Abstract
Morlin (7-ethoxy-4-methyl chromen-2-one) was discovered in a screen of 20,000 compounds for small molecules that cause altered cell morphology resulting in swollen root phenotype in Arabidopsis. Live-cell imaging of fluorescently labeled cellulose synthase (CESA) and microtubules showed that morlin acts on the cortical microtubules and alters the movement of CESA. Morlin caused a novel syndrome of cytoskeletal defects, characterized by cortical array reorientation and compromised rates of both microtubule elongation and shrinking. Formation of shorter and more bundled microtubules and detachment from the cell membrane resulted when GFP::MAP4-MBP was used to visualize microtubules during morlin treatment. Cytoskeletal effects were accompanied by a reduction in the velocity and redistribution of CESA complexes labeled with YFP::CESA6 at the cell cortex. Morlin caused no inhibition of mouse myoblast, bacterial or fungal cell proliferation at concentrations that inhibit plant cell growth. By contrast, morlin stimulated microtubule disassembly in cultured hippocampal neurons but had no significant effect on cell viability. Thus, morlin appears to be a useful new probe of the mechanisms that regulate microtubule cortical array organization and its functional interaction with CESA.
Keywords: Arabidopsis, cell wall, chemical genetics, cytoskeleton
Cell morphogenesis and diversity of cell shape form the fundamental building blocks for development of multicellular organisms (1). Plant cells are delimited by a rigid cell wall that resists internal turgor pressure but must extend in a controlled and organized manner to allow the cell to grow and acquire a specific shape (2). The major load-bearing constituent of the higher-plant cell wall is cellulose, which forms crystalline polymers that are often highly organized with respect to cell shape and growth pattern (3). Depletion of these organized cellulose microfibrils results in decreased anisotropy of cell wall expansion and an inability to achieve differentiated cell shape (4). The morphogenetic properties of the cell wall depend on cortical microtubule organization and function. Observations of coorientated cortical microtubules and cellulose (5), together with loss of directed cell expansion after disruption of microtubule organization (6), suggested that cortical microtubule organization guides the deposition and orientation of cellulose microfibrils. This hypothesis was tested recently by dynamic visualization of cellulose synthase (CESA) complexes in growing Arabidopsis cells (7). These experiments showed that labeled (YFP::CESA6) synthetic complexes appeared as discreet particles at the plasma membrane that colocalize with and move along linear paths coincident with underlying cortical microtubules (7). Reorganization of the cytoskeleton caused correlated reorganization of CESA complex distribution and trajectories. Although these results supported the microtubule guidance hypothesis for cellulose deposition, the relationship between cytoskeletal organization, cell wall biosynthesis, and cell morphogenesis remains a complex problem with many unanswered questions.
The fundamental mechanisms of cortical microtubule organization remain unknown, as do the mechanisms by which the cytoskeleton directs CESA to the plasma membrane and guides its movement once activated. Furthermore, CESA trajectories retain a high degree of transverse coorientation even in the absence of an intact microtubule cytoskeleton, raising questions about the mechanism of microtubule guidance as it relates to cellulose function and wall growth properties (8). Further development of the molecular toolbox to investigate the interaction between cellulose and microtubules and microtubules and the plasma membrane are needed to address these questions.
Based on the demonstrated utility of small-molecule inhibitors to dissect complex cellular processes (9), we conducted a screen of commercially available chemical libraries for compounds that inhibit cell wall synthesis. From this screen, we recovered a coumarin derivative that we named morlin. Analyses of morlin action, reported here, suggest that its target is required for normal microtubule organization and function and for CESA complex motility.
Results
Identification of Chemical Inhibitors of Cellulose Synthesis.
Inhibition of cellulose synthesis by mutations or inhibitors such as isoxaben, results in decreased anisotropy of cell expansion. Additionally, depending on the severity of inhibition, cells may expand manyfold in diameter relative to controls, resulting in swollen organs. Based on these phenotypes, two chemically diverse small-molecule libraries were visually screened for compounds that caused organ swelling. Arabidopsis seedlings were germinated in 20–50 μM solutions of each compound in 96-well plates. From a total of 20,000 compounds screened, 27 compounds (CBI1–27) that caused root or hypocotyl swelling were selected for further analysis. Compound CBI-21 (7-ethoxy-4-ethyl-chromen-2-one) was selected for detailed investigation. Germination on 5 μM CBI-21 caused severe cell and tissue swelling of the hypocotyl and upper root in both Arabidopsis and switchgrass (Panicum virgatum) seedlings, consistent with the effects expected from weakening of cell walls (Fig. 1A).
Fig. 1.
