Abstract
Although recent developments in MS have enabled the identification and quantification of hundreds of phosphorylation sites from a given biological sample, phosphoproteome analysis by MS has been plagued by inconsistent reproducibility arising from automated selection of precursor ions for fragmentation, identification, and quantification. To address this challenge, we have developed a new MS-based strategy, based on multiple reaction monitoring of stable isotope-labeled peptides, that enables highly reproducible quantification of hundreds of nodes (phosphorylation sites) within a signaling network and across multiple conditions simultaneously. We have applied this strategy to quantify temporal phosphorylation profiles of 222 tyrosine phosphorylated peptides across seven time points following EGF treatment, including 31 tyrosine phosphorylation sites not previously known to be regulated by EGF stimulation. With this approach, 88% of the signaling nodes were reproducibly quantified in four analyses, as compared with only 34% by typical information-dependent analysis. As a result of the improved reproducibility, full temporal phosphorylation profiles were generated for an additional 104 signaling nodes with the multiple reaction monitoring strategy, an 88% increase in our coverage of the signaling network. This method is broadly applicable to multiple signaling networks and to a variety of samples, including quantitative analysis of signaling networks in clinical samples. Using this approach, it should now be possible to routinely monitor the phosphorylation status of hundreds of nodes across multiple biological conditions.
Keywords: epidermal growth factor receptor, mass spectrometry, signal transduction, tyrosine phosphorylation
Ligand binding to cell surface receptors activates multiple protein tyrosine phosphorylation-mediated signaling cascades that regulate many cell biological processes, including proliferation, differentiation, migration, and cell death (1–3). To mechanistically define the relationship between signaling networks and downstream biological responses, it is necessary to quantify the dynamics of protein phosphorylation sites across multiple cell states and to correlate this information to quantitative phenotypic measurements. Until recently, antibody-based assays (e.g., FACS, Western blots, tissue microarrays) have been favored for most signaling network studies. These assays provide excellent quantitative information on the phosphorylation status of selected nodes within the network, but require a priori knowledge of the proteins and phosphorylation sites to be studied and are limited by the need to have high-quality, non-cross-reactive antibodies recognizing specific sites within the network. By comparison, analysis of protein phosphorylation by MS provides the capability of identifying novel phosphorylation sites on novel proteins within the network, requires minimal a priori knowledge, and is therefore compatible with both well and poorly characterized signaling networks. Recent developments in MS have also enabled the quantitative analysis of protein phosphorylation on hundreds of proteins, information that has been correlated to cell fate in recent studies (4–6).
In its current implementation, MS-based phosphoproteomics has many advantages (e.g., quantification accuracy, site specificity, sensitivity, broad coverage of signaling networks, discovery of novel signaling molecules) over antibody-based approaches. However, a chief disadvantage of employing MS to investigate signaling networks lies in the inconsistent reproducibility of the technique (6, 7). Run-to-run variability in peptide identification is due primarily to the information-dependent acquisition (IDA) mode in which MS data are usually collected. In this mode, the mass spectrometer continuously repeats a cycle consisting of a full-scan mass spectrum, followed by selection and fragmentation of the n (typically n = 1–10) most abundant m/z ratios for peptide identification and quantification. Although this mode of operation provides the potential to uncover novel sites and proteins, the complexity of the samples, coupled with inherent variability in automated peak selection, results in poor reproducibility in peptide identification and quantification.
As an alternative to a data-dependent operation of the mass spectrometer, it is also possible to program the instrument to continuously monitor selected precursor to fragment ion transitions that can be diagnostic for the presence of given compounds within the sample. Although full-scan MS/MS spectra are not acquired in this mode of operation [termed selected reaction monitoring (SRM) and, its extension, multiple reaction monitoring (MRM)], the method provides high selectivity by monitoring chromatographic coelution of multiple transitions for a given peptide. It is important to note that SRM and MRM have already been successfully applied to a variety of biological applications, including quantification of DNA adducts purified from tissue (8) and detection of doping substances in human urine and plasma (9–12). To date, the use of SRM and MRM in proteomics is less widespread, although MRM has been used to quantify protein expression (13), to find protein biomarkers for disease severity in rheumatoid arthritis (14), and for detection and quantitative analysis of protein phosphorylation (15–17).
