Abstract
Transcription, splicing, and translation are potentially coordinately regulatable in a temporospatial-dependent manner, although supporting experimental evidence for this notion is scarce. Yeast two-hybrid screening of a mammary gland cDNA library with human p21-activated kinase 1 (Pak1) as bait identified polyC-RNA-binding protein 1 (PCBP1), which controls translation from mRNAs containing the DICE (differentiation control element). Mitogenic stimulation of human cells phosphorylated PCBP1 on threonines 60 and 127 in a Pak1-sensitive manner. Pak1-dependent phosphorylation of PCBP1 released its binding and translational inhibition from a DICE-minigene. Overexpression of PCBP1 also inhibited the translation of the endogenous L1 cell adhesion molecule mRNA, which contains two DICE motifs in the 3′ untranslated region. We also found that Pak1 activation led to an increased nuclear retention of PCBP1, recruitment to the eukaryotic translation initiation factor 4E (eIF4E) promoter, and stimulation of eIF4E expression in a Pak1-sensitive manner. Moreover, mitogenic stimulation promoted Pak1- and PCBP1-dependent alternative splicing and exon inclusion from a CD44 minigene. The alternative splicing functions of PCBP1 were in turn mediated by its intrinsic interaction with Caper α, a U2 snRNP auxiliary factor-related protein previously implicated in RNA splicing. These findings establish the principle that a single coregulator can function as a signal-dependent and coordinated regulator of transcription, splicing, and translation.
Keywords: differentiation control element, eukaryotic translation initiation factor 4E, p21-activated kinase 1
The efficient control of cellular response to various stimuli relies on the coordination of multiple regulatory modules in the eukaryotic genetic architecture, including transcription, precursor mRNA processing, and translation of mature mRNA in the cytoplasm. In addition to inherent selective control at each level, these processes might be regulated in a coordinated manner. For example, steroid hormone receptors activated by cognate ligands or growth factors have been shown to functionally couple gene transcription activity and exon-content selection of pre-mRNA by recruiting coregulators (1, 2). These coordinated responses often involve posttranslational modifications of key components within their regulatory systems. Despite the growing realization of the significance of such delicate and efficient regulatory control of gene expression, the notion of signal-activated and temporospatial-dependent regulation of transcription, splicing, and translation lacks supporting proof-of-principle evidence.
In eukaryotic cells, multiple kinases provide obligatory and rapid regulatory switches for controlling mRNA transcription, splicing, and translation (3). P21-activated kinase 1 (Pak1), one of the evolutionarily conserved families of serine/threonine protein kinases, is a downstream effector of the activated Rho GTPases Rac1 and Cdc42, nonreceptor tyrosine kinases lipids, and growth factor receptor-generated mitogenic signals (4, 5). However, to date, Pak1 signaling has not been linked with the functions of RNA-binding proteins. To explore the biologic functions and to discover binding partners of Pak1, we performed a yeast two-hybrid screening of a human mammary gland cDNA library. PolyC-binding protein 1 (PCBP1) was identified as an interacting substrate for Pak1. PCBPs are ubiquitously expressed proteins that contain a highly conserved triple repeat of the KH domain, which is a distinctive feature of RNA-binding proteins. PCBPs participate in a variety of mRNA processing steps and have been implicated in posttranscriptional regulatory pathways, including translation silencing (6, 7). Although PCBP1 exists in both phosphorylated and unphosphorylated forms (8), no function of PCBP1 has yet been associated with its phosphorylation in cells, and no kinase has been shown to phosphorylate PCBP1. Our validation of PCBP1 as a Pak1-binding protein warranted further exploration of PCBP1 functions by this regulatory signaling kinase. Here, we investigated the functions and regulation of PCBP1, and identified a series of previously unrecognized roles for PCBP1, establishing the exciting concept that a single coregulator can function as a signal-dependent and coordinated regulator of transcription, splicing, and translation in eukaryotic cells.