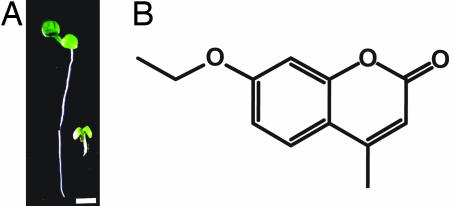
Morlin inhibits plant growth. (A) Arabidopsis seedling grown under continuous light for 5 d on MS agar containing morlin (5 μM) (Right) showed reduced growth compared with control plants (Left), (Scale bar, 2.5 mm.) (B) Morlin, 7-ethoxy-4-methyl chromen-2-one.
To identify the most active analogs of morlin, we conducted a structure-based search using webtools available through ChemMine (10) for commercially available analogs. Of the 19 analogs screened, the most potent inhibitors of plant root growth were 7-ethoxy-4-ethyl chromen-2-one, 7-ethoxy-4-methyl chromen-2-one, dehydrochromen-2-one, and 2H-chromen-2-one, all of which caused inhibition of root growth to <10% of the control at 25 μM [supporting information (SI) Fig. 6]. Several coumarins were found to cause either only a partial root swelling phenotype or no inhibition of seedling root growth (SI Fig. 6). Based on this analysis, we selected 7-ethoxy-4-methyl chromen-2-one for further analysis because it was commercially available in large quantities and highly active in root growth inhibition. We named the compound morlin (Fig. 1B) because, as noted later, it caused a cellular phenotype reminiscent of the mor1 mutation of Arabidopsis (11). The approximate concentration at which morlin inhibited root growth by ≈50% (the RI50) was 1 μM in both switchgrass and Arabidopsis (data not shown).
Inhibition of Cellulose Biosynthesis.
Morlin caused dose-dependent inhibition of cellulose synthesis, with significant inhibition of cellulose deposition observed at 1 μM (SI Fig. 7A). In addition to radial swelling in roots and hypocotyls, at sublethal concentrations (250 nM to 2.5 μM), morlin stimulated a right-handed helical root phenotype on plants grown on vertically oriented agar medium (data not shown). To quantify inhibition of cellulose synthesis, incorporation of [14C]glucose into the cellulose fraction of seedling cell walls was measured after a 2-h incubation period in the label. Seedlings that were preincubated for 1 h in 50 μM morlin exhibited a 23% decrease in [14C]glucose incorporation compared with a DMSO control (SI Fig. 7B). Although Morlin caused a marked decrease in cellulose production, after 2-h exposure to 25 μM, a 65% increase in callose biosynthesis was observed, demonstrating that the UDP-glucose pool was not depleted.
Inhibition of Microtubule and CESA Dynamics.
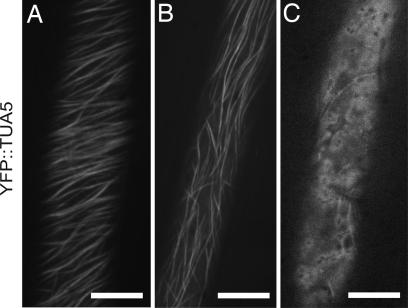
Loss of cell growth anisotropy and reduced cellulose content might be caused by either direct inhibition of cellulose biosynthesis or a failure to deliver and regulate CESA appropriately. To investigate these alternatives, we visualized both microtubules and CESA in living cells. In hypocotyl cells of dark-grown seedlings expressing microtubule markers, YFP::TUA5 (12) or GFP::MAP4 (13), morlin caused marked changes in cytoskeletal organization (Fig. 2–4 and SI Movies 1 and 2). As visualized with YFP::TUA5, the number of cells showing clear transverse-oriented cortical arrays dropped significantly in comparison with mock-treated controls, from 32% to 10% (P < 0.001 binomial test, SI Table 2). The remaining cells in both treatments had either longitudinal or disordered arrays. YFP::TUA5-labeled arrays in morlin-treated cells also appeared more sparse, with labeled microtubules occupying 23% less area in confocal images (P < 0.01, Student's t test, eight cells in seven plants for each treatment, see Methods for details of image analysis). The effect of morlin on cortical array organization was distinct from that of oryzalin; even at 50 μM morlin, the array was noticeably more intact than was the case after treatment with 20 μM oryzalin, a drug that binds tubulin dimers and interferes with dimer addition to polymer ends (Fig. 2C). YFP::TUA5 data were consistent with tubulin immunofluorescence studies in which intact microtubules were no longer detected in morlin-treated cells, and anti-tubulin labeling was primarily distributed in punctate beads at the cell cortex (SI Fig. 8 A and B).
Fig. 2.
Effect of morlin on microtubule organization. Confocal images of cortical microtubules labeled with YFP::TUA5 in hypocotyl cells of 3-d etiolated Arabidopsis seedlings. (A) DMSO control. (B) Morlin (50 μM; 2 h). (C) Addition of oryzalin (20 μM) for 2 h cleared most microtubules from the cortex. (Scale bars,10 μm.)
Fig. 4.