We now demonstrate integration of these two methods (IDA and MRM) to generate a strategy that provides the capability to identify hundreds of phosphorylation sites within a given signaling network, and to monitor these sites across multiple analyses and cell states with high reproducibility. Here we describe the application of this strategy to quantify high-resolution temporal dynamics of protein tyrosine phosphorylation within the epidermal growth factor receptor (EGFR) signaling network. The robustness of our method makes it ideal for the acquisition of large, consistent, statistically significant protein phosphorylation data sets across many conditions, data that are required for rigorous computational analysis and mathematical modeling.
Results
The strategy we have developed is an extension and improvement on our previously described liquid chromatography tandem MS (LC/MS/MS) technology for characterization and quantification of tyrosine phosphorylation events in cell signaling networks (18). In an effort to improve on the reproducibility of peptide quantification across multiple analyses, we have expanded our technology to combine an initial discovery phase (IDA mode) with a subsequent monitoring and quantification phase (MRM mode) (see Fig. 1). This combined technology incorporates the peptides identified from multiple IDA experiments to generate an MRM method that can be implemented by using a triple-quadrupole mass spectrometer (QTRAP 4000). We applied this method to investigate the temporal dynamics of these phosphorylation sites during a 7-point time course (0, 1, 2, 4, 8, 16, 32 min) of EGF stimulation of human mammary epithelial cells (HMECs) (19).
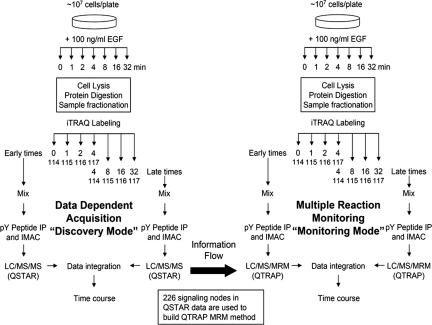
Fig. 1.
Schematic of the combined IDA/MRM method. Parental HMECs were treated with 100 ng/ml EGF for 0, 1, 2, 4, 8, 16, or 32 min after 12-h serum starvation. Samples were divided up into “early” or “late” halves of the time course, with the 4-min sample being present in both subsets as a normalization point. Multiple IDA analyses on the QSTAR mass spectrometer were used to identify nodes within the network and to obtain information needed to construct the MRM methods. After the MRM methods were constructed, identical samples from two biological replicates were analyzed by both IDA on the QSTAR and MRM on the QTRAP to compare results and estimate the quality of the MRM/QTRAP data set.
The MRM method was constructed from five parameters that are readily obtained from one or multiple IDA experiments: (i) chromatographic elution order of phosphorylated peptides, (ii) phosphorylated peptide (precursor ion) mass to charge ratio (m/z) and charge state (z), (iii) characteristic b- and y-type fragment ion m/z ratio, (iv) collision energy required for fragmentation to obtain b- and y-type fragment ions, and (v) collision energy required for fragmentation to obtain iTRAQ marker ions. As depicted in Fig. 2 for the tryptic peptide containing EGFR pY1173, the chromatographic retention time for each phosphorylated peptide of interest is extracted from the total ion chromatogram, and the precursor ion m/z ratio and charge state are extracted from the full-scan mass spectrum. Although the exact retention time is not critical, the time relative to other peptides enables the proper ordering of the list. Because the instrument software is currently limited to 300 MRM scans per cycle, it is necessary to arrange the peptides by elution order, breaking the list every 50 peptides, with a 3-peptide overlap between the lists. The MRM list also includes the m/z ratio for characteristic b- and y-type fragment ions, which were selected according to their signal intensity in MS/MS with the following constraints: each fragment ion should contain at least four amino acids, their added length should be at least 9 aa (when possible), and the fragments should not be complementary. Collision energy is also included in the MRM list and can be optimized; for instance, the intensity of b- and y-type ion fragments is greater at lower collision energy, whereas the intensity of iTRAQ marker ion fragments is greater at high-collision energies. The final MRM method is constructed by merging the data for all of the peptides, six rows per peptide, in order from first to last eluting peptide.
Fig. 2.