Results and Discussion
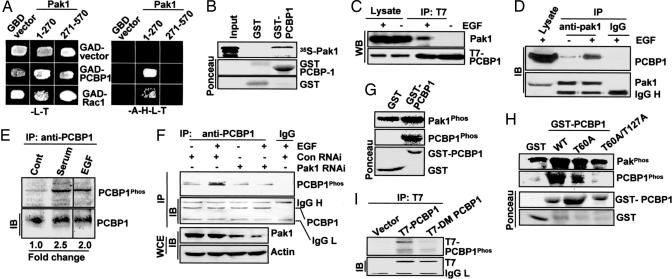
To explore the biologic function and to discover binding partners of Pak1, a major signaling nodule in the mammalian cells (4, 5), we performed a yeast two-hybrid screening of a human mammary gland cDNA library. We identified several RNA-binding proteins and splicing factors as Pak1-interacting proteins [see supporting information (SI) Table 1]. Because the role of Pak1 signaling in the biology of RNA-binding proteins is relatively unexplored, we decided to focus on PCBP1 (GenBank accession no. NM_006196), an evolutionarily conserved RNA-binding protein. PCBPs are ubiquitously expressed and contain a highly conserved triple repeat of the KH domain and participate in a variety of mRNA processing steps (7). The specificity of the Pak1–PCBP1 interaction was verified by cotransfection of Pak1 and PCBP1 plasmids into the yeast cells (Fig. 1A) as well as by the GST-PCBP1 fusion protein pull-down of in vitro translated Pak1 protein (Fig. 1B). Pak1 interacted specifically with amino acids 1–42 and 139–220 of PCBP1, regions overlapping with the first two KH domains of PCBP1; whereas PCBP1 interacted with amino acids 52–75 and 203–270 of Pak1, which contain the PBD and catalytic domains of Pak1 (SI Fig. 6). In vivo association of these two proteins was observed after growth factor stimulation of HeLa cells by coimmunoprecipitation of transient transfected T7-tagged PCBP1 with the endogenous Pak1 (Fig. 1C) and of endogenous Pak1 and PCBP1 (Fig. 1D). These results indicate that mitogen signal is sufficient to engage an in vivo association of Pak1 and PCBP1.
Fig. 1.
Mitogen-mediated phosphorylation of PCBP1 by Pak1. (A) Yeast two-hybrid identification of PCBP1 as Pak1-interacting partner. Yeast cells were cotransfected with a control GAD vector or GAD-PCBP1 along with a GBD vector, GBD-Pak1 constructs. Growth was recorded after 72 h on selection plates lacking leucine and tryptophan (−LT) or adenosine, histidine, leucine, and tryptophan (−AHLT). (B) In vitro GST-pull down for binding of Pak1 and GST-PCBP1. (C) In vivo interaction of T7-PCBP1 with Pak1 in HeLa cells. EGF, 100 ng/ml for 15 min. (D) Endogenous interaction between PCBP1 and Pak1 in HeLa cells. (E) Mitogenes phosphorylate PCBP1 in HeLa cells. Cell lysates were immunoprecipitated (IP) with goat PCBP1 Ab after [32P]orthophosphoric acid labeling and 10-min treatment with mitogens and blotted or autoradiographed as indicated. (F) HeLa cells were transfected with Pak1 or control siRNA, labeled with [32P]orthophosphoric acid, and treated with or without EGF. (G) In vitro phosphorylation of GST-PCBP1 by Pak1. (H) Pak1 phosphorylation of GST-PCBP1 mutant proteins. The two bottom images show the status of GST proteins by Ponceau staining. (I) Generation of MCF-7 pooled clones expressing T7-PCBP1-WT or T7-PCBP1-Mut. (Upper) Clones were metabolically labeled with [32P]orthophosphoric acid, and T7-PCBP1 was IP, and autoradiographed. (Lower) Western blotting of the above blot with an anti-T7 mAb to show equal IP of T7-tagged proteins. IB, immunoblotting.
We next determined whether PCBP1 is phosphorylated by mitogenic signals that activate the Pak1 kinase. We found that human EGF as well as serum stimulated the phosphorylation of the endogenous PCBP1 in HeLa cells (Fig. 1E). Moreover, transfection of Pak1-siRNA abolished EGF-mediated PCBP1 phosphorylation (Fig. 1F), suggesting a mediatory role of Pak1 signaling in growth factor-mediated phosphorylation of PCBP1. We next tested whether Pak1 can directly phosphorylate PCBP1. Pak1 enzyme efficiently phosphorylated GST-PCBP1 in vitro (Fig. 1G). Pak1 phosphorylation of various PCBP1 deletion constructs narrowed Pak1 phosphorylation sites to the N-terminal 1–91 and 91–139 aa fragments of PCBP1 (SI Fig. 7). Sequence analysis of the N-terminal PCBP1 revealed Thr 60 and 127 as potential Pak1 phosphorylation sites. We next substituted these threonines with alanine either individually or together. There was a substantial reduction in the single phosphorylation mutant of PCBP1 (PCBP1-T60A) (Fig. 1H). Pak1 failed to phosphorylate the double mutant PCBP1-T60A-T127A (PCBP1-Mut), thereby confirming that Pak1 phosphorylates PCBP1 on Thr-60 and Thr-127 (Fig. 1H). To show that Pak1 is also the enzyme responsible for Thr-60 and Thr-127 phosphorylation of PCBP1 in vivo, we next generated pooled MCF-7 clones stably expressing T7-PCBP1 or T7-PCBP1-T60A-T127A (PCBP1-Mut) (Fig. 1I). Labeling of these clones with [32P]orthophosphoric acid showed a substantial reduction of phosphorylation of PCBP1 in T7-PCBP1-Mut exponentially growing clones (Fig. 1I). These findings suggested that PCBP1 is a physiologic substrate of Pak1.
