Abstract
The intracellular events on the recycling pathway that lead from sorting endosomes to exocytosis at the plasma membrane are central to cellular function. However, despite intensive study, these processes are poorly characterized in spatial and dynamic terms. The primary reason for this is that, to date, it has not been possible to visualize rapidly moving intracellular compartments in three dimensions in cells. Here, we use a recently developed imaging setup in which multiple planes can be simultaneously imaged within the cell in conjunction with visualization of the plasma membrane plane by using total internal reflection fluorescence microscopy. This has allowed us to track and characterize intracellular events on the recycling pathway that lead to exocytosis of the MHC Class I-related receptor, FcRn. We observe both direct delivery of tubular and vesicular transport containers (TCs) from sorting endosomes to exocytic sites at the plasma membrane, and indirect pathways in which TCs that are not in proximity to sorting endosomes undergo exocytosis. TCs can also interact with different sorting endosomes before exocytosis. Our data provide insight into the intracellular events that precede exocytic fusion.
Keywords: immunoglobulin G, multifocal plane imaging, neonatal Fc receptor, sorting endosome, total internal reflection
Recycling and exocytosis of receptors and their cargo represent fundamental trafficking processes that have been intensively studied by using both biochemical and imaging techniques (1–3). The use of total internal reflection fluorescence microscopy (TIRFM) (4) has led to significant insight into the nature of exocytic events (5–10). In this technique, a thin layer of the cell adjacent to the coverglass is illuminated, which allows for high-sensitivity imaging of events on the plasma membrane. However, the illumination of only a thin section of the cell, including the plasma membrane, has the limitation that events cannot be visualized that precede exocytosis at sites deeper within the cell (11). As a consequence, there is uncertainty concerning the nature of the processes that lead to exocytic release (12–15). For example, the origin of the exocytosing vesicles or tubules is typically difficult to determine, and it therefore remains a matter of controversy as to whether recycling from sorting endosomes involves direct fusion of endosomes or some vesicular/tubular intermediates (16). Resolving such issues is of central importance to understanding intracellular trafficking.
To address such uncertainties, we have introduced a microscope configuration that permits the simultaneous imaging of multiple focal planes within a cell (17). In particular, this microscopy setup allows for the imaging of doubly labeled cellular components on the plasma membrane by using total internal reflection illumination and, at the same time, in the interior of the cell by using epifluorescence. In this way, dynamic processes such as the movement of transport containers (TCs) from the cell interior to the plasma membrane that would be difficult to track by using other imaging methods can be visualized.
In this study, we have analyzed events preceding exocytosis in the recycling pathway taken by the MHC class I-related receptor, FcRn (18). Through its ability to transport IgGs within and across cells, this Fc receptor is known to play a central role in the maintenance of serum IgG levels and in the trafficking of IgG molecules across cellular barriers (19–26). The available data support the model (27) that IgGs are taken up by cells through fluid-phase pinocytosis. The pH dependence of the IgG–FcRn interaction is such that binding is relatively strong at pH 6.0 but becomes negligible as near-neutral pH is approached (28–31). Following uptake into cells, IgG can therefore bind to FcRn in slightly acidic sorting endosomes (32). FcRn subsequently transports bound IgG from the sorting endosomes to the plasma membrane, where cargo is released at near-neutral pH during exocytic events (10).
Our analyses provide insight into the intracellular processes that lead to exocytosis of FcRn and its IgG cargo. In particular, we describe distinct classes of events that precede exocytic fusion. Representing a direct pathway from sorting endosomes to exocytosis, we observe tubular TCs that in some cases appear to form contiguous connections between the endosome and plasma membrane. Before exocytosis, TCs can also arrest for varying time periods at intracellular locations that are distal to sorting endosomes. We therefore observe direct and indirect pathways to exocytosis and provide dynamic insight into the intracellular events that constitute these pathways. Visualization of these pathways provides insight into how FcRn functions as a salvage receptor within cells to regulate IgG transport and levels.
Results
The primary focus of this study was to investigate intracellular trafficking events that precede exocytosis. We have shown previously that human FcRn sorts bound IgG into a recycling pathway that overlaps with that taken by the transferrin receptor (32). This sorting occurs in early endosomes that, based on transferrin and dextran trafficking, can be defined as sorting endosomes. Sorting is followed by exocytic events at the plasma membrane that involve FcRn (10). To analyze FcRn trafficking, we have used endothelial cells transfected with FcRn–fluorescent protein constructs in combination with a multiplane imaging setup, which allows the simultaneous visualization of fluorescently labeled cellular components in several planes within the cell by using epifluorescence and on the surface of the cell by using TIRFM (17). To detect FcRn, we have used constructs in which FcRn is tagged with fluorescent proteins at either the N or C terminus. The location of the fluorescent protein with respect to FcRn does not appear to affect the results. To facilitate the visualization of exocytic events in most of the experiments, a pH-sensitive variant of GFP [ecliptic pHluorin (33)] that has substantially higher fluorescence emission at near-neutral pH relative to acidic pH has been used [see supporting information (SI) Text, Sections 1 and 5]. For the visualization of events by using simultaneous TIRF (membrane plane) and epifluorescence (upper planes) imaging, cells were cotransfected with constructs encoding FcRn tagged with GFP/pHluorin and mRFP (see SI Text, Section 5 for the rationale for the different experimental setups used).
