Abstract
Germ cells are required for the successful propagation of sexually reproducing species. Understanding the mechanisms by which these cells are specified and how their totipotency is established and maintained has important biomedical and evolutionary implications. Freshwater planarians serve as fascinating models for studying these questions. They can regenerate germ cells from fragments of adult tissues that lack reproductive structures, suggesting that inductive signaling is involved in planarian germ cell specification. To study the development and regeneration of planarian germ cells, we have functionally characterized an ortholog of nanos, a gene required for germ cell development in diverse organisms, from Schmidtea mediterranea. In the hermaphroditic strain of this species, Smed-nanos mRNA is detected in developing, regenerating, and mature ovaries and testes. However, it is not detected in the vast majority of newly hatched planarians or in small tissue fragments that will ultimately regenerate germ cells, consistent with an epigenetic origin of germ cells. We show that Smed-nanos RNA interference (RNAi) results in failure to develop, regenerate, or maintain gonads in sexual planarians. Unexpectedly, Smed-nanos mRNA is also detected in presumptive testes primordia of asexual individuals that reproduce strictly by fission. These presumptive germ cells are lost after Smed-nanos RNAi, suggesting that asexual planarians specify germ cells, but their differentiation is blocked downstream of Smed-nanos function. Our results reveal a conserved function of nanos in germ cell development in planarians and suggest that these animals will serve as useful models for dissecting the molecular basis of epigenetic germ cell specification.
Keywords: epigenesis, germ cell specification, RNA interference, Schmidtea mediterranea, Platyhelminthes
Germ cells provide intriguing examples of cellular differentiation, in which highly specialized cells retain their totipotency (1). In metazoans, two apparently distinct modes of germ cell specification are observed: determinate specification (or preformation), in which maternally supplied, localized cytoplasmic determinants act early in embryogenesis; and epigenetic specification, in which inductive interactions between cells specify germ cell fate later in embryogenesis (2–4). Surveys of germ cell specification mechanisms throughout metazoan phyla revealed that epigenesis appears to be more widely distributed than determinate specification (2–4). Furthermore, epigenesis is observed in basal metazoans (e.g., Porifera and Cnidarians), whereas determinate specification is typically seen in more derived lineages, leading to the hypothesis that epigenesis is ancestral (4, 5).
Much of our understanding of the mechanisms underlying germ cell specification is based on genetic analyses in Drosophila melanogaster and Caenorhabditis elegans, both of which use determinate specification (for reviews, see refs. 6 and 7). Studies aimed at deciphering the mechanisms of epigenesis have been limited almost exclusively to mouse (8, 9). Understanding the extent to which germ cell specification mechanisms have been conserved or have diverged among disparate groups requires analyzing these mechanisms in additional organisms.
Planarian flatworms (10, 11) are well suited to serve as a simple model for studying epigenesis. Classic histological analyses suggested that these animals specify germ cells postembryonically: no obvious primordial germ cells or germ plasm are observed during embryogenesis, and the animals do not develop reproductive organs until after hatching (12; for references to early literature, see ref. 13). In addition, they possess amazing regenerative abilities that permit them to regenerate the germ cell lineage in fragments of tissue that lack reproductive organs (14). A population of stem cells (neoblasts) maintained during the course of the animal's life is the likely source of the regenerated germ cell lineage (15). Two strains with distinct reproductive strategies are observed in the planarian Schmidtea mediterranea: a sexual strain that reproduces as cross-fertilizing hermaphrodites, and an asexual strain that reproduces by transverse fission (10). These strains can be distinguished by a chromosomal translocation observed in asexual individuals (16). The nature of the asexuals (e.g., whether they produce germ cells that are defective in some way or fail to specify germ cells altogether) remains an open question (17).
To analyze germ cell formation in planarians, we have identified and functionally characterized a nanos ortholog from S. mediterranea (Smed-nanos). In Drosophila, nanos is required for abdominal segmentation (18, 19) as well as for germ cell differentiation (20, 21) and maintenance (22, 23). Conserved functions in germ cell maintenance have been reported for nanos orthologs in C. elegans (24), zebrafish (25), and mouse (26, 27). nanos is expressed in primordial germ cells in widely divergent metazoans, ranging from cnidarians to humans (4, 28–31); however, little is known about the function of nanos outside of the major genetic model organisms. A nanos homolog (Djnos) that is expressed in presumptive germ cells in both sexual and asexual individuals of the planarian Dugesia japonica was recently described; however, no functional data were reported (32). Here we show that Smed-nanos function is required for proper germ cell development, regeneration, and maintenance in both sexual and asexual planarians.
Results and Discussion
Identification of Smed-nanos and Its Expression in Intact and Regenerating Sexual Planarians.