PCBP1 Phosphorylation Releases Its Translational Inhibition Activity on L1CAM.
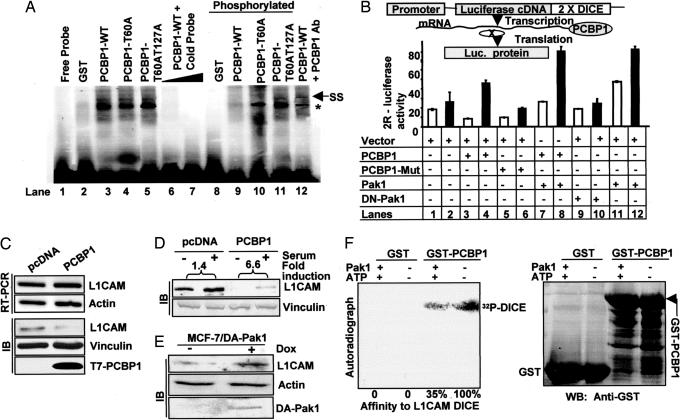
Because PCBP1 binds to the DICE-containing RNA sequence (9), we examined the influence of Pak1 phosphorylation of PCBP1 on the binding affinity of PCBP1 to the DICE sequences from rabbit 15-lipoxygenase (LOX) mRNA, 4R-DICE (9), by EMSA. Purified PCBP1 was bound to 4R-DICE, and PCBP1 phosphorylation by Pak1 enzyme abolished this association (Fig. 2A, lanes 3 and 9). In contrast, there was no change in the binding affinity of PCBP1 with mutant Pak1 phosphorylation sites to the 4R-DICE sequence (Fig. 2A, lanes 4, 5, 10, and 11). The PCBP1 binding to the 4R-DICE sequence was supershifted by an anti-PCBP1 antibody (Fig. 2A, lane 12). Thus, PCBP1 phosphorylation regulates its binding affinity to the DICE sequence.
Fig. 2.
Phosphorylation of PCBP1 by Pak1 relieves translational repression on DICE-containing RNA. (A) EMSA using labeled RNA 4R-DICE with PCBP1-WT and PCBP1-T60A and PCBP1-T60AT127A that were phosphorylated by Pak1 enzyme. ∗, Shifted band of 4R-DICE by PCBP1; SS, super shift with anti-PCBP1 Ab. (B) MDA-435 cells were transfected with Luc-2R luc reporter and with indicated plasmids. (C) Expression of L1CAM mRNA and protein in MCF-7 clones expressing empty vector or T7-PCBP1. (D) MCF-7 clones expressing pcDNA or T7-PCBP1 were serum stimulated for 12 h. (E) Expression of L1CAM protein in MCF-7/DA-Pak1 cells treated with or without doxycycline for 24 h. (F) Membrane with GST, GST-PCBP1, and Pak1 phosphorylated GST, GST-PCBP1, were probed with 32P-DICE of human L1CAM in vitro transcripts (SI Methods).