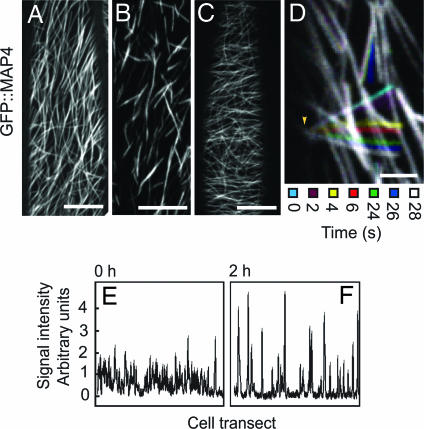
Microtubules show increased bundling and deattachment from the plasma membrane in GFP::MAP4 transgenic plants treated with morlin. Confocal images of GFP::MAP4 in 4-d etiolated hypocotyl cells. (A) DMSO control; 2 h. (B) Morlin (2 h) causes GFP::MAP4 microtubules to assume a short and bundled configuration that is reversible upon washout (C) in same seedling experiments. Each image is an average of 60 frames taken at 5-s intervals on the same Z plane. (Scale bars, 10 μm.) (D) Color-coded montage of GFP::MAP4 over 28 s shows freely pivoting polymer movement indicative of partial deattachment of microtubule bundles from the plasma membrane. The stationary microtubule end is indicated by the arrowhead. (Scale bar, 2.5 μm.) (E and F) Cross sectional intensity profiles across a representative GFP::MAP4 cell treated with 50 μM morlin at t = 0 (E) and t = 2 h (F). Higher intensity peaks in the 2-h treatment indicate that the drug causes increased apparent bundling of GFP::MAP4-labeled microtubules. Similar results were obtained in >12 cells.
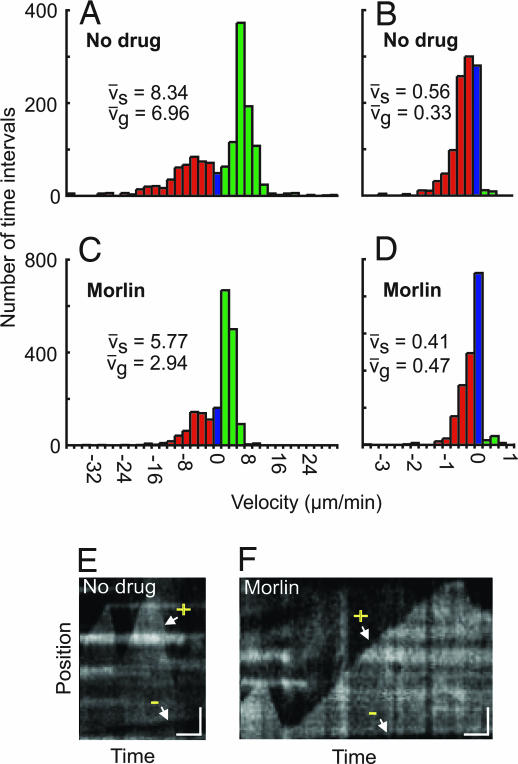
Measurement of microtubule polymerization dynamics showed that morlin caused a significant decrease in both elongation and shrinkage rates at the plus end (Fig. 3 A and C and Table 1) and in shrinkage rate at the minus end (Fig. 3 B and D and Table 1) as compared with control cells (polymerization events at the minus end were too small for meaningful comparison). The relative reduction in mean elongation rate (−58%) was nearly twice that of the relative reduction in shrinking rate (−31%), consistent with the observation of less microtubule area in the presence of the drug. A change in the proportion of time spent either in elongation or shrinking does not explain loss of polymer after drug treatment, because morlin treatment actually appeared to increase slightly the amount of time spent in growth and reduce modestly the amount of time spent shrinking (Table 1). Transition frequencies were not calculated because of the limited amount of time that the ends of any single treadmilling microtubule could be observed in the relatively dense cortical arrays of epidermal cells in etiolated tissue. Consistent with measurement of reduced elongation and shrinkage velocities (Fig. 3 E and F), the dynamicity (total polymer gain and loss per unit time) was significantly lower in morlin-treated cells (Table 1). As was reported previously (12), a small net gain of polymer was observed in each treatment (Table 1), suggesting that either these arrays were not at a steady state at the time of observation or there was a sampling bias in the observed microtubule ends. Taken together, measurements of polymerization dynamics suggest that a primary effect of morlin on the microtubule cytoskeleton is to reduce polymer dynamicity by inhibiting the rates of both dimer addition and loss at polymer ends.
Fig. 3.
