and time-dependent manner (15 min after stimulation). (F) Ulk1 and Ulk2 can be coimmunoprecipitated with endogenous TrkA from DRG neurons, as can p62, TRAF6, and p75. Preimmune sera of anti-Ulk1 and anti-Ulk2 did not detect any bands on the same blot.
We next asked whether NGF treatment could induce ubiquitination of Ulk1 and recruitment of polyubiquitinated-Ulk1 to the TrkA receptor complex. We transfected HEK 293 cells with FLAG-Ulk1, TrkA, p62 and HA-Ub. Interestingly, the presence of TrkA significantly reduced the basal level ubiquitination of FLAG-Ulk1. This allowed us to test the effect of adding NGF directly to the culture. The transfected cells were stimulated with NGF for various times. Ulk1 was maximally ubiquitinated within 15 min after addition of 50 ng/ml NGF and the ubiquitination disappeared after 30 min (Fig. 4C). As expected, overexpression of the K63R ubiquitin mutant blocked the polyubiquitination of Ulk1, whereas the K29R and K48R mutants did not (data not shown).
To examine whether and when the polyubiquitinated-Ulk1 (polyUb-Ulk1) was recruited to the TrkA receptor complex, HEK cells were cotransfected with HA-TrkA or FLAG-Ulk1, with or without myc-p62. Upon NGF stimulation for 15 min, and only in the presence of p62, Ulk1 was coimmunoprecipitated with TrkA (IP with anti-TrkA antibody) (Fig. 4 D and E). These results demonstrate that NGF induced the transient formation of a large complex containing TrkA, p62, and polyUb-Ulk1.
To further substantiate these biochemical interactions in sensory neurons, we performed coimmunoprecipitation with anti-TrkA antibody from fresh isolated embryonic day (E) 12.5 DRG. Both Ulk1 and Ulk2 were coimmunoprecipitated with the TrkA receptor (Fig. 4F). Other proteins known to interact with TrkA were also present in the co-IP products, particularly, p62, p75 and TRAF6 (Fig. 4F). This experiment provided evidence for in vivo interaction of Ulk1/2 with the TrkA receptor complex.
Enhanced Phosphorylated-Akt Signal in Ulk1/2 Knockdown Neurons.
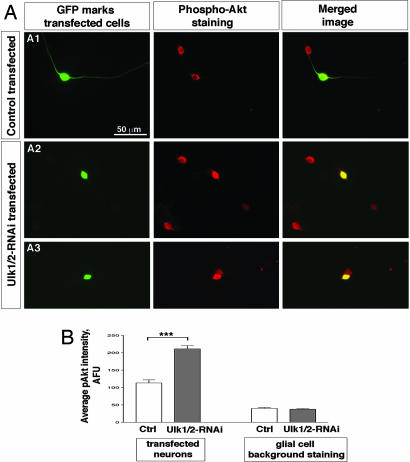
We propose that the association of Ulk1/2 with TrkA receptor and the Ulk1/2-mediated non-CVV endocytosis attenuate/terminate TrkA signaling, and reducing Ulk1/2 results in elevated TrkA signaling. In sensory neurons, NGF/TrkA activates the PI3-kinase (PI3K) signaling pathway and Akt is a major downstream target of PI3K. Our observed phenotype in Ulk1/2 knockdown neurons resembled the phenotype induced by overexpressing an constitutively active form of Akt, which was also found to increase the extent of axon branching (22). We compared the level of phosphorylated-Akt (pAkt) in control and in Ulk1/2 knockdown neurons. A moderate pAkt signal is observed in GFP (or scrambled-RNAi-transfected) neurons (Fig. 5A1 and data not shown), whereas a much brighter pAkt signal is detected in Ulk1/2-RNAi-transfected neurons (Fig. 5 A2 and A3). Quantification of the average fluorescence intensity (arbitrary unit, AFU) revealed approximately a 2-fold increase of the pAkt signal in Ulk1/2 knockdown neurons compared with that in controls (Fig. 5B): 211.6 ± 8.9 AFU in Ulk1/2-RNAi neurons (n = 20) versus 113.3 ± 8.8 AFU in GFP plasmid-transfected neurons (n = 22, P < 0.001, Fig. 5B). The pAkt intensity in scrambled-RNAi-transfected neurons was 95.6 ± 8.4 AFU (n = 25). As an additional control for our parallel staining procedure, we quantified the weak pAkt staining in glial cells growing on the same cover slips as the transfected neurons and found no statistically significant differences between the Ulk1/2-RNAi-transfected cultures and the control cultures (Fig. 5B). These results were repeated in three independent sets of experiments. Thus we conclude that reducing the level of Ulk1/2 proteins resulted in enhanced activation of Akt in sensory neurons, probably because of prolonged TrkA signaling (see Discussion).