To study the biologic consequences of PCBP1 phosphorylation by Pak1, we examined the effects of Pak1 and of WT-PCBP1 or its double-phosphorylation mutant on mRNA translation by using a previously characterized luciferase reporter luc-2R that contains two copies of rabbit LOX-DICE sequences in the 3′ UTR of the luciferase cDNA (10) (Fig. 2B). We found that transient expression of PCBP1 in MDA-435 cells inhibited the translation of reporter luciferase mRNA (Fig. 2B, lanes 1 and 3). However, EGF treatment of PCBP1-transfected cells was accompanied by an enhancement of luciferase mRNA translation (Fig. 2B, lanes 3 and 4). The translation-inhibitory effect of mutant PCBP1 by growth factor was modestly derepressed compared with that of cells transfected with WT-PCBP1 (Fig. 2B, lanes 4 and 6). Because mutant PCBP1 interacted with the DICE-RNA sequence (Fig. 2A), the mutant PCBP1 also repressed the translation of reporter luciferase mRNA (Fig. 2B, lanes 5 and 6). Cotransfection of Pak1, but not kinase-dead Pak1 (DN-Pak1), also released the inhibitory translation effect of PCBP1 (Fig. 2B, lanes 7–12). As expected from the above results, inclusion of Pak1 stimulated the basal reporter activity (compare lanes 11 and 12 with lanes 1 and 2). These findings revealed a role for the Pak1-PCBP1 pathway in modulating the DICE sequence-mediated translation.
We next examined the involvement of the Pak1–PCBP1 interaction in regulating the translation status of a natural DICE sequence-containing gene, LICAM, which has been predicted to contain DICE sequences (11) in its 3′ UTR (SI Fig. 8). Although there were no differences in the L1CAM transcripts of the clones expressing the WT or Mut PCBP1, the L1CAM protein level was reduced in MCF-7/PCBP1 cells as compared with MCF-7/pcDNA cells (Fig. 2C). The mitogenic stimulation of cells modestly induced the level of L1CAM protein MCF-7/pcDNA clone (1.4-fold over unstimulated), whereas the amount of LICAM protein was 6.6-fold increased in MCF-7/PCBP1 cells compared with basal level (Fig. 2D). Furthermore, expression of HA-tagged catalytically active Pak1 under an inducible Tet-on promoter in MCF-7/DA-Pak1 cells (12) resulted in an increased level of L1CAM expression as compared with DA-Pak1 cells without induction of Pak1 expression (Fig. 2E). To demonstrate that the inhibitory effect of PCBP1 on L1CAM translation was mediated by means of its DICE sequence, we cloned the natural 2× DICE sequence from the 3′ UTR region of human L1CAM (39 bp, from residues 3822 to 3860 of L1CAM, accession No. M77640) into pBluescript KS+. The DICE sequence was in vitro transcribed and radiolabeled and tested for its binding affinity to PCBP1 by a Northwestern assay (SI Methods). PCBP1 binds to L1CAM DICE, and preactivation of PCBP1 by Pak1 enzyme significantly reduced its association with the L1CAM DICE (Fig. 2F). These findings indicate that activation of Pak1, a kinase important in cellular signaling (4), could modulate expression level of L1CAM, an adhesion molecule that plays important roles in cell migration (13). These results suggest an essential role of Pak1-PCBP1 signaling in regulating the translation of endogenous L1CAM RNA, a DICE sequence containing PCBP1 target.
Pak1 Signaling Is Required for PCBP1 Localization in the Nucleus.
Because PCBP1 contains nuclear localization signals (14), and because it has been shown to function as a transcription factor for mouse mμ-opioid receptor gene (15), we hypothesized that Pak1 signaling might be important for PCBP1's nuclear localization and, consequently, its putative nuclear functions. We found that transiently expressed T7-PCBP1 was localized in both the cytoplasmic and nuclear compartments, whereas T7-PCBP1-Mut was predominantly localized in the cytoplasm (SI Fig. 9A). Because some of the mut-PCBP1 appears to be in the nucleus, it is possible that there could be more potential in vivo phosphorylation sites in PCBP1 other than T60 and T127 that are Pak1-sensitive. Transfection of Pak1-siRNA in HeLa cells resulted in an increased cytoplasmic PCBP1 (SI Fig. 9B), suggesting a role for Pak1 phosphorylation in PCBP1 nuclear localization. Biochemical fractionation of MCF-7 clones stably expressing either T7-PCBP1 or T7-PCBP1-Mut confirmed an increased accumulation of T7-PCBP1 in the nucleus as compared with the nuclear level of T7-PCBP1-Mut (Fig. 3A), thus revealing a role of Pak1 signaling in compartmental retention of PCBP1 to the nucleus.
Fig. 3.