To characterize the process of germ cell formation in the planarian S. mediterranea, we identified sequences encoding the highly conserved NANOS zinc finger motif in whole-genome shotgun sequence data from the hermaphroditic strain. Using these sequences, we isolated full-length nanos cDNAs (839 bases) from a sexual S. mediterranea cDNA library (33) [see supporting information (SI) Methods]. Northern blot analysis revealed a single Smed-nanos transcript of ≈0.8 kb in total RNA from sexual planarians (SI Fig. 7). Smed-nanos (for brevity, referred to as nanos) encodes a predicted protein of 233 aa, containing two conserved zinc finger domains at the C terminus (SI Fig. 7).
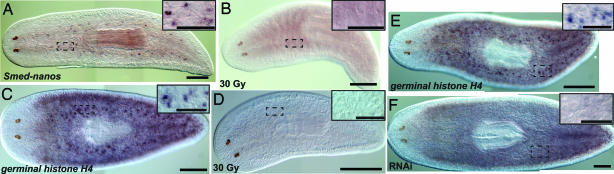
To examine the spatial expression pattern of nanos, we performed in situ hybridization on S. mediterranea hermaphrodites (Fig. 1A) at various stages after hatching from the egg capsule. Hybridizations to hatchlings fixed within 12 h of emergence (D1 hatchlings) failed to detect nanos expression in 89% of the samples (34 of 38; Fig. 1B and SI Fig. 8C). Controls using germinal histone H4 (33) (see below) and the neural marker anosmin-1 (34) as probes showed that newly emerged hatchlings were not refractory to in situ hybridization (SI Fig. 8 A and B). In hatchlings examined on the 3rd (D3, 48–60 h) and 7th days (D7, 144–156 h) after hatching, nanos RNA was detected dorsolaterally, in positions corresponding to presumptive testes primordia (17 of 21 D3 hatchlings were nanos-positive and 9 of 9 D7 hatchlings were nanos-positive; SI Fig. 8 D–F). To address whether the development of nanos-positive cells in hatchlings requires cell proliferation, we irradiated D1 hatchlings with 30 Gy, a dose sufficient to eliminate all neoblasts from adults (35), and fixed them 2 days later. After irradiation, we could not detect dorsolateral clusters of nanos-positive cells in irradiated D3 hatchlings (n = 13), whereas control D3 hatchlings were nanos-positive (n = 5; SI Fig. 8 G and H). These results suggest that the postembryonic development of nanos-positive germ cell precursors requires either neoblasts or cell division.
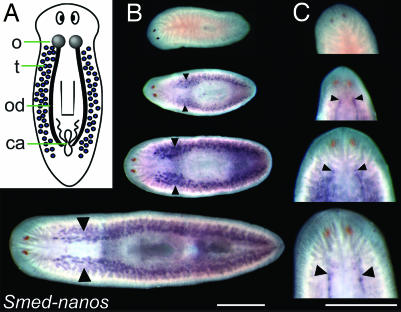
Fig. 1.
Smed-nanos is expressed in the testes and the ovaries of juvenile and mature S. mediterranea hermaphrodites but not in young hatchlings. (A) Diagram of S. mediterranea hermaphrodite illustrating the reproductive organs. o, ovaries; t, testes; od, oviducts; ca, copulatory apparatus. (B) Whole-mount in situ hybridization showing nanos expression in testes (arrowheads); expression is not detected in newly hatched animals. From Top to Bottom: hatchling (34 of 38 lacked detectable Smed-nanos mRNA), juvenile (n = 4), smaller (n = 5), and larger mature worms (n = 11) (dorsal views). (C) Ventral views of the worms in B, showing nanos expression in the ovaries (arrowheads). (Scale bars, 1 mm.)
Later during development, nanos RNA was detected in the developing testes of juveniles (planarians lacking completely developed reproductive structures) and fully mature worms (Fig. 1B). nanos RNA was also detected in ovaries of both juvenile and mature worms but not in hatchlings (Fig. 1C). nanos expression was detected much earlier during gonad development than previously examined markers of the reproductive organs (33).
As an additional marker for studying the appearance of germ cells in young hatchlings, we used germinal histone H4 (germinal H4) (33), a transcript that labels presumptive germ cells as well as somatic neoblasts (see below). As observed with nanos, the majority of D1 hatchlings (6 of 7) did not have obvious germinal H4 labeling outside of the neoblast population (SI Fig. 8I). Whereas 4 of 6 D3 hatchlings had clusters of dorsolateral cells with a greater intensity of germinal H4 signal (SI Fig. 8J, arrows), these clusters were reminiscent of the nanos-positive cells observed in D3 hatchlings. Given the great variability in the length of time from egg capsule deposition to hatching as well as variability in the size of the individuals emerging from the same egg capsule, it seems likely that the few animals in which nanos and germinal H4 mRNAs were detected at D1 were either precocious developers, had relatively delayed hatching, or had spent the most time between hatching and fixation (up to ≈12 h).