Fig. 5.
Increased phosphorylated Akt signal in Ulk1/2 knockdown neurons. (A) pAkt staining. (A1) Representative image of pAkt staining in cultures containing GFP-transfected neurons. In the same image, a transfected (GFP-positive) and a nontransfected (GFP-negative) neuron can be seen simultaneously, and the pAkt signals appear similar in both cells. (A2 and A3) Representative image of pAkt staining in cultures containing Ulk1/2-RNAi-transfected neurons. In both images, the transfected neuron (GFP-positive) seems to have a stronger pAkt signal than the neighboring untransfected cells. (B) Quantitative analyses of the pAkt staining intensity (AFU) averaged over the area of the cell body. The average signal in GFP-transfected neurons was 113.3 ± 8.8 AFU (n = 22); in Ulk1/2-RNAi-expressing neurons it was 211.6 ± 8.9 AFU (n = 20, P < 0.001). The background staining in glia cells was 40.3 ± 2.7 AFU for GFP-transfected cultures (n = 10) and 37.7 ± 2.2 AFU (n = 12, P > 0.4) for Ulk1/2-RNAi-transfected cultures.
Discussion
We demonstrated that Ulk1/2 proteins are required for efficient endocytosis of NGF and for suppressing excessive filopodia extension and axon branching in sensory neurons during NGF-induced axon outgrowth. How might Ulk1/2 exert their functions? In embryonic DRG neurons, it is known that NGF stimulates filopodia extension and the advancing growth cone by binding to TrkA receptors and activating various intracellular signaling pathways. In this study, we found that NGF also induces K63-polyubiquitination of Ulk1 (and perhaps Ulk2), where TRAF6 is a potential candidate for the E3 ligase. This K63-polyubiquitination allows the binding of Ulk1/2 to the p62 protein via its UBA-domain. The binding of p62 with activated TrkA recruits Ulk1/2 to the active TrkA complex. Previous work demonstrated that Ulk1/2 interacts with syntenin and SynGAP, proteins that regulate endocytosis (13). We propose that these interactions allow Ulk1/2 to traffic the NGF-bound TrkA receptors into an endocytic compartment that attenuates NGF signaling, thereby allowing filopodia to withdraw even in the continued presence of NGF. This model predicts that reducing the activities of Ulk1/2 will lead to prolonged TrkA signaling and impaired filopodia retraction, which is exactly what we observed in Ulk1/2 knockdown neurons, i.e., increased activation/phosphorylation of Akt and an excessive number of filopodia and axon branches. These data support the functional involvement of a phylogenetically conserved Ulk1/2-mediated endocytic pathway in modulating the signaling of neurotrophins in sensory neurons.
It was known that active TrkA receptors can also be endocytosed via CVV into signaling endosomes that continue to transduce the NGF signal in a retrograde manner back to cell bodies in DRG neurons (23, 24). Recently, Pincher-mediated macroendocytosis of TrkA and the formation of multivesicular bodies were shown to be the primary pathway responsible for retrograde neurotrophin signaling in sympathetic neurons (25). Therefore, endocytosis per se does not necessarily result in termination of active TrkA signaling. It is interesting to note that Ulk1/2 puncta, presumably representing the Ulk1/2-associated endocytic vesicles, do not colocalize with clathrin in the growth cone (Fig. 3 E and F). Thus the segregation of Ulk1/2-mediated endocytosis from the clathrin-mediated pathway may allow sensory neurons to use the Ulk1/2 pathway for internalization of TrkA into signaling-inactive endosomes and either recycles the TrkA back to the cell surface or directs it to the degradation pathway, while using the clathrin pathway for the formation of retrograde signaling endosomes. Future work is needed to further characterize the molecular and cellular nature of the Ulk1/2–mediated endocytic pathway, as well as the fates of the Ulk1/2-mediated endocytosed vesicles and the internalized TrkA receptors (i.e., degraded or recycled). Suffice it to say, our study does highlight the importance of endocytic processes in regulating diverse signaling events during axon elongation and neuronal morphogenesis.