Morlin inhibits microtubule dynamicity. (A–D) Growth (vg) and shrinkage (vs) velocities for the leading (A and C) and lagging ends (B and D) of single microtubules in 3-d etiolated hypocotyl cells treated with 0.2% DMSO (A and B; 34 and 21 polymer ends tracked, respectively, n = 1,442 and 2,127 2-s time intervals, respectively) or 0.2% DMSO plus 50 μM morlin (C and D; 27 and 19 polymer ends tracked, respectively, n = 1,966 and 2,045 2-s time intervals, respectively). Velocities were calculated by kymograph analysis. Histograms are color coded for growth (green), pause (blue), and shrinkage (red). Mean velocity measurements are noted within each figure component. Distributions of vg and vs at both the leading and lagging ends were significantly different between control and morlin treatment (P < 0.001), as calculated by using the Wilcoxon rank-sum test. (E and F) Representative kymographs for control (E) and morlin treatment (F). After morlin treatment, the rates of both plus end (+) growth and shrinking decrease, as does the rate of minus end (−) shrinking, resulting in inhibition of dynamicity at both polymer ends. (Vertical scale bar, 2 μm; horizontal bar, 0.5 min.)
Table 1.
In vivo measurements of single microtubule growth phases, dynamicity, and net polymer gain/loss in no drug versus morlin-treated cells
| Time in phase | No drug |
Morlin |
||
|---|---|---|---|---|
| (+) | (−) | (+) | (−) | |
| Growth, % | 62.4 | 1.9 | 69.1 | 4.0 |
| Pause, % | 1.0 | 20.5 | 2.9 | 45.3 |
| Shorten, % | 36.6 | 77.6 | 28.0 | 50.7 |
| Dynamicity, μm/min | 7.4 ± 4.9 | 0.4 ± 0.4 | 3.5 ± 2.7 | 0.3 ± 0.3 |
| Net polymer gain, μm/min | 1.2 | −0.4 | 0.4 | −0.2 |
Data are derived from the same population of measurements as described in Fig. 3. (+) denotes the presumed plus end, and (−) denotes the presumed minus end.
In addition to experiments with YFP::TUA5, we also visualized microtubules by expression of GFP::MAP4 (Fig. 4A). As with YFP::TUA5, reduction in the rates of microtubule elongation and shrinking were readily observed in morlin-treated cells by time-lapse imaging (SI Movie 2). However, additional changes in microtubule organization were also evident. Microtubules became arranged into short segments of similar lengths (Fig. 4B). The high degree of labeling indicated that these segments were polymer bundles, and analysis of intensity distribution among these segments suggested that the frequency and degree of polymer bundling increased after morlin treatment (Fig. 4 E and F). These bundles often lost positional stability and showed translocation behaviors that suggested detachment from the cell membrane (SI Movie 2). In many cases, pivoting movements of bundles about one end were observed, suggesting that bundles often remained attached to the cell membrane at one end (SI Movie 2 and Fig. 4D). Some bundles showed complete detachment from the cortex, resulting in a microtubule moving in the underlying cytoplasmic stream (SI Movie 2).
The origin of 20 of these short and mobile microtubule bundles was traced in time-lapse movies acquired during morlin application. Observation of new bundle creation during the period from the onset of morlin effects (17–47 min) showed that short MAP4-labeled bundles did not arise by severing or depolymerizing existing bundles. Rather, these structures appeared to arise de novo, initially being composed of short and dynamic polymers that stabilized at a fixed length after several minutes. Stabilization of microtubule turnover was supported by florescence recovery after photobleaching analysis, in which GFP::MAP4 microtubules showed recovery T1/2 values of ≈1 min, whereas, after treatment with morlin, T1/10 was >15 min (data not shown). Both polymer bundling and membrane detachment were reversed by drug washout (Fig. 4C).
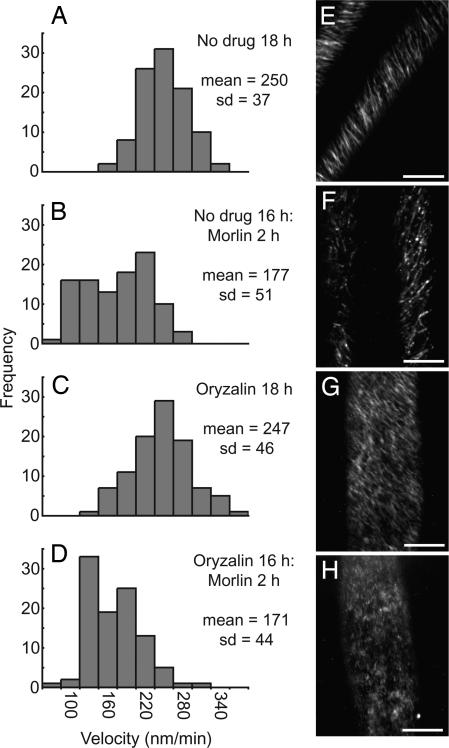
The inhibition of cellulose synthesis by morlin did not follow a classical inhibition curve, but a clear decrease in the cellulose content and 14C incorporation into cellulose was measured (SI Fig. 7 A and B). To explore further how morlin inhibits cellulose synthesis, we took advantage of a functional live cell tag for the cellulose biosynthetic complex, YFP::CESA6 (7), to ask whether morlin affects localization or dynamic behavior of the complex. Consistent with a reduction in cellulose content, time-lapse confocal imaging of YFP::CESA6 1 mm below the apical hook of 2- to 3-d-old etiolated seedlings showed that morlin treatment reduced the velocity of YFP::CESA6 complexes in the plasma membrane (control: 250 ± 37 nm/min; morlin: 177 ± 51 nm/min; Fig. 5B and SI Movie 3). In addition to a reduction in YFP::CESA6 complex velocity, the distribution of label was altered by morlin. Although labeled particles remained organized into obliquely linear arrays at the cell cortex, particles were fewer in number and often higher in label intensity (SI Movie 3).