The planarian reproductive system can regenerate after amputation. Using nanos as a germ cell marker, we analyzed head pieces that were amputated anterior to the ovaries; such fragments devoid of reproductive tissues ultimately regenerate reproductive organs, suggesting that germ cells can be derived from somatic cells (14). After amputation, we used fluorescent in situ hybridization (FISH) and confocal microscopy to detect nanos-positive cells in the regenerating head pieces (Fig. 2). Seven days after amputation, 7 of 14 head fragments lacked any detectable nanos expression (Fig. 2 A and B, Top); based on the clustering of nanos-positive cells and their appearance at the posterior-most extent of the uninjured tissue, the nanos-positive cells observed at this stage were likely residual germ cells derived from amputation sites that included portions of the ovaries and/or testes (data not shown). By day 14, all animals examined were positive for nanos expression (Fig. 2 A and B, Middle). By day 21, nanos-positive cell clusters increased in both number and size (Fig. 2B, Bottom). Similar results were reported for the reappearance of Djnos expression in head fragments of D. japonica (32), although the colorimetric staining may have made it difficult to visualize the earliest appearance of Djnos-positive cells. These results, together with the lack of detectable nanos RNA in the vast majority of D1 hatchlings, support the view that germ cells in planarians can be specified postembryonically.
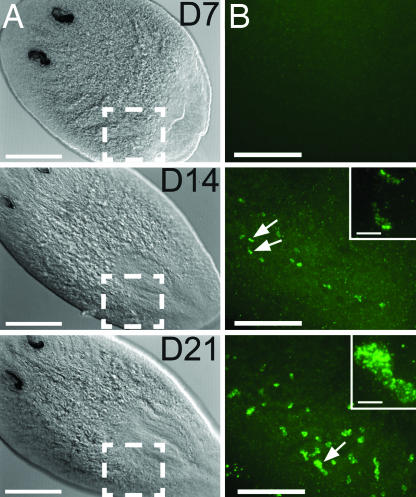
Fig. 2.
Smed-nanos expression in regenerating head fragments amputated anterior to the ovaries. (A) Differential interference contrast microscopic images of regenerating heads fixed 7, 14, or 21 days after amputation (animals were ≥1.2 cm when amputated). The numbers of animals in which nanos mRNA was detected were 7 of 14 at 7 days, 8 of 8 at 14 days, and 8 of 9 at 21 days. (Scale bars, 250 μm.) (B) Confocal projections corresponding to the boxed regions in A showing nanos mRNA detected by FISH. (Scale bars, 100 μm.) Arrows indicate nanos-positive cells shown at higher magnification in the Insets. (Inset scale bars, 10 μm.)
Amputation behind the ovaries leads to regression of the testes in the decapitated posterior fragment; after regeneration of the cephalic ganglia, the testes regenerate (36). The processes of regression and regeneration of the testes can be monitored by using in situ hybridization to detect T-plastin mRNA (33), a transcript expressed abundantly in spermatocytes and spermatids (data not shown). One week after amputation behind the ovaries, T-plastin expression was not detected in regenerating tail fragments (Fig. 3A); after 15 days of regeneration, the pattern of testes expression of T-plastin was restored (Fig. 3A). A sublethal dose of γ-irradiation (10 Gy) also led to degeneration of the testes followed by their regeneration (37) in a time course similar to that shown for transverse amputation behind the ovaries (data not shown). In contrast to the regression and regeneration of the testes observed by morphological criteria (36, 37) and by T-plastin in situ hybridization, expression of nanos mRNA persisted throughout the process of testes regression and regeneration after amputation (Fig. 3B) and sublethal doses of γ-irradiation (data not shown). Combined FISH and confocal microscopy showed the changes in the distribution of nanos-positive cells during testes regeneration (Fig. 3C). In intact animals, nanos-positive cells were detected around the periphery of the testes lobes. Three days after amputation, nanos appeared to be up-regulated in the testes, and positive cells were still distributed around the periphery (Fig. 3C, arrows). By 7 days, the central portion of the testes was no longer visible (Fig. 3C), consistent with the disappearance of T-plastin expression (Fig. 3A). By 15 days, a pattern very similar to that observed in intact animals was reestablished (Fig. 3C).
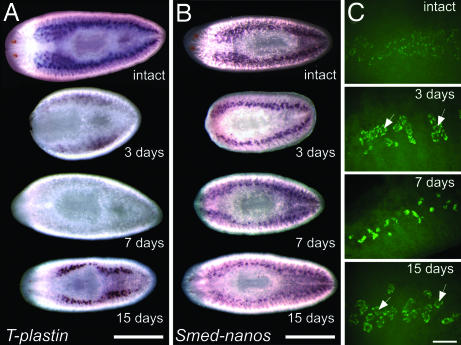
Fig. 3.