PCBP1 phosphorylation controls its nuclear localization and eIF4E transactivation. (A) Nuclear and cytoplasmic extracts from MCF-7 clones expressing pcDNA or T7-PCBP1 were blotted with indicated antibodies. (B) Induction of DA-Pak1 in MCF-7 cells up-regulates eIF4E mRNA (Left), and Pak1siRNA down-regulates eIF4E mRNA in MCF-7 cells (Right). (C) eIF4E mRNA in MCF-7 cells stably expressing PCBP1 or PCBP1-Mut. (D) Levels of eIF4E mRNA in HeLa cells transfected with PCBP1 siRNA for 3 d. (E) HeLa cells were treated with serum or EGF for 45 min, and ChIP analysis was performed on an eIF4E promoter fragment (678–873 bp, GenBank accession no. NM_001968). (F) MCF-7 cells expressing T7-PCBP1 or T7-PCBP1-Mut were treated with EGF and were subjected to ChIP on eIF4E promoter. (G) HeLa cells treated with con- or PCBP1-siRNA, transfected with eIF4E core promoter p543-Luc, and stimulated with EGF for 12 h before the luc assay. (H) MCF-7/PCBP1 clones were transfected with p543-Luc and stimulated with EGF.
PCBP1 Is a Transcriptional Activator of eIF4E.
Because PCBP1 acts as a transcription activator in the neuronal system (15), we explored the effect of PCBP1 on the transcription of specific genes. While our study was in progress, Lynch et al. (16) demonstrated that hnRNP K, another PCBP family member, specifically binds to and transactivates the basal element of the TATA-less proximal promoter of eIF4E. To test whether the Pak1-PCBP1 pathway participates in the regulation of eIF4E transcription, we evaluated the effect of modifying Pak1 status on eIF4E transcription. Expression of catalytically active Pak1 resulted in an increased the level of eIF4E mRNA; conversely, depletion of endogenous Pak1 expression by siRNA in MCF-7 cells led to reduced levels of eIF4E mRNA (Fig. 3B). Importantly, MCF-7 cells stably expressing T7-PCBP1, but not T7-PCBP1-Mut, exhibited an increased level of eIF4E mRNA (Fig. 3C). In contrast, ablation of PCBP1 by siRNA decreased the level of eIF4E mRNA in HeLa cells (Fig. 3D).
To understand the mechanism of PCBP1-mediated transactivation of eIF4E, we carried out ChIP analysis in HeLa cells. We found an increased recruitment of PCBP1 to the eIF4E promoter region that contains eIF4E basal element (4EBE), in cells stimulated with serum or EGF (Fig. 3E). The necessity of Pak1 phosphorylation of PCBP1 in growth factor-activated targeting of PCBP1 to the eIF4E promoter was demonstrated by the inability of EGF to further stimulate PCBP1-Mut recruitment to the eIF4E promoter over detectable basal levels (Fig. 3F). To demonstrate the inherent role of PCBP1 in stimulating the transcription of eIF4E, we found that PCBP1 siRNA reduces promoter activity measured by an eIF4E promoter luciferase plasmid containing 4EBE sequence (Fig. 3G). Also, transfection of WT-PCBP1, but not Mut-PCBP1, in MCF-7 cells led to a reproducible increase in eIF4E promoter activity (Fig. 3H). These findings suggested that PCBP1 and its phosphorylation by growth factors play a regulatory role in eIF4E transcription in growth factor-stimulated cells.
Pak1-PCBP1 Signaling Nodule Regulates mRNA Splicing.
Because Pak1-interacting proteins identified in the yeast two-hybrid screening also included several splicing factors (SI Table 1), we hypothesized that Pak1 signaling and the downstream substrate PCBP1 also could affect pre-mRNA splicing. In examining this hypothesis, we showed that most of the nuclear PCBP1 in HeLa cells colocalized with the sc-35 splicing factor in the nucleus (SI Fig. 10), thus, highlighting the possibility of a role for PCBP1 in pre-mRNA splicing. Next, we determined whether Pak1 or its upstream stimulators (such as serum or EGF) are involved in pre-mRNA splicing by using the luciferase-based splice reporter system (17) pETCatEBLucv5, which, upon transient expression, can generate two possible mature mRNAs depending on inclusion of the CD44 v5 exon in the reporter gene mRNA during posttranscriptional splicing. HeLa cells transfected with pETCatEBLucv5 were stimulated with growth factors, and luciferase activity was measured. The splicing activity on CD44 v5 exon in pETCatEBLucv5 was enhanced on EGF stimulation (Fig. 4A). Up-regulation of luciferase fusion gene mRNA because of enhanced specific splicing of CD44 v5 exon was shown by RT-PCR (Fig. 4A Inset). We found that Pak1 promoted splicing activity on the pETCatEBLucv5 minigene in a dose-responsive manner (Fig. 4B); kinase-dead mutant Pak1-K299R failed to stimulate minigene splicing activity; however, kinase-active Pak1-T423E promoted minigene splicing activity (Fig. 4C). To validate these results, we used another CD44 splicing minigene reporter (18) and found a 4-fold enhancement of CD44 v4 and v5 exon splicing in the cells cotransfected with Pak1 and HSV-CD44 (Fig. 4D). In brief, Pak1 signaling promotes alternative RNA splicing, presumably because of phosphorylation of its substrates with a putative role in splicing.