Direct Delivery of Transport Containers from Sorting Endosomes to Exocytic Sites.
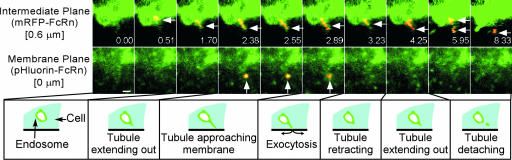
We observed that multiple exocytic events can occur in close proximity to sorting endosomes (Fig. 1, see also SI Figs. 4 and 5 and SI Movies 1–5). After visual inspection, exocytic events were verified by using quantitative methods analogous to those described in refs. 6 and 7 (see also SI Text, Section 6). In some cases, tubular TCs can be seen extending from sorting endosomes and migrating toward the plasma membrane (Fig. 1). In the data presented in Fig. 1, the tip of the tubule (≈1.5 μm long) is seen to merge with the plasma membrane in an exocytic event that lasts for ≈0.3 s. In the example shown, exocytic fusion is followed by retraction of the tubule back to the sorting endosome (within ≈2 s after exocytosis). In a second example, two sequential fusion events (≈9 s apart) involve a tubule that extends from a sorting endosome and appears to fragment before the second fusion event, so that the “free” TC is involved in exocytosis (SI Fig. 6 and SI Movies 6 and 7). In contrast, the data shown in Fig. 1 suggest that the tubule is connected to the sorting endosome during the exocytic event. However, whether this connection is maintained continuously throughout the analysis period is uncertain because of the limitations of the sensitivity of fluorescence imaging. In addition, tubular TCs can fuse along their “edges” with the plasma membrane rather than at their tips (SI Fig. 7 and SI Movies 8–10), consistent with earlier studies involving TIRFM imaging of the biosynthetic pathway (34).
Fig. 1.
Partial exocytic fusion of a tubule extending from a sorting endosome. HMEC-1 cells cotransfected with pHluorin-FcRn and mRFP-FcRn were imaged. Individual images showing an area of interest of a cell are presented with the time (in seconds) at which each image was acquired (first image is arbitrarily set to time 0). FcRn is shown in green, and the events of interest are highlighted in red (resulting in yellow–orange overlay). A tubule (leftward arrows) extends from a sorting endosome in the intermediate plane (0.51–2.38 s) and approaches the plasma membrane (2.38–2.55 s, upward arrows). Partial fusion (upward arrow) is observed on the membrane plane (2.89 s) at the tip of this tubule while it appears to remain connected to the sorting endosome. Immediately after exocytosis, this tubule retracts toward the sorting endosome (3.23 s), subsequently detaches, and moves away from the sorting endosome. Images shown are individual frames of SI Movie 1. The images have been presented without highlighting and also as surface displays in SI Fig. 4 (with corresponding SI Movies 2 and 3, respectively). (Scale bar, 1 μm.)
Exocytic Events Involving Transport Containers That Are Distal to Sorting Endosomes.