Construction of the MRM method from IDA data. Elution time of the peptides is obtained from total ion chromatogram (TIC) (Top Left), parent ion m/z from the corresponding full-scan mass spectrum (Middle Left), and the m/z ratio for b- and y-type ions is obtained from the appropriate MS/MS scan (Top and Middle Right). Peptides are ordered according to their LC elution profiles, and CEs are calculated as a linear function of parent ion mass-to-charge ratio (Bottom). To favor marker ion detection, iTRAQ CE is calculated with a 25% increase in slope and an increase of 6 eV in the baseline with respect to b- and y-type ion fragmentation. Accumulation times can also be adjusted, but 35 msec was used for all selected fragments. The table shows the necessary information to detect EGFR pY1173 in MRM mode, including its expected relative elution order with respect to other peptides.
MRM/QTRAP Data Output.
As described in Fig. 1, the MRM/QTRAP method was used to quantify 226 phosphorylated peptides covering 208 phosphorylation sites in 143 proteins across seven time points of EGF stimulation. For each half of the time course, a single analysis consisted of five separate MRM methods, with 300 precursor-fragment ion transitions for the first four methods and 1,428 total MRM transitions. Following acquisition, each data set was processed to extract elution profiles for all peptides in the MRM methods. The elution profile of the tryptic peptide containing EGFR pY1173 is shown in Fig. 3a, where the left column shows detection of b5, y4 ions, and iTRAQ marker ions 114.1–117.1 for the first half of the time course and the right column shows detection of these same ions for the second half of the time course in a subsequent analysis. Note that in both columns, b5, y4, and iTRAQ marker ions coelute, a necessary condition given that each of these ions represents a fragmentation event from a single precursor ion eluting from the LC column. By normalizing the data in Fig. 3a to the overlapping (4-min) time point and combining data from the early and late time points, a single time course covering all seven time points was constructed (error bars are from two biological replicates).
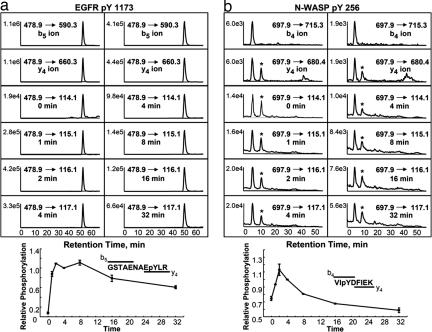
Fig. 3.
Protein phosphorylation detection and quantification by MRM. MRM elution profiles of EGFR pY1173 and N-WASP pY256 with respective phosphorylation time courses. (a) (Left) Coelution of the b5, y4, and iTRAQ ions identifying EGFR pY1173 and quantifying its phosphorylation after 0, 1, 2, and 4 min of EGF stimulation. (Right) Coelution of these same ions, quantifying phosphorylation after 4, 8, 16, and 32 min of EGF stimulation. The data were normalized to 4-min stimulation to couple together both halves of the time course. Error bars are from two biological replicates. (b) Quantification of the time course for N-WASP pY256. Note that detection and coelution of b4 and y4 ions are required to ensure correct peptide identification vs. alternate ion with similar precursor and fragment ion m/z ratio (marked by ∗).
A key requirement of the MRM/QTRAP method is the ability to distinguish between correct identification events and false positives. Because each fragmentation event provides information about one position in the peptide, by requiring coelution of two sequence-specific fragment ion transitions, the method is highly specific, providing confidence in the results even in the absence of a full MS/MS spectrum. An example of this specificity is demonstrated by N-WASP pY256 (Fig. 3b), for which all precursor-fragment ion transitions coelute in the first peak, corresponding to N-WASP pY256, whereas the second peak is absent from the precursor-b4 transition channel and therefore represents a false positive.
MRM/QTRAP Quantification.
To confirm the accuracy of quantification results obtained with the MRM method, we compared these results to IDA-based analysis of the same samples. In this comparison, we selected 903 iTRAQ peaks with intensities between 50 and 1,500 counts (to avoid low signal-to-noise measurements or detector saturation) from IDA data and compared them to the same iTRAQ peaks as detected by MRM; all data from both methods are relative to the 4-min time point for the given time course. As demonstrated in Fig. 4a, a very good correlation was achieved between MRM and IDA quantification, with an offset of 0 and a slope of nearly 1, confirming that MRM quantification is comparable to optimal results obtained by using IDA. To further compare the MRM and IDA data sets, the ratio between the MRM and IDA quantification value was calculated for each data point, and several statistical distributions (e.g., Gaussian, Laplace, and Weibull, among others) were tested for closeness of fit (data not shown). As shown in Fig. 4b, the data closely match the Laplace distribution; parameters were estimated with the maximum likelihood estimation procedure. The resulting Laplace location and scaling parameter estimates were 1.01 [95% confidence interval (C.I.): (1.006 1.013)] and 0.09 [95% C.I.: (0.084 0.096)], respectively, indicating a strong agreement between IDA and MRM quantification results.