Fig. 4.
Pak1 and PCBP1 regulate pre-mRNA splicing. (A) MCF-7 cells transfected with pETCatEBLucv5 were treated with serum or EGF for 16 h, and minigene transcripts and splicing activity was assayed. RT-PCR was performed to analyze splicing products. Lower band is the splicing product with CD44 v5 skipped, and the upper band is the product with v5 included. (B) MCF-7 cells were cotransfected with pETCatEBLucv5 and Pak1 plasmid, and luc activity was measured. (C) MCF-7 cells were transfected with indicated plasmids along with pETCatEBLucv5, and luc activity was measured. (D) HeLa cells were cotransfected with HSV-CD44 splicing minigene and Pak1 and were subjected to RT-PCR analysis of minigene transcripts. Splicing activity on this minigene was represented by the ratio of transcripts of included exons (bands a and b) to transcripts of both exons excluded (band c). (E) MCF-7 cells were cotransfected with pETCatEBLucv5 and different amounts of PCBP1-WT or PCBP1-Mut and were assayed for the luc activity. (F) MCF-7 cells were cotransfected with HSV-CD44 and PCBP1-WT or PCBP1-Mut plasmids and subjected to RNA isolation and RT-PCR analysis of minigene transcripts. (G) MCF-7 clones were transfected with pETCatEBLucv5 splicing minigene reporter, and luc activity was assayed. (H) HeLa cells cotransfected with PCBP1-siRNA with Pak1 or control plasmid and pETCatEBLucv5 minigene and assayed for the luc-activity. (I) HeLa extracts were immunoprecipitated with a Caper α antibody (Left) or PCBP1 antibody (Right) and were followed by IB with indicated antibodies. (J) HeLa cells were treated with con or Caper α-specific siRNA, cotransfected with pcDNA-PCPB1 and pETCatEBLucv5 splicing minigene, and luc activity was measured.
We next explored whether PCBP1 could act as a mediator of Pak1-mediated pre-mRNA splicing. We found that coexpression of PCBP1-WT, but not PCBP1-Mut, enhanced pre-mRNA splicing-generated luciferase activity (Fig. 4E). Similar results were obtained when the HSV-CD44 minigene was used (Fig. 4F). Accordingly, there was a significant reduction in the splicing of the minigene in MCF-7/PCBP1-Mut clone as compared with MCF-7/PCBP1 clones (Fig. 4G). Further selective knockdown of PCBP1 in HeLa cells also prevented the ability of Pak1 to stimulate splicing activity on the pETCatEBLucv5 minigene (Fig. 4H). Together, these findings suggested a role for PCBP1 in the modulation of cellular pre-mRNA splicing.
Caper α Is a Downstream Effector of PCBP1-Mediated PremRNA Splicing.
Evidence does not exist for a direct role of PCBP1 in modifying the recruitment or assembly of spliceosome components to pre-mRNA splicing sites. Consequently, the splicing effect of PCBP1 that we discovered might be mediated indirectly by other splicing factors. Recently we found that an endogenous PCBP family protein (19) coimmunoprecipitates with a U2 snRNP auxiliary factor-related protein Caper-α implicated in splicing (20). Consequently, we explored whether PCBP1 interacts with Caper α and whether this interaction contributes to the splicing-promoting activity of PCBP1. In this context, we found that endogenous PCBP1 interacts with Caper α in HeLa cells (Fig. 4I). We next showed that PCBP1 directly binds to Caper α and uses its two binding regions, namely amino acids 56–91 (overlapping with the first KH domain) and 220–356 (full third KH domain) to interact with Caper α (SI Fig. 11). Not surprisingly, PCBP1 binds to the C-terminal RNA recognition motif of Caper α (SI Fig. 11). To demonstrate the significance of the interaction between PCBP1 and Caper α in PCBP1-mediated pre-mRNA splicing in HeLa cells, we knocked down Caper α and found a reduction in the RNA splicing-promoting activity of PCBP1 on the minigene (Fig. 4J). These observations demonstrated an inherent role of Caper α, at least in-part, in the regulation of Pak1-PCBP1-mediated pre-mRNA splicing activation.