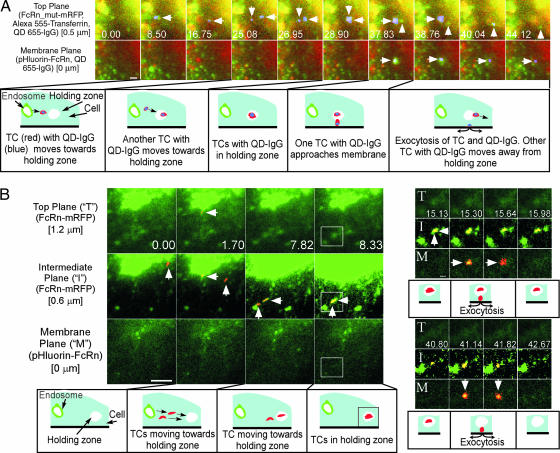
In contrast to the data described above, exocytosis can also involve TCs that are not in proximity to sorting endosomes (Fig. 2 A and B; see also SI Fig. 8 and SI Movies 11–14). In these events, TCs can travel a significant distance within the cell before becoming stationary above the plasma membrane. The TCs typically stay in these “holding zones” for significant amounts of time (mean: 7.7 ± 4.8 s, n = 18) before exocytosing. The existence of such a holding zone can be observed above the focal plane in Fig. 2A, where QD655-labeled IgG is seen leaving a sorting endosome in an FcRn-positive TC (<0.5 μm in diameter). In these analyses, a mutated human IgG1 variant (MST-HN) that has higher affinity relative to wild-type IgG1 (35) was used in combination with a mutated variant of human FcRn (FcRn_mut) (36) tagged with mRFP. The FcRn_mut:MST-HN interactions at both pH 6.0 and near-neutral pH have substantially higher affinities relative to the corresponding wild-type human FcRn:human IgG1 interactions [at pH 6.0, KD1 = 0.7 nM for FcRn_mut:MST-HN vs. 528 nM for the wild-type molecules; at pH 7.2, KD1 = 4.5 nM for FcRn_mut:MST-HN vs. immeasurably low affinity for the wild-type molecules; (35) and SI Text, Section 2]. This allows efficient uptake of QD655–IgG complexes in medium at near-neutral pH when IgG concentrations of 10 nM are used. In addition, flow cytometry and microscopy analyses indicate that QD655–IgG (MST-HN) uptake is specific and FcRn-dependent (see SI Text, Sections 3 and 4). Importantly, the involvement of cargo (IgG) in this series of events indicates that this trafficking is on the recycling, rather than the biosynthetic, pathway. After the departure of the TC from the sorting endosome, a second compartment containing QD-labeled IgG approaches and intersects with the earlier compartment (Fig. 2A). Subsequently, (≈9 s after intersection of TCs), one TC is seen exocytosing, whereas the other TC (visible in a focal plane that is 0.5 μm above the plasma membrane) moves away from the trafficking intersection.
Fig. 2.
TCs can migrate into “holding zones” before exocytosis. Individual images showing areas of interest of transfected HMEC-1 cells are presented with the time (in seconds) at which each image was acquired (first image is arbitrarily set to time 0). (A) Cells cotransfected with pHluorin-FcRn and FcRn_mut-mRFP (36) were incubated in medium (≈pH 7.3) with QD655-human IgG1 mutant [MST-HN (35)] complexes and Alexa Fluor 555-labeled transferrin before and during imaging. FcRn and transferrin (detected in the same channel) are indicated by green and QD-IgG complexes by red. The events of interest are highlighted in blue. An FcRn/transferrin-positive TC with IgG (leftward arrow) leaves a sorting endosome at 8.50 s. Later, another TC containing IgG (downward arrows) enters the same holding zone (28.90 s). One of the TCs exocytoses (38.76 s), releasing IgG (rightward arrows) on the membrane plane. The second TC (upward arrows) moves away from the holding zone in the top plane. The images have been presented as surface displays in SI Fig. 8A and B (with corresponding SI Movie 12). (Scale bar, 1 μm.) (B) HMEC-1 cells cotransfected with pHluorin-FcRn and FcRn-mRFP were imaged. FcRn is shown in green, and the events of interest are highlighted in red (resulting in yellow–orange overlay). A tubule (leftward arrows) leaves a sorting endosome (1.70 s) and enters a holding zone (7.82–8.33 s) in the intermediate plane in proximity to a second TC (upward arrows). One of the TCs fuses with the plasma membrane at 15.30–15.64 s (rightward arrows) and the second TC subsequently exocytoses at 41.14–41.82 s (downward arrows). Rectangles marked at 8.33 s are subsequently presented as cropped images. Focal planes marked as “T,” “I,” and “M” denote top plane, intermediate plane, and membrane plane, respectively, in the cropped images. Individual images in A and B are from SI Movies 11 and 13, respectively. The images have been presented without highlighting in SI Fig. 8C (with corresponding SI Movie 14). [Scale bars: 5 μm (Bottom Left) and 1 μm (Upper Right, in frame marked with “M”).]
Similar intersection of TCs can also be seen where there is a significant delay (≈7 s) between the “aggregation” event and exocytic events (Fig. 2B). In Fig. 2B, two FcRn+ TCs (≈1 and 2 μm long) enter a holding zone that can be visualized in the upper focal plane. One of these TCs originates (visible in two focal planes that are 0.6 and 1.2 μm above the plasma membrane) from a sorting endosome before traveling ≈6 μm within the cell, again demonstrating that the recycling pathway is being analyzed. The movement of TCs into the holding zone is followed by two exocytic events ≈7 and 33 s later. In addition, the behavior associated with holding zones can be more complex than the events shown in Fig. 2, with multiple entering and leaving events of TCs (SI Fig. 9 and SI Movies 15 and 16).
Importantly, the data in Fig. 2A also show that the immediate fate of the TC in the holding zone is not predetermined, i.e., it can exocytose directly below the holding zone or it can travel above the plasma membrane away from the holding zone. In addition, the different timing and destinations of the TCs entering and leaving holding zones is an indication that the TCs do not fuse with each other at these sites.