Fig. 4.
Quantification of MRM/QTRAP and IDA/QSTAR data. (a) Linear regression comparing quantification results obtained by MRM and IDA methods. Each data point represents the level of phosphorylation for a given phosphorylated peptide at a given time point, relative to 4-min stimulation for that same peptide, quantified by either MRM or IDA. Regression parameters show a strong correlation R2 = 0.94. The slope is 1 and offset is 0, indicating that the MRM method does not introduce bias and MRM-based quantification is equivalent to IDA-based quantification. (b) Distribution of the MRM/IDA quantification ratio. The data follow a Laplace distribution with location parameter of 1.01 and scaling parameter of 0.09. (c) Phosphorylation time courses for six phosphorylation sites (Cbl pY552, EGFR pY1173, ERK2 pT184/pY186, PLC-γ pY1253, N-WASP pY256, and STAT3-1 pY705) measured by using MRM (blue) and IDA (red), respectively. Error bars are from two biological replicates.
To further visualize the quantification accuracy between the two methods, time courses derived from MRM (blue) and IDA (red) analyses were selected for tryptic phosphopeptides from six different proteins (Fig. 4c). Interestingly, the error bars for the two methods overlap for all time points except for the 2-min time point in Cbl pY552 and EGFR pY1173. The error in these points may be due to the iTRAQ 116 peak and its associated contaminant ion (see Discussion). However, other than these two measurements, the remaining 40 time points show remarkable agreement between MRM and IDA quantification.
Reproducibility.
By joining the MRM methodology to our previous IDA-only technology, we were able to significantly improve the reproducibility of our quantitative data for the 7-point time course of EGF stimulation. From the list of 226 peptides previously found in various QSTAR analyses and built into the MRM method, 223 (99%) were quantified in at least two of the four MRM analyses, 217 (96%) were quantified in at least three analyses, and 199 were found in all four MRM analyses, for a final 88% reproducibility (Fig. 5a). By comparison, only 186 (82%) of these 226 peptides were detected and quantified in any of the four analyses by using IDA/QSTAR. Of these 186 peptides quantified by IDA, 148 (80%) were found in at least two of the four IDA analyses, 104 (56%) were found in at least three analyses, and only 63 peptides were found in all four IDA analyses, for a final 34% reproducibility (Fig. 5b). It is possible to fit the MRM and IDA data to a binomial distribution, with success probability P of 0.88 [95% C.I.: (0.831, 0.920)] for MRM and 0.34 [95% C.I.: (0.271, 0421)] for IDA. To estimate the quality of data generated by replicate MRM analyses, we quantified the mean signal intensities for each precursor-iTRAQ ion transition and compared these data to the relative SD of the replicate analyses (Fig. 5c). The vast majority of peptide transitions have a medium- to high-intensity signal (i.e., >1,000 counts, with noise levels typically present at <100 counts) and a small SD, indicative of high-quantification accuracy of replicate MRM analysis.
Fig. 5.
Reproducibility of MRM/QTRAP and IDA/QSTAR. (a) Venn diagram quantifying the intersection of data from four MRM analyses. From the original list of 226 peptides, 223 were identified by the MRM method, and 199 peptides (88%) were found in all of the MRM analyses. (b) Venn diagram quantifying the intersection of data from four IDA analyses. A total of 186 peptides (of the list of 226 used to construct the QTRAP MRM method) were identified by IDA, but only 63 (34%) were found in all of the IDA analyses. (c) Relative SD was compared with mean intensity for each precursor-iTRAQ transition at each time point for all peptides in the QTRAP data set. Most transitions have high signal intensity and low SD, indicating the high quantification accuracy of replicate MRM analysis. (d) Complete time course could be constructed for 222 of the 223 (99%) peptides detected by MRM (for one biological replicate, whereas 199 of the 223 were quantified in both biological replicates). Complete time courses could be constructed for only 118 of the 186 (63%) peptides detected by IDA (for one biological replicate, whereas only 63 were found in both biological replicates). Early time points (0, 1, 2, 4 min) for the first (I) and second (II) biological samples are represented by A and B; late time points (4, 8, 16, 32 min) for the two biological replicates are represented by C and D.