Materials and Methods
Molecular Biology.
Ulk1 and Ulk2 cDNA were cloned by RT-PCR (using total RNA prepared from E12.5 DRG neurons) into the pCAGGS vector that has a chicken β-actin promoter. The hairpin sequences of the oligonucleotides chosen for Ulk1 and Ulk2 knockdown experiments in neurons are 5′-GATCCCCAGACTCCTGTGACACAGATTTCAAGAGAATCTGTGTCAC A G G A GTCTTTTTTA-3′ for Ulk1-RNAi and 5′-GATCCCCTGC-CTAGTATTCCCAGAGATTCAAGAGATC T C T G G G A A T ACTAGGCATTTTTA-3′ for Ulk2-RNAi. The sequence of the scrambled oligonucleotide control is 5′-GATCCCCCATAGCATGCGTATC A T G C TTCAAGAGAGCATGATACGCAT G C T A T GTTTTTA-3′.
Antibodies and Immunofluorescence.
Anti-TrkA antibody was kindly provided by Dr. Louis Reichardt (University of California, San Francisco). Rabbit polyclonal anti-Ulk1 antibody was raised against a peptide with a sequence of SSSPSPSGRPGPFSS. Rabbit polyclonal anti-Ulk2 antibody was raised against a peptide with a sequence of GTSPTKHTGSSPRNS (Covance, Richmond, CA). Anti-clathrin (Covance), anti-phosphorylated Akt (Cell Signaling Technology, Danvers, MA), and secondary antibodies (Invitrogen, Carlsbad, CA) were purchased. Quantification of pAkt staining is described in SI Materials and Methods.
Cy3-NGF and Alexa Fluor 568-Transferrin Internalization Assay and Quantification.
Cy3-conjugation of NGF and measurement of its activity have been described (19). Alexa Fluor 568-Transferrin was purchased from Invitrogen. Detailed procedure for endocytosis assays and quantification can be found in SI Materials and Methods.
Other Methods.
Procedures for neuronal culture and transfection, immunoprecipitation, Western blot, and GST pull-down assay can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Bao-Xia Han, Molly Heyer, and Zhonghua Lu for help with experiments; Dr. Louis Reichardt for anti-TrkA antibody; Dr. Mary Beth Hatten for advice; and Drs. Brigid Hogan, Guoping Feng, Dan Tracey, Michael Ehlers, Vann Bennett, Avraham Yaron, Hiroshi Hasegawa, and, particularly, the late Lawrence Katz for critically reading the manuscript. F.W. is supported by the Whitehall Foundation, the Esther A. and Joseph Klingenstein Fund, an Alfred Sloan Scholarship, and the National Institutes of Health (NIH) (National Institute of Dental and Craniofacial Research Grant DE016550). S.d.S. is supported by the Foundation of Science and Technology Portugal and the Ph.D. program of the University of Coimbra (Coimbra, Portugal). M.W.W. is supported by the NIH (National Institute of Neurological Disorders and Stroke Grant 33661).
Abbreviations
- NGF
nerve growth factor
- DRG
dorsal root ganglion
- Ulk
Unc-51-like kinase
- AFU
arbitrary fluorescence unit
- CVV
clathrin-coated vesicle
- En
embryonic day n.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701402104/DC1.