Small Vesicles Can Be Associated with the Triggering of Exocytic Events.
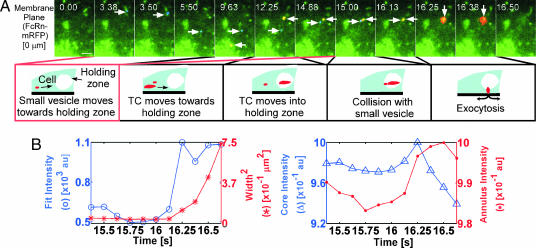
The question arises as to what determines whether and when a TC that is in a holding zone will exocytose. Interestingly, in some cases small vesicles can be observed that collide with larger TCs, and these interactions can be rapidly followed by exocytic events. In Fig. 3(see also SI Fig. 10 and SI Movies 17 and 18), movement of the TC (≈1 μm long) in the membrane plane is initially observed, followed by a stationary phase and interaction with a small vesicle that appears to trigger exocytosis. Fig. 3B shows the intensity width-square and core annulus intensity plots for this exocytic event (see SI Text, Section 6.3 for details of quantitative methods used). In a second example of such a “triggering” event (SI Fig. 11 and SI Movies 19 and 20), a TC is observed that moves from the upper plane (0.6 μm) to the membrane plane, where it arrests for ≈10 s. A small vesicle (<0.5 μm in diameter) moves from the upper plane to the membrane plane and appears to interact with this TC. This interaction is followed within ≈0.3 s by an exocytic event.
Fig. 3.
Exocytosis of a TC can be preceded by interaction with a smaller vesicle. HMEC-1 cells cotransfected with FcRn-GFP and FcRn-mRFP were imaged. (A) Individual images showing area of interest of a cell are presented with the time (in seconds) at which each image was acquired (first image is arbitrarily set to time 0). FcRn is shown in green, and events of interest are highlighted in red (resulting in yellow–orange overlay) or blue. A small vesicle (highlighted in blue, rightward arrows) moves away from a tubule in the membrane plane (3.38–5.50 s) and then appears to be stationary for ≈7 s. A TC (highlighted in red, leftward arrows) moves ≈6 μm in the membrane plane (9.63–12.25 s) before becoming stationary (12.25–14.88 s). Subsequently, the vesicle (highlighted in blue) moves and appears to interact with the TC at 15.00 s. After this collision, the TC fuses (within ≈1 s) with the plasma membrane in an exocytic event (16.25–16.38 s, downward arrows), which lasts for ≈0.1 s. Images shown are individual frames of SI Movie 17. The images have been presented without highlighting in SI Fig. 10 (with corresponding SI Movie 18). (Scale bar, 2 μm.) (B) Quantitative analysis of the exocytic fusion event. (Left) The fit intensity width-square plot as a function of time. (Right) The core annulus intensity plot as a function of time for the same TC. SI Text, Section 6.3 describes further details of the analytical approaches used to analyze exocytic events.
Such “triggering” vesicles are not seen before all exocytic events. However, the vesicles that can be detected are small with very low signal levels and may therefore be below the level of detection in some cases. It is, however, also possible that the “triggering” vesicles are not causally related to the exocytic process and are therefore not associated with every exocytic event.
TCs Can Have Indirect Itineraries.
TCs, some of which are seen to leave sorting endosomes, can take different intracellular pathways that involve their interaction with distinct sorting endosomes. In SI Fig. 12 (see also SI Movies 21 and 22) a tubular TC (≈1.5 μm long, initially visible in a focal plane that is 1.3 μm above the plasma membrane), leaves a sorting endosome and then travels to the membrane plane, where it remains for ≈20 s before moving to a higher focal plane, where it appears to fuse with a second sorting endosome. Alternatively, SI Fig. 13 (see also SI Movies 23 and 24) shows a tubular TC (≈2 μm long, initially visible in two focal planes that are 0.5 μm and 1.1 μm above the plasma membrane) that interacts with an extended tubule from a sorting endosome and then travels ≈4 μm to a holding zone, where it remains for ≈5 s before it moves to the plasma membrane and exocytoses. Collectively, the data show that TCs can interact with multiple sorting endosomes within a cell and, most importantly, that the pathway of a TC to exocytosis can involve more than one interaction with sorting endosomes.
Discussion
Recycling of receptors and their cargo is a fundamental aspect of cell biological processes. FcRn-mediated recycling of IgG represents an example of a salvage pathway where cargo interacts with cognate receptor in sorting endosomes and is subsequently transported to the membrane, where exocytic release occurs (10, 32). These exocytic events can be visualized by using TIRFM (10). However, to date, there is a paucity of data concerning the nature of the intracellular trafficking events that comprise the recycling pathway and lead to exocytosis of FcRn and other recycling receptors. As a result, the nature of these processes has been the subject of controversy (12–16). In this study, we have investigated these pathways using a multiplane imaging modality that allows for the simultaneous observation of events in multiple focal planes within and at the surface of a cell. As such, this permits the tracking of rapidly moving intracellular compartments, which might not be possible to image by using more conventional approaches.