Fig. 5.
Morlin causes inhibition of CESA motility in the plasma membrane. Time-lapse confocal images of YFP::CESA6 in hypocotyl cells of 3-d-old etiolated plants were used to measure CESA particle velocity. (A and E) Controls. (B and F) Morlin (50 μM) for 2 h. (C and G) Oryzalin (20 μM) for 18 h. (D and H) Oryzalin (20 μM) for 16 h, followed by 20 μM oryzalin and 50 μM morlin for 2 h. (A–D) Histograms of particle velocities calculated from kymograph analysis of 100 particles from five cells per treatment. Only particles displaying constant velocity were measured. Statistical significance of distribution shifts was calculated by using the Wilcoxon rank sum test for control and morlin (P < 0.001); control and oryzalin (P = 0.692); control and morlin plus oryzalin (P < 0.001); morlin and oryzalin (P < 0.001); morlin and morlin plus oryzalin (P = 0.204); and oryzalin and morlin plus oryzalin (P < 0.001). (E–H) Average time projections of 61 frames representing 5 min. (Scale bars, 10 μm.)
To determine whether the effects of morlin on YFP::CESA6 distribution and behavior depended on the microtubule cytoskeleton, we pretreated seedlings with oryzalin before morlin application to deplete the cell cortex of microtubules (Fig. 5G). After 16-h incubation in oryzalin, depolymerization of all detectable cortical microtubules was achieved, and, as reported (7), a more uniform dispersion of CESA6 particles was observed, with individual particles tracking in linear and oblique trajectories across the cell cortex. We measured the rate of YFP::CESA translocation and found that oryzalin had no measurable effect on YFP::CESA6 complex velocity (Fig. 5C), yet, when cells were treated further with morlin for 2 h, the velocities of YFP::CESA6 complexes were significantly reduced (Fig. 5H) to values similar to those measured after treatment with morlin alone (Fig. 5F, 247 ± 46 nm/min to 171 ± 44 nm/min). This experiment was repeated by using shorter oryzalin exposure times with similar results (data not shown). Therefore, it appears that inhibition of CESA complex velocity by morlin does not depend on the interaction of CESA with an intact microtubule cytoskeleton. By contrast, the effect of morlin on the distribution of YFP::CESA6 label depended highly on the presence of detectible microtubules. Whereas morlin treatment on its own caused the redistribution of YFP::CESA6 into fewer and brighter particles as described above (SI Movie 3B), this morlin-dependent redistribution was not observed when microtubules were depolymerized by oryzalin (SI Movie 3D). Instead, YFP::CESA6 distribution appeared much as it did with oryzalin treatment alone (SI Movie 3C).
To test for other effects of morlin treatment, a variety of fluorescent protein markers were assayed for alterations in organelle morphology or protein distribution. Markers highlighting the endoplasmic reticulum, actin filaments, Golgi apparatus, the vacuole, and the plasma membrane (marker lines are summarized in refs. 14 and 15) did not show any pronounced changes in response to drug treatment that could not be reasonably accounted for by changes in microtubule organization (SI Fig. 9).
Growth of Escherichia coli, Pichia pastoris, and Mammalian Cell Lines Were Not Inhibited by Morlin.
Proliferation of E. coli in liquid culture was not inhibited by addition of 10 or 20 μM morlin (SI Fig. 10). Similarly, addition of 20 μM morlin did not inhibit growth of the yeast P. pastoris. Mouse myoblast C2C12 cells treated with 25 μM morlin for 5 h and then stained for α-tubulin also showed no changes in microtubule organization (Tim Stearns, personal communication).
Morlin Induces Neuritic Beading in Hippocampal Neurons.
Immunolabeling of β-tubulin in neuronal processes showed that morlin caused severe microtubule disarray in high-density hippocampal neuronal cultures, characterized by an increase in free tubulin, microtubule disassembly, and neuritic beading (SI Fig. 11 and SI Materials and Methods). Microtubule disarray caused by morlin (25 μM) depended on the time of exposure to the drug (SI Fig. 11) and the neuronal stage of development, because DIV15 neurons exhibited a smaller degree of susceptibility to the drug compared with DIV5 neurons. Morlin further caused disruption of vesicular transport as observed by the accumulation of the vesicular glutamate transporter VGLUT1, a synaptic marker for excitatory glutamatergic neurons (SI Fig. 11). Despite the disruption of cytoskeletal structure, the same concentration of the drug had no significant effect on cell viability, as determined morphologically, by analysis of nuclear condensation and by MTT reduction assay (SI Fig. 11).