References
- 1.Cowan CW, Shao YR, Sahin M, Shamah SM, Lin MZ, Greer PL, Gao S, Griffith EC, Brugge JS, Greenberg ME. Neuron. 2005;46:205–217. doi: 10.1016/j.neuron.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 2.Fournier AE, Nakamura F, Kawamoto S, Goshima Y, Kalb RG, Strittmatter SM. J Cell Biol. 2000;149:411–422. doi: 10.1083/jcb.149.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jurney WM, Gallo G, Letourneau PC, McLoon SC. J Neurosci. 2002;22:6019–6028. doi: 10.1523/JNEUROSCI.22-14-06019.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamiguchi H. Mol Neurobiol. 2003;28:219–228. doi: 10.1385/MN:28:3:219. [DOI] [PubMed] [Google Scholar]
- 5.Nishimura T, Fukata Y, Kato K, Yamaguchi T, Matsuura Y, Kamiguchi H, Kaibuchi K. Nat Cell Biol. 2003;5:819–826. doi: 10.1038/ncb1039. [DOI] [PubMed] [Google Scholar]
- 6.Piper M, Salih S, Weinl C, Holt CE, Harris WA. Nat Neurosci. 2005;8:179–186. doi: 10.1038/nn1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmer M, Palmer A, Kohler J, Klein R. Nat Cell Biol. 2003;5:869–878. doi: 10.1038/ncb1045. [DOI] [PubMed] [Google Scholar]
- 8.Ginty DD, Segal RA. Curr Opin Neurobiol. 2002;12:268–274. doi: 10.1016/s0959-4388(02)00326-4. [DOI] [PubMed] [Google Scholar]
- 9.Howe CL, Mobley WC. Curr Opin Neurobiol. 2005;15:40–48. doi: 10.1016/j.conb.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Ogura K, Wicky C, Magnenat L, Tobler H, Mori I, Muller F, Ohshima Y. Genes Dev. 1994;8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- 11.Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M. Nature. 2005;436:78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- 12.Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME. Neuron. 1999;24:833–846. doi: 10.1016/s0896-6273(00)81031-4. [DOI] [PubMed] [Google Scholar]
- 13.Tomoda T, Kim JH, Zhan C, Hatten ME. Genes Dev. 2004;18:541–558. doi: 10.1101/gad.1151204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McIntire SL, Garriga G, White J, Jacobson D, Horvitz HR. Neuron. 1992;8:307–322. doi: 10.1016/0896-6273(92)90297-q. [DOI] [PubMed] [Google Scholar]
- 15.Geetha T, Wooten MW. J Biol Chem. 2003;278:4730–4739. doi: 10.1074/jbc.M208468200. [DOI] [PubMed] [Google Scholar]
- 16.Samuels IS, Seibenhener ML, Neidigh KB, Wooten MW. J Cell Biochem. 2001;82:452–466. doi: 10.1002/jcb.1177. [DOI] [PubMed] [Google Scholar]
- 17.Pridgeon JW, Geetha T, Wooten MW. Biol Proced Online. 2003;5:228–237. doi: 10.1251/bpo66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 19.Tani T, Miyamoto Y, Fujimori KE, Taguchi T, Yanagida T, Sako Y, Harada Y. J Neurosci. 2005;25:2181–2191. doi: 10.1523/JNEUROSCI.4570-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geetha T, Wooten MW. FEBS Lett. 2002;512:19–24. doi: 10.1016/s0014-5793(02)02286-x. [DOI] [PubMed] [Google Scholar]
- 21.Wooten MW, Seibenhener ML, Mamidipudi V, Diaz-Meco MT, Barker PA, Moscat J. J Biol Chem. 2001;276:7709–7712. doi: 10.1074/jbc.C000869200. [DOI] [PubMed] [Google Scholar]
- 22.Markus A, Zhong J, Snider WD. Neuron. 2002;35:65–76. doi: 10.1016/s0896-6273(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 23.Howe CL, Valletta JS, Rusnak AS, Mobley WC. Neuron. 2001;32:801–814. doi: 10.1016/s0896-6273(01)00526-8. [DOI] [PubMed] [Google Scholar]
- 24.Delcroix JD, Valletta JS, Wu C, Hunt SJ, Kowal AS, Mobley WC. Neuron. 2003;39:69–84. doi: 10.1016/s0896-6273(03)00397-0. [DOI] [PubMed] [Google Scholar]
- 25.Valdez G, Akmentin W, Philippidou P, Kuruvilla R, Ginty DD, Halegoua S. J Neurosci. 2005;25:5236–5247. doi: 10.1523/JNEUROSCI.5104-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.