We show that different types of intracellular events can precede exocytosis. The events can be broadly classified into direct and indirect pathways: the direct pathway involves exocytic sites that are proximal to sorting endosomes. In some of these exocytic events, tubular TCs can be observed that extend from the sorting endosome and subsequently undergo exocytosis. Interestingly, in some cases, these tubular extensions appear to form a continuous connection between the sorting endosome and the plasma membrane. The second type of event (“indirect” events) occurs at sites that are more distal to sorting endosomes. In these cases, the TCs can travel relatively long distances within the cell before exocytosis. These TCs can pass through holding zones, where they can accumulate and sequentially exocytose. In some cases, not all TCs in the holding zone undergo exocytosis, indicating that intracellular location at such a zone alone is insufficient to ensure exocytic fusion. The holding of TCs at these sites before their exocytosis suggests that a trigger is needed to prompt subsequent exocytosis. In some cases, we observe that exocytosis of a TC can be preceded by the movement of a relatively small vesicle in close proximity, suggesting that this vesicle might endow the TC with the necessary complement of components to trigger exocytosis.
Based on the kinetics and cell biology of constitutive (e.g., transferrin receptor) and regulated recycling, slow and fast pathways have been described (8, 13, 37, 38). However, the intracellular processes in terms of TCs and their itineraries that lead to these two pathways are not well characterized. This is particularly so for the direct pathway from sorting endosome to plasma membrane, where mechanisms ranging from the shuttling of TCs between endosomes and the plasma membrane to direct (lysosomal-like or exosomal) (39) fusion of the sorting endosome have been suggested (16). Here, we present data that demonstrate types of TC behavior that might be representative of kinetically distinct pathways. Our data are also consistent with recycling models that invoke distinct intracellular pathways (8, 13, 37, 38, 40), rather than models involving different rates along the same pathway (12, 41).
Observation of the exocytic events described in this article by using only TIRFM could lead to ambiguity. For example, multiple exocytic release events shown in Fig. 2B might be interpreted in several ways: they might be due to sequential, partial release events from the same TC or emanate from discrete TCs. Our multiplane imaging setup allows us to define the origin and nature of TCs that subsequently fuse with the plasma membrane. Similarly, the event in which an extended tubule on the direct pathway fuses with the plasma membrane at its tip and then retracts might be interpreted as a kiss-and-run type of exocytic event (42, 43) if visualized by using only TIRFM. Our methodology therefore allows significant insights concerning the dynamics of intracellular trafficking in three dimensions. However, this approach also has limitations. For example, TIRFM is a more sensitive imaging approach for fluorescence detection relative to epifluorescence microscopy. Therefore TCs that are not brightly labeled might be visualized by using TIRFM near the plasma membrane, but may be undetectable by using epifluorescence and consequently not be trackable back to the interior of the cell. In addition, photobleaching also poses a significant problem and limits the time for which the TCs can be tracked. Furthermore, by comparison with the field of view in our imaging setup, HMEC-1 cells can be very large. As a result, TCs may become untrackable because of departure from the field of view.
Our results raise questions concerning the role of different intracellular trafficking pathways, such as why does not all recycling occur by the apparently more efficient route from sorting endosomes to the plasma membrane? It has been suggested that the slow recycling pathway might be involved in more effective segregation of receptors for transcytosis (13) and may therefore be particularly relevant for polarized cells. We also observe that the pathway of exocytosing TCs can involve more than one interaction with sorting endosomes. Such processes may allow the same TC to iteratively collect cargo and thereby increase the efficiency of recycling.
In summary, we have elucidated aspects of the recycling/exocytic pathway. This has been achieved by using a recently developed multiplane imaging setup. Our data define distinct intracellular trafficking routes for which visualization would be difficult, if not impossible, by using other imaging approaches. These studies have relevance to understanding how FcRn functions as a salvage receptor to maintain IgG transport and levels in addition to being more generally applicable to recycling receptors.
Materials and Methods
Plasmid Constructs.
Plasmid constructs to express human FcRn fused at the C terminus to enhanced GFP (in pEGFP-N1; Clontech, Mountain View, CA; FcRn-GFP) and human β2-microglobulin (β2m) have been described (32). Similar constructs to express a FcRn-monomeric RFP (FcRn-mRFP) fusion protein were generated by first recloning the mRFP gene (generously provided by Roger Tsien, University of California at San Diego, La Jolla, CA) (44) as a BamHI-NotI fragment to replace eGFP in pEGFP-N1 and then recloning the FcRn gene as an EcoRI fragment into this vector. To generate FcRn_mut-mRFP, this construct was subsequently modified by replacing wild-type FcRn codons with corresponding codons encoding the 79–89/136–147 mutation that confers higher-affinity binding to IgGs on human FcRn (36).