Discussion
The mechanisms of plant microtubule cortical array organization and functional interaction of the cortical array with CESA remain unknown. The discovery of the effects of morlin provides a tool for investigating these questions. Morlin is an analog of a class of bioactive small molecules, the coumarins, that have long been implicated in both the inhibition of cellulose deposition in plants and the inhibition of microtubules in mammalian cells (16–22).
Morlin stimulated morphological defects, such as right handed helical root growth at low concentrations and radial swelling in elongating roots, and hypocotyls at high concentrations (>5 μM). In agreement with previous reports of inhibition of cellulose synthesis by various coumarins (16–19), morlin caused a dose-dependent decrease in [14C]glucose incorporation into cellulose, and cell wall neutral sugar analysis revealed analogous decreases. Microtubule dynamicity at both polymer ends was inhibited, and cortical array organization was altered by morlin (Fig. 3 and Table 1), with further affects on polymer bundling and attachment to the cell membrane being observed in GFP::MAP4 transgenics (Fig. 4B). The observation that morlin also caused cytoskeletal defects in neuronal cells indicates that there may be a common target affecting cytoskeletal organization in both plant and animal cells. In addition to changes in microtubule behavior and organization, morlin caused a reduction in the velocity of CESA complexes in the plasma membrane and also altered the number and spatial distribution of labeled complexes. Although both of these effects may have contributed to and help explain the reduction in cellulose synthesis, complex redistribution caused by morlin depended on the intact microtubule array, whereas inhibition of complex velocity did not. Thus, if morlin has a single primary target, it must alter a cellular component that interacts with or impinges on both CESA and microtubules.
To test whether morlin has other effects on cell growth and metabolism, we examined what effect, if any, morlin had on proliferation of bacterial, fungal, and HEK1 cells and on cytoskeletal organization in mouse myoblast and primary neuronal culture cells. The only effect observed was concentration-dependent depolymerization of microtubules in primary neuronal cultures (SI Fig. 10). Morlin caused clear depolymerization of neuronal microtubules but no measurable effect on cell body morphology or apoptosis, even at relatively high doses (250 μM). Additionally, in plant cells, morlin did not disrupt endoplasmic reticulum morphology, inhibit Golgi streaming, or cause F actin disassembly. Similarly, previous Northern blot analysis showed no measurable effect on plant gene expression after treatment with coumarin at concentrations up to 1 mM (23). Thus, we consider it likely that the effects on cellulose synthesis and microtubule dynamics are due to a single primary effect rather than to a nonspecific effect of morlin.
The observation that morlin is active in neuronal cells but not in mouse C2C12 myoblast cells indicates that the drug target(s) are cell-type specific. Target specificity was also investigated by analysis of structural analogs. Avers and Goodwin (16) showed that coumarin, but not the structural analog scopoletin, caused individual cells to swell after 1.5–2 h. We extended this study to 19 structural analogs and found that activity was modified with only simple changes in structure; for instance an ethoxy versus methoxy group at carbon-7 resulted in embryo lethality compared with near-normal plant growth (SI Fig. 6). These results suggest that morlin's activity depends on fine details of its structure and thus is not likely to act by a nonspecific mechanism.
Three hypotheses might serve to explain how multiple changes in microtubule and CESA behavior, organization, and function result from morlin treatment. First, it is possible that morlin has separate effects on cellulose synthesis and microtubule dynamics. However, the probability of a drug acting separately on two of two processes studied is low, and the structural and cell-type specificity of morlin action supports the argument against multiple protein targets. Second, it is possible that multitarget inhibition could arise through targeting a signaling protein such as a protein kinase (22) that coordinates microtubule and CESA function. By modulating the phosphorylation of multiple effector proteins, a variety of changes in molecular behaviors might be explained, such as reduced polymerization and depolymerization rates, alteration of membrane association, and microtubule bundling as observed in GFP::MAP4 transgenic plants as well as limitations in CESA motility. Signaling cascades that modulate multiple pathways have been shown in many systems (24) and are likely to be involved in plants. A third possibility is that morlin targets a structural protein that interacts with both microtubules and the CESA complex. We measured a significantly reduced rate of both dimer addition and removal at the ends of microtubules, suggesting that the target of morlin may be a facilitator of dimer addition and removal. An example of such a protein is XMAP215, which has been shown to facilitate the addition or removal of oligomers of tubulin polymer to the growing and shrinking ends of microtubules in vitro (25, 26). The propensity for microtubules to form shorter, bundled, and less dynamic polymers in the GFP::MAP4 background was indeed reminiscent of the MOR1–1 mutant (11), which is a homologue of XMAP215.