To construct complete time courses from these data sets (see Fig. 5d), the same peptide must be identified in both halves of a time course. When viewed in this fashion, 98% (222/226) of the peptides analyzed by the MRM method were quantified in complementary analyses, compared with only 63% (118/186) of the peptides detected by the IDA method. Significantly, the MRM method enabled quantification of the full temporal dynamics for 104 additional signaling nodes within the EGFR signaling network, an increase of 88% over that possible with the IDA method. Also, with the MRM method, an additional 136 peptides were quantified in both biological replicates, providing much greater confidence in the quantification results.
Improved Temporal Resolution.
By using our combined IDA- and MRM-based method to generate higher temporal resolution data in this signaling network, we are now able to differentiate temporal phosphorylation profiles that had previously been clustered together based on low-resolution temporal data (18). Interestingly, although the phosphorylation profile for each of these sites was almost identical with low-resolution temporal data, improved temporal resolution clearly distinguishes the profiles for these sites [see supporting information (SI) Fig. 6]. As the high-resolution data show, phosphorylation of these four sites happens much more rapidly than the previous data suggested. In fact, c-Cbl pY552 and MARVEL D2 pY23 reach maximal phosphorylation within 2 min after EGF stimulation, ERK1 pY204 is maximally phosphorylated between 4 and 5 min, and phosphorylation of EGFR pY1148 increases rapidly in the first minute and then plateaus until 8 min of stimulation. Significantly, EGFR pY1148, ERK1 pY204, c-Cbl pY552, and MARVEL D2 pY23 can also be distinguished from each other based on their dephosphorylation after the 8-min time point, arguing for the presence of different mechanisms of dephosphorylation and/or degradation for these proteins.
The improved temporal resolution in the current data set also highlights the diversity of the immediate early response of the EGFR signaling network. For instance, a significant number of protein tyrosine phosphorylation sites increase dramatically (>5-fold) and achieve >90% of their maximum phosphorylation level within the first minute. Several of these proteins are expected to be phosphorylated immediately after stimulation, including EGFR and proteins known to interact with EGFR (e.g., SHC, Cbl, PLC-γ, HER2). However, many novel and/or unexpected proteins also demonstrate a similar rapid response, including EHM2, Marvel D2, or novel phosphorylation sites on AHNAK and hypothetical protein FLJ00261. The current study does not identify a function for these proteins, but highlights specific phosphorylation sites as targets for future, in-depth biological studies to better characterize the EGFR signaling network and/or construct models predictive of cell function (20).
Discussion
We have developed a new method for highly reproducible quantitative analysis of protein phosphorylation-mediated signaling networks. As described above, the method combines an initial discovery phase (IDA/QSTAR) with a subsequent monitoring and quantification phase (MRM/QTRAP). Because selected phosphopeptides are specifically targeted and quantified in the second phase of the method, the reproducibility of the method has increased significantly, from 34% (IDA) to 88% (MRM) (peptides quantified in four analyses). The increase in reproducibility has dramatically improved our ability to quantify the temporal dynamics of phosphorylation within the EGFR signaling network, enabling the acquisition of 222 full temporal profiles (MRM/QTRAP), as compared with only 118 with the previous IDA/QSTAR method. It is worth noting that the recently developed “8-plex” iTRAQ from Applied Biosystems (Foster City, CA) will enable analysis of 7 or 8 time-point data in a single experiment, diminishing the adverse effects of poor reproducibility of the IDA method in analyzing larger time courses. However, because biological replicates still need to be analyzed, the high reproducibility of the MRM method will significantly improve the data output.