N-terminal fusions of fluorescent protein genes were made by inserting a KpnI site, followed by codons encoding a Gly-Gly-Ser linker between the 3′ end of the human FcRn leader peptide and sequence encoding the mature N terminus of FcRn (45) by using splicing by overlap extension (46). This gene was assembled into the NheI and SalI sites of a variant of pEGFP-C1 in which the fluorescent protein gene and KpnI site had been deleted. Genes encoding either mRFP or pHluorin (ecliptic) were appended with KpnI sites by using designed oligonucleotides and the PCR and were then inserted in-frame into the KpnI site. The generation of a gene encoding ecliptic pHluorin, and expression and characterization of the recombinant protein are described in SI Text, Section 1. All constructs were made by using standard methods of molecular biology and were sequenced before use in these studies.
Antibodies and Reagents.
Alexa Fluor 555-labeled transferrin and Quantum dot 655 (QD655)-streptavidin conjugates were obtained from Molecular Probes and Quantum Dot, respectively (both currently part of Invitrogen, Carlsbad, CA). To express the MST-HN mutant [human IgG1-derived; (35)] as an antibody with a site-specific biotinylation peptide at the C terminus, codons encoding Gly-Ser-Leu-His-His-Ile-Leu-Asp-Ala-Gln-Lys-Met-Val-Trp-Asn-His-Arg were appended to the 3′ end of the CH3 domain by using two rounds of PCR with overlapping oligonucleotides and an oligonucleotide close to the 5′ end of the CH3 domain gene. The resulting product was digested with XmaI (by using an internal XmaI site overlapping codons 14–16 of the CH3 domain gene and a 3′ primer encoded site) and used to replace the wild-type gene in a human Fc construct (47, 48). The final heavy-chain expression construct was assembled as described (47) by using a vector (48) generously provided by J. Foote (Arrowsmith Technologies, Seattle, WA). This construct was transfected into an NSO transfectant expressing an anti-hen egg lysozyme light-chain [(48) generously provided by J. Foote], and transfectants were selected as described (47, 48). Recombinant protein was purified by using lysozyme-Sepharose. To site-specifically biotinylate the antibody, the protein (≈2 mg/ml) was dialyzed into 20 mM Tris·HCl, 50 mM NaCl, pH 8.0, and then incubated with BirA (18 units per mg of antibody; Avidity, Denver, CO) for 16 h at room temperature (49). The protein was then dialyzed into PBS, pH 7.2. Biotinylation and binding activity of the treated MST-HN antibody were analyzed by using ELISA and surface plasmon resonance, respectively. QD–IgG complexes were made by mixing streptavidin-coated QD655 and site-specifically biotinylated human IgG1 mutant [MST-HN (35)] at a 1:0.5 molar ratio of QD to mutated IgG1.
Cells and Transfections.
The human endothelial cell line HMEC-1 was generously provided by F. Candal at the Centers for Disease Control (Atlanta, GA). HMEC-1 cells were maintained in MCDB 131 medium, and two combinations of expression constructs were used to transiently transfect these cells by using Nucleofector technology (Amaxa Biosystems, Cologne, Germany) (32). The first combination comprised pHluorin (ecliptic)-FcRn (2.4–2.6 μg), and β2m (1–2 μg) expression plasmids plus either FcRn-mRFP (3–4 μg), mRFP-FcRn (2.6 μg), or FcRn_mut-mRFP (2.4–4 μg). The second combination comprised FcRn-GFP (1 μg), FcRn-mRFP (4 μg) and β2m (1 μg) expression plasmids. Immediately after transfection, cells were plated on coverslips in phenol red-free Ham's F-12K medium. Cells were imaged 15–47 h after transfection. For imaging FcRn and IgG simultaneously in three dimensions, the cells were incubated in medium (≈pH 7.3) with QD655-human IgG1 mutant (11 nM with respect to IgG) and Alexa Fluor 555-labeled transferrin (130 nM).
Microscopy.