In summary, we have defined a signal-dependent mechanism that engages multiple distinct, regulatable cellular molecules in the eukaryotic gene expression machinery in response to environmental growth signals. We propose a model that consists of three nodes (i.e., transcription, splicing, and translation), each one of which is efficiently modulated by PCBP1 (Fig. 5). Our findings suggest that growth factor signaling could simultaneously target various pools of PCBP1 to each of the three levels of the gene expression machinery. Moreover, in this model, modulation of one node (e.g., transcription) by PCBP1 attunes the productivity of another node (e.g., translation). We show that growth signals activate Pak1, which in turn phosphorylates PCBP1 on T60 and T127 and contradicts PCBP1's inhibitory function on mRNA translation. Phosphorylated PCBP1 then in turn stimulates transactivation of the eIF4E gene promoter. Thus, growth factor signaling-dependent phosphorylation of PCBP1 coordinately regulates the stimulation of the eIF4E promoter while derepressing translation inhibition through release of the inhibitory binding of unphosphorylated PCBP1 to mRNA templates containing the DICE sequence. Finally, a potential for promotion of alternative RNA splicing by phosphorylated PCBP1 was revealed by demonstrating alternative splicing from a CD44 minigene, aiding the accelerated accumulation of mature mRNAs that can serve as templates for translation. This type of coordinated regulation of distinct cellular compartmental functions would not only ensure efficient cellular response to environmental changes, but perhaps more importantly, the rapidity and reversibility of regulation of phosphorylation underscores the economy of this mechanism in coordinating the actions of three modules of the gene expression machinery. These findings substantiate the concept that a single coregulator molecule can function as a signal-dependent and coordinated regulator of transcription, splicing, and translation in eukaryotic cells.
Fig. 5.
Working model for signaling-dependent alterations of the nuclear and cytoplasmic activities of PCBP1. Upstream activators of the Pak1 pathway induce Pak1 kinase activity wherein Pak1 exists in an autophosphorylated state. The active Pak1 exerts both cytoplasmic and nuclear functions. PCBP1 gets phosphorylated by Pak1 in the cytoplasmic compartment, and phosphorylation reduces its RNA-binding capabilities, thus releasing the translational repression on specific target mRNAs. In the nucleus, the phosphorylated PCBP1 is involved in splicing and transcriptional activities. Whether PCBP1 is phosphorylated by Pak1 in the nucleus after translocation or the phosphor-PCBP1 moves to the nucleus from the cytoplasm is still not known. Phosphorylation induced by Pak1 results in enhanced binding to recently transcribed mRNA in the nucleus to influence the splicing machinery via CAPER α. Furthermore, phosphorylated PCBP1 in the nucleus also gets recruited to promoters of target genes and regulates transcriptional activity of the gene. Broken lines represent the events that are not fully understood.
Methods
Cell Cultures and Reagents.
Cells were maintained in DMEM/F-12 (1:1) supplemented with 10% FBS. EGF and antibodies against vinculin, actin, and SC-35 were from Sigma (St. Louis, MO). Antibodies against PCBP1 were from Santa Cruz Biotechnology (Santa Cruz, CA), Caper α was from Bethyl Laboratories (Montgomery, TX), and Pak1 was from Cell Signaling Technology (Beverly, MA). SMART POOL siRNA was purchased from Dharmacon (Lafayette, CO).
Two-Hybrid Library Screening and Mutagenesis.
Pak1 cDNA sequences for amino acids 1–270 and amino acids 271–545 were cloned into the Gal4-binding domain vector pGBD as baits to screen a mammary gland cDNA library fused to the Gal4 activation domain. Cloning and site-specific mutagenesis and deletion of various complimentary DNA was done by using primers as described in SI Table 2.
Biochemical Assays.
The GST pull down, Western blotting, immunoprecipitation, splicing and luc-reporter assay, Pak1 activity, EMSA, ChIP and Northwestern assays were performed as described in SI Methods.
Immunofluorescence and Confocal Studies.
The cellular location of proteins was determined by using indirect immunofluorescence as described earlier (21) and summarized in SI Methods.