Although the main advantage of the MRM/QTRAP method lies in the increased reproducibility, performing MRM-type analyses on a triple-quadrupole instrument also provides significant improvement in sensitivity (see SI Figs. 7 and 8) and much greater dynamic range (≈5 orders of magnitude on the QTRAP vs. two orders of magnitude on the QSTAR) as compared with full MS/MS on a QSTAR. The improvement in sensitivity is primarily associated with increased detection time per fragment ion transition, which results in an increase in the signal-to-noise ratio. To further improve the results, the MRM method can be customized by increasing the detection time to provide improved signal for low-abundance peptides or decreasing the detection time for high-abundance peptides. These modifications translate into more accurate quantification for both low-abundance peptides (better signal-to-noise ratio) and very high-abundance peptides (increased linear dynamic range) when using the MRM method. Unfortunately, improvements in sensitivity and dynamic range are offset in part by decreased mass resolution on the triple quadrupole relative to the QqTof, such that the QTRAP instrument is unable to distinguish iTRAQ peaks from adjacent contaminant ions at the same nominal m/z ratio. Although the appearance of contaminant ions is infrequent, the high-resolution QSTAR instrument is able to resolve contaminant ions (most frequent and abundant at 116.07 Da) from the iTRAQ ions (e.g., at 116.1 Da) and provides accurate quantification of the iTRAQ marker ions. We have attempted to correct for contaminant ion contribution by using QSTAR data to identify peptides most affected by the appearance of the contaminant ions and to determine a correction factor (relative peak area of contaminant ions to iTRAQ marker ions). As demonstrated in Fig. 4, there is still a small amount of error associated with contamination of the 116 iTRAQ marker ion (e.g., 2-min time point EGFR pY1173), indicating that a more accurate correction factor needs to be implemented for these selected peptides.
From our analysis of temporal dynamics of tyrosine phosphorylation in the EGFR signaling network, it is clear that using the MRM/QTRAP method has resulted in robust quantification of a large number of nodes across multiple time points and biological replicates. This data set can be compared with other recently published analyses of phosphorylation dynamics within the EGFR signaling network (21, 22). For instance, Blagoev et al. (21) used antiphosphotyrosine antibodies to immunoprecipitation (IP) proteins from SILAC-encoded HeLa cells stimulated with EGF for 0, 1, 5, 10, or 20 min, quantifying temporal changes for 81 proteins. Although only 26 of these proteins are contained in our analysis, this disparity is most likely due to protein IP and quantification in the Blagoev et al. study vs. peptide IP and site-specific quantification in our method. More recently, Olsen et al. (22) identified and quantified >6,000 phosphorylation sites in EGF-stimulated HeLa cells. Of the 103 tyrosine phosphorylation sites in their data set, 23 are also contained within our data, and most of these yield similar temporal phosphorylation profiles. Interestingly, the principal disparity occurs at the zero time point, indicating that the HMECs may have lower basal phosphorylation levels and respond more vigorously to EGF stimulation relative to the HeLa cells, reflecting the EGF dependence of the HMECs. The difference in identified sites is most likely due to the specific isolation of tyrosine phosphorylation sites in our study, compared with the global approach used by Olsen et al. From these analyses, it is clear that we have not comprehensively identified the EGFR signaling network, and that multiple methods can be used to yield increased coverage of the network.
It is important to note that the specific MRM method we have generated in this study should be directly applicable to EGFR signaling network analysis in a broad variety of biological samples, including additional cell lines, primary cells, and tissues. In this application, following sample processing [including iTRAQ labeling, peptide immunoprecipitation, and immobilized metal affinity chromatography (IMAC) enrichment of phosphorylated peptides], samples will be directly analyzed by (LC-ESI)-MRM/QTRAP, and 208 nodes (226 peptides) within the EGFR signaling network will be automatically monitored, providing relative quantification across a broad sampling of the network. The application of this technology to the study of clinical biopsies will allow the consistent monitoring of phosphorylation levels at hundreds of signaling nodes in tumor samples. These data will greatly improve our understanding of the protein biochemistry underlying cancer biology and will facilitate recognition of dysregulated pathways and potential protein therapeutic targets on a patient-to-patient basis.
This method is also easily extended to additional signaling networks: for each network of interest, the discovery phase consists of several IDA analyses to determine the key signaling nodes. These nodes are then used to construct the MRM method that can be applied for a variety of biological samples.