Images were acquired by using Axiovert microscopy (Zeiss, Thornwood, NY) imaging stations that were modified to image different focal planes simultaneously (17, 50) with a ×100 Zeiss αPlan-Fluar 1.45N.A. objective. This was achieved here by using up to four cameras and by placing the cameras at specific, calibrated distances from the tube lens. For imaging FcRn only, mRFP- or pHluorin-labeled FcRn was imaged in the membrane plane by using TIRFM, with GFP- or mRFP-labeled FcRn imaged in the higher focal planes by using epifluorescence (with one camera per focal plane). To image FcRn and IgG ligand in two focal planes, pHluorin-labeled FcRn was imaged by using TIRFM in the plasma membrane plane. Simultaneously, QD-labeled IgG was imaged in the plasma membrane plane by using both TIRFM and epifluorescence, and QD-labeled IgG plus mRFP-labeled FcRn (and Alexa Fluor 555-Transferrin) were imaged by using epifluorescence in the upper plane (with two cameras per focal plane). Acquired data were processed (adjusted for magnification differences, images from different cameras temporally and spatially aligned by using bead samples (50), piecewise linear intensity adjusted, overlaid, and annotated) with custom-written routines in Matlab (Mathworks, Natick, MA) and visualized by using the Microscopy Image Analysis Tool (MIATool) software package (www4.utsouthwestern.edu/wardlab/miatool). Exocytic events were visually detected and verified as in refs. 6 and 7 (see also SI Text, Section 6). Events of interest were highlighted for presentation by applying local thresholding. The colors of the images were chosen according to the proteins that are displayed. Additional details concerning multifocal plane microscopy, image processing, analysis, and display of the data can be found in SI Text (Sections 5 and 6).
Supplementary Material
Acknowledgments
We thank R. Guevara, A. Pickl-Herk, L. Veereshlingam, J. Zhou, and Q. Tang for their assistance with molecular biology experiments and sample preparation and S. Sanjee, A. Abraham, P. Gehalot, and H. Nguyen for help with data analysis and the preparation of figures. This work was supported by National Institutes of Health Grants R01 AI039167 and R01 GM071048.
Abbreviations
- FcRn
neonatal Fc receptor
- TIRFM
total internal reflection fluorescence microscopy
- TC
transport container
- QD
quantum dot.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700337104/DC1.
References
- 1.Steyer JA, Almers W. Nat Rev Mol Cell Biol. 2001;2:268–275. doi: 10.1038/35067069. [DOI] [PubMed] [Google Scholar]
- 2.Maxfield FR, McGraw TE. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Boulan E, Kreitzer G, Musch A. Nat Rev Mol Cell Biol. 2005;6:233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- 4.Axelrod D. In: Light Microscopy in Biology. Lacey AJ, editor. Oxford: Oxford Univ Press; 1999. pp. 399–423. [Google Scholar]
- 5.Toomre D, Steyer JA, Keller P, Almers W, Simons K. J Cell Biol. 2000;149:33–40. doi: 10.1083/jcb.149.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmoranzer J, Goulian M, Axelrod D, Simon SM. J Cell Biol. 2000;149:23–32. doi: 10.1083/jcb.149.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zenisek D, Steyer JA, Almers W. Nature. 2000;406:849–854. doi: 10.1038/35022500. [DOI] [PubMed] [Google Scholar]
- 8.Lampson MA, Schmoranzer J, Zeigerer A, Simon SM, McGraw TE. Mol Biol Cell. 2001;12:3489–3501. doi: 10.1091/mbc.12.11.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuboi T, Zhao C, Terakawa S, Rutter GA. Curr Biol. 2000;10:1307–1310. doi: 10.1016/s0960-9822(00)00756-9. [DOI] [PubMed] [Google Scholar]
- 10.Ober RJ, Martinez C, Lai X, Zhou J, Ward ES. Proc Natl Acad Sci USA. 2004;101:11076–11081. doi: 10.1073/pnas.0402970101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutter GA, Hill EV. Physiology. 2006;21:189–196. doi: 10.1152/physiol.00002.2006. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh RN, Gelman DL, Maxfield FR. J Cell Sci. 1994;107:2177–2189. doi: 10.1242/jcs.107.8.2177. [DOI] [PubMed] [Google Scholar]
- 13.Sheff DR, Daro EA, Hull M, Mellman I. J Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oheim M. Biophys J. 2004;87:1403–1405. doi: 10.1529/biophysj.104.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickman DK, Horne JA, Meinertzhagen IA, Schwarz TL. Cell. 2005;123:521–533. doi: 10.1016/j.cell.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 16.van Dam EM, Ten BT, Jansen K, Spijkers P, Stoorvogel W. J Biol Chem. 2002;277:48876–48883. doi: 10.1074/jbc.M206271200. [DOI] [PubMed] [Google Scholar]