Finally, experiments with morlin show that altered microtubule properties can change the distribution of CESA in the plasma membrane, supporting the idea that there is a functional interaction between the cortical cytoskeleton and CESA. However, it is clear from these experiments that the velocity of CESA complexes remains independent of an intact cortical microtubule cytoskeleton. The unusual and pronounced effect of morlin on microtubule attachment to the plasma membrane in GFP::MAP4-expressing cells suggests that the target for morlin may be involved in mediating this attachment. However, because membrane deattachment was not evident with either morlin treatment of YFP::TUA5-expressing cells or with GFP::MAP4 expression on its own, this result also indicates that multiple mechanisms for membrane association may exist. Identification of the target for morlin may provide insight into the mechanisms of cortical microtubule interaction with the cell membrane and the cell wall biosynthetic machinery.
Materials and Methods
Plant Material and Growth Conditions.
All Arabidopsis lines used were of the Columbia ecotype. Seedlings were germinated and grown under continuous light (200 μmol/m2/s) or in the dark at 21°C on plates containing 0.5× Murashige and Skoog (MS) mineral salts (Sigma, St. Louis, MO) and 0.8% agar. Inhibitors were solubilized in dimethyl sulfoxide (DMSO) or methanol (MeOH) and added to media so that solvent concentrations never exceeded 0.2%. Florescent protein experiments were performed on seedlings prepared essentially as described (7, 12, 13).
Chemical Screens.
Two 96-well format DIVERSet libraries (ChemBridge, San Diego, CA), each consisting of 10,000 small synthetic molecules solubilized in DMSO were screened; this represented 250 plates containing 80 compounds each (plates 10,126–10,250 and 10,626–10,750). Each compound was added to 100 μl of MS agar media to a final concentration of 20–40 μM. Approximately 10 sterilized Arabidopsis seeds were sown per well, and the plates were incubated in continuous fluorescent light (50 μE/m2/s) at 21°C for 3 d. Plates were visually scored after 3 d of growth by inspecting each well with a dissecting microscope. Effects of inhibitors on growth were tested by measuring root length of ≈50 Arabidopsis seedlings grown for 5 d on vertically oriented MS-agar plates containing 5 or 20 μM solutions of each compound in both light and dark conditions.
Confocal Microscopy.
Seeds were germinated on 0.5× MS agar for 2–4 d in darkness at 21°C. Seedlings were mounted in control or drug solutions between two 48 × 60 mm cover glasses or placed on a thin pad of 1.5% agar containing the appropriate control or drug and sandwiched between two coverslips. Once mounted, specimens were incubated for defined times in the dark before imaging. For incubations longer than 2 h, seedlings were submerged in solution and incubated in darkness before mounting. Imaging was performed on a purpose-built spinning-disk confocal microscope (7) using a Leica ×63 N.A. = 1.4 oil-immersion objective and a Roper Cascade 512b EMCCD camera. EGFP and YFP were excited at 488 nm, and fluorescence was collected through a 525/50 nm band-pass filter (Chroma Technologies, Brattleboro, VT).
Image Analysis.
All image processing was performed by using Metamorph (Molecular Dynamics, Sunnyvale, CA) and ImageJ (W. Rasband, National Institutes of Health, Bethesda, MD) software. Kymograph analysis to estimate particle velocities was performed by using the Walking Average (three-frame window) and Multiple Kymograph plug-ins for ImageJ (J. Rietdorf and A. Seitz, European Molecular Biology Laboratory, Heidelberg, Germany). To estimate the relative area occupied by microtubule-localized signal in confocal images, pixels defining labeled microtubules were thresholded above the local background, then the relative area occupied by these pixels was measured in a fixed 40 × 450-pixel region centered on the midline of each cell image. Image segmentation was performed by first applying a rolling-ball filter with a diameter of eight pixels to remove smoothly changing features in the image background (http://rsb.info.nih.gov/ij/docs/menus/process.html) and then applying a common threshold value to all measured images (see SI Fig. 12 for example).
Callose Synthase.
To visualize callose deposition in whole plants, seedling were cleared with 95% ethanol and stained with analine blue. Treatments were dH2O, 2-6-dichlobenzonitrile (20 μM), morlin (20 μM), and 2-6-dichlobenzonitrile and morlin (20 μM). Individual plants were then mounted in water and examined under UV illumination.
Whole-Plant in Situ Tubulin Antibody Studies.
Intact roots of Arabidopsis plants grown on MS-agar media containing 25 μM morlin were fixed, digested, and assayed by using a tubulin antibody essentially as described (27). Secondary antibody was visualized at 488 nm by confocal microscopy.