Conceptually, our use of the MRM method to investigate the EGFR signaling network is analogous to performing, in a single 2-h analysis, 206 simultaneous, highly specific, quantitative, reproducible, and high-throughput Western blots with antibodies targeted to specific phosphorylation sites. This technique should significantly improve our ability to accurately quantify signaling networks across a variety of conditions, bringing routine application of phosphoproteomics closer to reality.
Methods
Technical details for all experiments are available in the SI Methods.
Cell Culture and Stimulation.
184A1 HMECs (19) were maintained in DFCI-1 medium supplemented with 12.5 ng/ml EGF, as in ref. 23. Cells were washed with PBS and incubated for 12 h in serum-free media (DFCI-1 without EGF, bovine pituitary extract, or FBS) after 80% confluence was reached in 15-cm plates (≈107 cells). Synchronized cells were then stimulated with 100 ng/ml EGF in serum-free media for 0, 1, 2, 4, 8, 16, or 32 min.
Sample Preparation and Peptide IP.
Samples were prepared as described previously (18), with the following exceptions: 10 μg of protein G Plus-agarose beads (Calbiochem, San Diego, CA) was incubated with 12 μg of each anti-phosphotyrosine antibody [pTyr100 (Cell Signaling Technology, Danvers, MA) and PT-66 (Sigma–Aldrich, St. Louis, MO)] in 200 μl of IP buffer (100 mM Tris, 100 mM NaCl, 1% Nonidet P-40, pH 7.4) for 8 h at 4°C.
IMAC and IDA MS Analysis.
To remove nonspecifically retained nonphosphorylated peptides, phosphopeptide enrichment by IMAC was performed as described (18). Peptides were analyzed by ESI LC/MS/MS on a QqTof (QSTAR XL Pro, Applied Biosystems) operated in IDA mode, as described in ref. 18.
MRM MS Analysis.
Following IMAC enrichment, peptides were analyzed by ESI LC/MS/MS on a triple-quadrupole (Qq linear ion trap) mass spectrometer (QTRAP 4000, Applied Biosystems). The instrument was set up to cycle through one full-scan mass spectrum [enhanced MS (EMS), with Q0 trapping activated] followed by all MRM transitions (typically 300 per method) in a given method, for a total cycle time of ≈11.6 sec (1.1 sec per EMS scan followed by 300 MRM scans of 35 msec each). In EMS mode, masses between 370 and 1,500 atomic mass units (amus) were scanned at a scan rate of 1,000 amus per cycle. For MRM mode, Q1 resolution was set to high (resolution = 2,500, FWHM = 0.4 Da at m/z = 1,000) and Q3 resolution was set to unit (resolution = 1,700, FWHM = 0.6 Da at m/z = 1,000). Collision energies (CEs) were maintained from the QSTAR for y- and b-type ion transitions, whereas iTRAQ ion transitions were increased by 25% in the slope and six units in the offset (CE parameter tables available in SI Tables 1 and 2).
Phosphopeptide Sequencing, Data Clustering, and Analysis.
For IDA experiments, MS/MS spectra were extracted and searched against human protein database by using ProQuant (Applied Biosystems) as described by the manufacturer. Phosphotyrosine-containing peptides identified by ProQuant were manually validated. ProQuant quantification results were corrected by removing contaminant signals near iTRAQ tag peaks. To correct for variations in the amount of sample analyzed for each condition, the iTRAQ quantitation data were further corrected with values generated from the peak areas of nonphosphorylated peptides from analysis of the supernatant from the peptide IP. Data from each analysis were normalized to the 4-min peak area for time-course integration.
For MRM experiments, peak detection and automatic quantification methods were built for each method by using Analyst 1.4.1 (Applied Biosystems). Peak detection was inspected visually to ensure correct peak identification (coelution) and shape (smoothness). For quantifying peptides eluting at the very beginning or end of an MRM method, the manual quantification option of Analyst 1.4.1 was used. As an alternative, peptides were also quantified directly from the MRM raw data in the original acquisition file (.wiff). iTRAQ marker ion intensities were averaged for three to five MRM cycles at the apex of the chromatographic elution profile. Automatic quantification was generally in close agreement with manual quantification values. Peak detection was problematic for low signal-to-noise ratio chromatographic elution profiles; as a result, manual results were used to quantify these time points. Data were corrected by analyzing nonphosphorylated peptides from the supernatant of the IP for possible variations in the starting amounts of sample for each time point. All data from each analysis were normalized to the 4-min peak area for time-course integration. All MS data are available in SI Tables 3 and 4.