Supplementary Material
Acknowledgments
We thank Mike Kiledjian (Rutgers University, Piscatway, NJ) for providing pGEX6P-α CP1, Rau Andino (University of California, San Francisco, CA) for pMAL-PCBP1 plasmids, Mathias W. Hentze (European Molecular Biology Laboratory, Heidelberg, Germany) for Luc-2R plasmid, S. Weg-Remers (Institute of Toxicology and Genetics, Karlsruhe, Germany) for pETCatEBlucv5 plasmid, Muxiang Zhou (Emory University School of Medicine, Atlanta, GA) for eIF4E promoter luc plasmid p543-luc, Vance Lemon (Case Western Reserve University, Cleveland, OH) for providing the L1CAM antibody, Christopher Barnes for confocal staining, and Joseph Mascarenhas for cloning the L1CAM DICE sequence. This study was supported by National Institutes of Health Grants CA90970 and CA80066 (to R.K.).
Abbreviations
- Pak1
p21-activated kinase 1
- PCBP1
poly-C-RNA-binding protein 1
- eIF4E
eukaryotic translation initiation factor 4E
- DICE
differentiation control element.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701065104/DC1.
References
- 1.Auboeuf D, Honig A, Berget SM, O'Malley BW. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 2.Masuhiro Y, Mezaki Y, Sakari M, Takeyama K-i, Yoshida T, Inoue K, Yanagisawa J, Hanazawa S, O'Malley B, Kato S. Proc Natl Acad Sci USA. 2005;102:8126–8131. doi: 10.1073/pnas.0503197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin C, Manley JL. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 4.Bokoch GM. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 5.Kumar R, Gururaj AE, Barnes CJ. Nat Rev Cancer. 2006;6:459–470. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 6.Kiledjian M, DeMaria CT, Brewer G, Novick K. Mol Cell Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong J, Ji X, Liebhaber SA. Mol Cell Biol. 2003;23:1125–1134. doi: 10.1128/MCB.23.4.1125-1134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leffers H, Dejgaard K, Celis JE. Eur J Biochem. 1995;230:447–453. [PubMed] [Google Scholar]
- 9.Barnhart KM, Kim CG, Banerji SS, Sheffery M. Mol Cell Biol. 1988;8:3215–3226. doi: 10.1128/mcb.8.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 11.Reimann I, Huth A, Thiele H, Thiele BJ. J Mol Biol. 2002;315:965–974. doi: 10.1006/jmbi.2001.5315. [DOI] [PubMed] [Google Scholar]
- 12.Vadlamudi RK, Adam L, Wang RA, Mandal M, Nguyen D, Sahin A, Chernoff J, Hung MC, Kumar R. J Biol Chem. 2000;275:36238–36240. doi: 10.1074/jbc.M002138200. [DOI] [PubMed] [Google Scholar]
- 13.Gavert N, Conacci-Sorrel M, Gast D, Schneider A, Altevogt P, Brabletz T, Ben-Ze'ev A. J Cell Biol. 2005;168:633–642. doi: 10.1083/jcb.200408051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chkheidze AN, Liebhaber SA. Mol Cell Biol. 2003;23:8405–8415. doi: 10.1128/MCB.23.23.8405-8415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SS, Pandey KK, Choi HS, Kim SY, Law PY, Wei LN, Loh HH. Mol Pharmacol. 2005;68:729–736. doi: 10.1124/mol.105.012245. [DOI] [PubMed] [Google Scholar]
- 16.Lynch M, Chen L, Ravitz MJ, Mehtani S, Lorenblat K, Pazin MJ, Schmidt EV. Mol Cell Biol. 2005;25:6436–6453. doi: 10.1128/MCB.25.15.6436-6453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weg-Remers S, Ponta H, Herrlich P, Konig H. EMBO J. 2001;20:4194–4203. doi: 10.1093/emboj/20.15.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auboeuf D, Honig A, Berget SM, O'Malley BW. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 19.O'Malley BW. 2003 Nursa database http://www.nursa.org/corip.cfm?detail=assay&dataset=25&ex_no=25.1.
- 20.Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O'Malley BW. Mol Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 21.Gururaj AE, Singh RR, Rayala SK, Holm C, den Hollander P, Zhang H, Balasenthil S, Talukder AH, Landberg G, Kumar R. Proc Natl Acad Sci. 2006;103:6670–6675. doi: 10.1073/pnas.0601989103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.