Supplementary Material
Acknowledgments
We thank Craig Garner (Stanford University) for timed-pregnant rats, Sergio Leal-Ortiz for technical assistance in preparing neuronal cultures, and Tim Stearns for assay of mouse myoblast cells. C.V.M. was supported by the Calouste Gulbenkian Foundation. This work was supported in part by U.S. Department of Energy Grant DOE-FG02-03ER20133 (to C.S.) and National Science Foundation Grant NSF-0524334 (to D.W.E.).
Abbreviations
- CESA
cellulose synthase
- MAP
microtubule associated protein.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700789104/DC1.
References
- 1.Green PB. Science. 1962;138:1404–1405. doi: 10.1126/science.138.3548.1404. [DOI] [PubMed] [Google Scholar]
- 2.Brown RM. J Polymer Sci. 2004;42:487–495. [Google Scholar]
- 3.Somerville CR, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab, et al. Science. 2004;306:2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- 4.Heim DR, Skomp JR, Tschabold EE, Larrinua IM. Plant Physiol. 1990;93:695–700. doi: 10.1104/pp.93.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hepler PK, Palevitz BA. Annu Rev Plant Physiol Plant Mol Biol. 1974;25:309–362. [Google Scholar]
- 6.Baskin TI, Beemster GTS, Judy-March JE, Marga F. Plant Physiol. 2004;135:2279–2290. doi: 10.1104/pp.104.040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paredez A, Somerville C, Ehrhardt D. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 8.Paredez A, Wright A, Ehrhardt DW. Curr Opin Plant Biol. 2006;9:571–578. doi: 10.1016/j.pbi.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Peterson JR, Mitchison TJ. Chem Biol. 2002;9:1275–1285. doi: 10.1016/s1074-5521(02)00284-3. [DOI] [PubMed] [Google Scholar]
- 10.Girke T, Cheng LC, Raikhel N. Plant Physiol. 2005;138:573–577. doi: 10.1104/pp.105.062687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO. Nature. 2001;411:610–613. doi: 10.1038/35079128. [DOI] [PubMed] [Google Scholar]
- 12.Shaw SL, Kamyar R, Ehrhardt DW. Science. 2003;300:1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- 13.Marc J, Granger CL, Brincat J, Fisher DD, Kao TH, McCubbin AG, Cyr RJ. Plant Cell. 1998;10:1927–1939. doi: 10.1105/tpc.10.11.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh S, André A, Edwards H, Ehrhardt DW, Somerville S. Plant J. 2005;44:516–529. doi: 10.1111/j.1365-313X.2005.02545.x. [DOI] [PubMed] [Google Scholar]
- 15.Sedbrook JC, Ehrhardt DW, Fisher SE, Scheible WR, Somerville CR. Plant Cell. 2004;16:1506–1520. doi: 10.1105/tpc.020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avers CJ, Goodwin RH. Am J Bot. 1956;43:612–620. [Google Scholar]
- 17.Hara M, Noriharu U, Miyamoto C, Tamari K. Plant Cell Physiol. 1973;14:11–28. [Google Scholar]
- 18.Hopp HE, Romero PA, Pont Lezica R. FEBS Lett. 1978;86:259–262. doi: 10.1016/0014-5793(78)80575-4. [DOI] [PubMed] [Google Scholar]
- 19.Montezinos D, Delmer DP. Planta. 1980;148:305–311. doi: 10.1007/BF00388116. [DOI] [PubMed] [Google Scholar]
- 20.Madari H, Panda D, Wilson L, Jacobs RS. Cancer Res. 2003;63:1214–1220. [PubMed] [Google Scholar]
- 21.Buey RM, Barasoain I, Jackson E, Meyer A, Giannakakou P, Paterson I, Mooberry S, Andreu JM, Diaz JF. Chem Biol. 2005;12:1269–1279. doi: 10.1016/j.chembiol.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Han S, Zhou V, Pan S, Liu Y, Hornsby M, McMullan D, Klock H, Lesley SA, Gray N, Caldwell J, Gu X. Bioorg Med Chem Lett. 2005;15:5467–5473. doi: 10.1016/j.bmcl.2005.08.097. [DOI] [PubMed] [Google Scholar]
- 23.Abenavoli MR, Cacco G, Sorgonà A, Marabottini R, Paolacci AR, Ciaffi M, Badiani M. J Chem Ecol. 2006;32:489–506. doi: 10.1007/s10886-005-9011-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou FQ, Snider WD. Science. 2005;308:211–214. doi: 10.1126/science.1110301. [DOI] [PubMed] [Google Scholar]
- 25.Kerssemakers JWJ, Munteanu LE, Laan L, Noetzel TL, Janson ME, Dogterom M. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 26.Cassimeris L, Gard D, Tran PT, Erickson HP. J Cell Sci. 2001;114:3025–3033. doi: 10.1242/jcs.114.16.3025. [DOI] [PubMed] [Google Scholar]
- 27.Sugimoto K, Williamson RE, Wasteneys GO. Plant Physiol. 2000;124:1493–1506. doi: 10.1104/pp.124.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.