Supplementary Material
Acknowledgments
The authors thank Terry Kehoe (Applied Biosystems, Foster City, CA) and Yi Zhang, Neil Kumar, and other members of the White and Lauffenburger laboratories for helpful discussions. This work was supported by National Cancer Institute Grant CA96504, National Institutes of Health Grant P50-GM68762, National Institute of Allergy and Infectious Diseases Grant R33-AI65354, the David Koch Research Fund, Biocentrum Helsinki (S.H.), and a Ludwig Fellowship (to A.W.-Y.).
Abbreviations
- CE
collision energy
- EGFR
epidermal growth factor receptor
- EMS
enhanced MS
- HMECs
human mammary epithelial cells
- IDA
information-dependent acquisition
- IMAC
immobilized metal affinity chromatography
- IP
immunoprecipitation
- LC/MS/MS
liquid chromatography tandem MS
- MRM
multiple reaction monitoring
- SRM
selected reaction monitoring.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608638104/DC1.
References
- 1.Blume-Jensen P, Hunter T. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 2.Pawson T, Nash P. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y, Sliwkowski MX. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 4.Kim JE, White FM. J Immunol. 2006;176:2833–2843. doi: 10.4049/jimmunol.176.5.2833. [DOI] [PubMed] [Google Scholar]
- 5.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Science. 2005;308:1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 6.Wolf-Yadlin A, Kumar N, Zhang Y, Hautaniemi S, Zaman M, Kim HD, Grantcharova V, Lauffenburger DA, White FM. Mol Syst Biol. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elias JE, Haas W, Faherty BK, Gygi SP. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 8.Koc H, Swenberg JA. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;778:323–343. doi: 10.1016/s1570-0232(02)00135-6. [DOI] [PubMed] [Google Scholar]
- 9.Guan F, Uboh CE, Soma LR, Luo Y, Rudy J, Tobin T. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;829:56–68. doi: 10.1016/j.jchromb.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 10.Ho EN, Leung DK, Wan TS, Yu NH. J Chromatogr A. 2006;1120:38–53. doi: 10.1016/j.chroma.2006.03.089. [DOI] [PubMed] [Google Scholar]
- 11.Thevis M, Opfermann G, Schanzer W. Biomed Chromatogr. 2001;15:393–402. doi: 10.1002/bmc.87. [DOI] [PubMed] [Google Scholar]
- 12.Herrin GL, McCurdy HH, Wall WH. J Anal Toxicol. 2005;29:599–606. doi: 10.1093/jat/29.7.599. [DOI] [PubMed] [Google Scholar]
- 13.Kirkpatrick DS, Gerber SA, Gygi SP. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Liao H, Wu J, Kuhn E, Chin W, Chang B, Jones MD, O'Neil S, Clauser KR, Karl J, Hasler F, et al. Arthritis Rheum. 2004;50:3792–3803. doi: 10.1002/art.20720. [DOI] [PubMed] [Google Scholar]
- 15.Cox DM, Zhong F, Du M, Duchoslav E, Sakuma T, McDermott JC. J Biomol Tech. 2005;16:83–90. [PMC free article] [PubMed] [Google Scholar]
- 16.Unwin RD, Griffiths JR, Leverentz MK, Grallert A, Hagan IM, Whetton AD. Mol Cell Proteomics. 2005;4:1134–1144. doi: 10.1074/mcp.M500113-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Zappacosta F, Collingwood TS, Huddleston MJ, Annan RS. Mol Cell Proteomics. 2006;5:2019–2030. doi: 10.1074/mcp.M600238-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Stampfer MR, Bartley JC. Proc Natl Acad Sci USA. 1985;82:2394–2398. doi: 10.1073/pnas.82.8.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar N, Wolf-Yadlin A, White FM, Lauffenburger DA. PLoS Comput Biol. 2006 doi: 10.1371/journal.pcbi.0030004. e4.eor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blagoev B, Ong SE, Kratchmarova I, Mann M. Nat Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 22.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 23.Hendriks BS, Opresko LK, Wiley HS, Lauffenburger D. Cancer Res. 2003;63:1130–1137. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.