- 17.Prabhat P, Ram S, Ward ES, Ober RJ. IEEE Trans Nanobiosci. 2004;3:237–242. doi: 10.1109/tnb.2004.837899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simister NE, Mostov KE. Nature. 1989;337:184–187. doi: 10.1038/337184a0. [DOI] [PubMed] [Google Scholar]
- 19.Ghetie V, Hubbard JG, Kim JK, Tsen MF, Lee Y, Ward ES. Eur J Immunol. 1996;26:690–696. doi: 10.1002/eji.1830260327. [DOI] [PubMed] [Google Scholar]
- 20.Junghans RP, Anderson CL. Proc Natl Acad Sci USA. 1996;93:5512–5516. doi: 10.1073/pnas.93.11.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Israel EJ, Wilsker DF, Hayes KC, Schoenfeld D, Simister NE. Immunology. 1996;89:573–578. doi: 10.1046/j.1365-2567.1996.d01-775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickinson BL, Badizadegan K, Wu Z, Ahouse JC, Zhu X, Simister NE, Blumberg RS, Lencer WI. J Clin Invest. 1999;104:903–911. doi: 10.1172/JCI6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy KM, Yoong Y, Simister NE. J Cell Sci. 2000;113:1277–1285. doi: 10.1242/jcs.113.7.1277. [DOI] [PubMed] [Google Scholar]
- 24.Spiekermann GM, Finn PW, Ward ES, Dumont J, Dickinson BL, Blumberg RS, Lencer WI. J Exp Med. 2002;196:303–310. doi: 10.1084/jem.20020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claypool SM, Dickinson BL, Yoshida M, Lencer WI, Blumberg RS. J Biol Chem. 2002;277:28038–28050. doi: 10.1074/jbc.M202367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellinger I, Schwab M, Stefanescu A, Hunziker W, Fuchs R. Eur J Immunol. 1999;29:733–744. doi: 10.1002/(SICI)1521-4141(199903)29:03<733::AID-IMMU733>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 27.Ghetie V, Ward ES. Annu Rev Immunol. 2000;18:739–766. doi: 10.1146/annurev.immunol.18.1.739. [DOI] [PubMed] [Google Scholar]
- 28.Rodewald R. J Cell Biol. 1976;71:666–669. doi: 10.1083/jcb.71.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace KH, Rees AR. Biochem J. 1980;188:9–16. doi: 10.1042/bj1880009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raghavan M, Bonagura VR, Morrison SL, Bjorkman PJ. Biochemistry. 1995;34:14649–14657. doi: 10.1021/bi00045a005. [DOI] [PubMed] [Google Scholar]
- 31.Popov S, Hubbard JG, Kim J, Ober B, Ghetie V, Ward ES. Mol Immunol. 1996;33:521–530. doi: 10.1016/0161-5890(96)00004-1. [DOI] [PubMed] [Google Scholar]
- 32.Ober RJ, Martinez C, Vaccaro C, Zhou J, Ward ES. J Immunol. 2004;172:2021–2029. doi: 10.4049/jimmunol.172.4.2021. [DOI] [PubMed] [Google Scholar]
- 33.Miesenbock G, De Angelis DA, Rothman JE. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 34.Schmoranzer J, Simon SM. Mol Biol Cell. 2003;14:1558–1569. doi: 10.1091/mbc.E02-08-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaccaro C, Zhou J, Ober RJ, Ward ES. Nat Biotechnol. 2005;23:1283–1288. doi: 10.1038/nbt1143. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J, Mateos F, Ober RJ, Ward ES. J Mol Biol. 2005;345:1071–1081. doi: 10.1016/j.jmb.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Ren M, Xu G, Zeng J, Lemos-Chiarandini C, Adesnik M, Sabatini DD. Proc Natl Acad Sci USA. 1998;95:6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fevrier B, Raposo G. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Gruenberg J, Maxfield FR. Curr Opin Cell Biol. 1995;7:552–563. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 41.French AR, Lauffenburger DA. Biotechnol Bioeng. 1996;51:281–297. doi: 10.1002/(SICI)1097-0290(19960805)51:3<281::AID-BIT4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 42.Fesce R, Meldolesi J. Nat Cell Biol. 1999;1:E3–E4. doi: 10.1038/8950. [DOI] [PubMed] [Google Scholar]
- 43.Valtorta F, Meldolesi J, Fesce R. Trends Cell Biol. 2001;11:324–328. doi: 10.1016/s0962-8924(01)02058-x. [DOI] [PubMed] [Google Scholar]
- 44.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Story CM, Mikulska JE, Simister NE. J Exp Med. 1994;180:2377–2381. doi: 10.1084/jem.180.6.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 47.Firan M, Bawdon R, Radu C, Ober RJ, Eaken D, Antohe F, Ghetie V, Ward ES. Int Immunol. 2001;13:993–1002. doi: 10.1093/intimm/13.8.993. [DOI] [PubMed] [Google Scholar]
- 48.Foote J, Winter G. J Mol Biol. 1992;224:487–499. doi: 10.1016/0022-2836(92)91010-m. [DOI] [PubMed] [Google Scholar]
- 49.Radu CG, Anderton SM, Firan M, Wraith DC, Ward ES. Int Immunol. 2000;12:1553–1560. doi: 10.1093/intimm/12.11.1553. [DOI] [PubMed] [Google Scholar]
- 50.Prabhat P, Ram S, Ward ES, Ober RJ. Proc SPIE. 2006;6090:115–121. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.