Testes regeneration in S. mediterranea hermaphrodites. (A) Expression pattern of a T-plastin homolog (DN311193) in intact and regenerating animals (n = 3 per stage). Days after amputation are indicated below each animal. (Scale bar, 1 mm.) (B) Expression pattern of Smed-nanos in intact (n = 2) and regenerating planarians (n = 4 per stage). (Scale bar, 1 mm.) (C) Confocal projections showing FISH to detect nanos mRNA in intact (n = 4) and regenerating planarians (n = 4 per stage). Arrows indicate the central portion of testes lobes. (Scale bar, 100 μm.)
Smed-nanos RNAi Results in Failure to Regenerate or Develop the Gonads.
RNA interference (RNAi) (38) is a powerful tool for dissecting gene function in planarians, but previous RNAi experiments have been limited to studies of asexual planarians (35, 39–44); therefore, we tested the efficacy of gene knockdown in S. mediterranea hermaphrodites after feeding bacterially expressed dsRNAs (42, 45). After two feedings with dsRNA targeting nanos, T-plastin, or Smedwi-2 [expressed in neoblasts (ref. 43) as well as ovaries and testes (ref. 33)], we found that the target RNAs were dramatically reduced 10 days after the last feeding (SI Fig. 9 A–F). Thus, RNAi can be used to inhibit gene expression in the planarian germ cells.
To assay the effects of nanos RNAi knockdown on the regeneration of the reproductive organs, mature animals were fed twice with nanos dsRNA and then amputated posterior to the ovaries. Feeding of dsRNA was resumed 2 weeks after amputation to allow the animals to grow and undergo sexual maturation; they were analyzed after 2–3 months of weekly dsRNA feedings. Control planarians fed bacteria containing vector alone regenerated normally, and the ventral gonopore (opening to the copulatory apparatus) was usually observed within 1 month after amputation (data not shown). Worms fed nanos dsRNA regenerated their somatic tissues normally; however, they did not form gonopores until 3 months after amputation. These pores appeared to open into an empty cavity, and no copulatory apparatus was formed (data not shown).
We analyzed the developmental state of the testes in these animals by using several different markers (Fig. 4). The condensing nuclei in clusters of spermatocytes, spermatids, and spermatozoa enabled us to use DAPI staining to visualize the testes dorsolaterally in whole-mount preparations of control planarians (Fig. 4A); such dorsolateral clusters were absent from nanos RNAi knockdown animals (Fig. 4B). Histological sections of control animals showed normal testes morphology (SI Fig. 9G); no testes were observed in sections of nanos knockdown animals (SI Fig. 9H). Microtubules are prominent components of the structure of planarian spermatozoa (46–48); therefore, we used anti-tubulin immunofluorescence to visualize clusters of spermatocytes, spermatids, and spermatozoa within the testes of control animals (Fig. 4 C and G). These anti-tubulin-positive clusters were not observed in animals fed nanos dsRNA (Fig. 4 D and H); the ciliated ducts of the excretory system were the only anti-tubulin-positive structures observed within the mesenchyme. In planarian testes, dividing spermatogonia undergo three rounds of incomplete cytokinesis to generate a cyst of 8 primary spermatocytes; after meiosis and spermiogenesis, 32 spermatids are produced (48). Clusters of dividing cells within these cysts were labeled with anti-phospho-histone H3-S10 (H3-S10P) antibodies (49) (Fig. 4 E and G). Anti-H3-S10P-positive clusters were not seen in nanos RNAi knockdown worms; the only mitotic figures observed in these animals corresponded to unclustered, dividing neoblasts typically observed in the mesenchyme of asexual planarians (Fig. 4 F and H) (35, 42–44, 50).
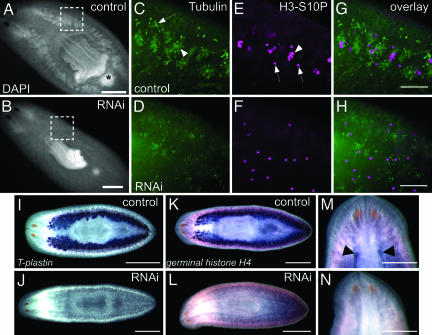
Fig. 4.
Smed-nanos RNAi worms fail to regenerate testes or ovaries after amputation. (A and B) DAPI staining of animals fixed 3 months after amputation, showing normal testes and copulatory apparatus (asterisk) in controls (A; n = 7) but not in nanos RNAi animals (B; n = 8). (Scale bars, 1 mm.) (C–H) Immunofluorescent images of the boxed regions in A and B. (C and D) Anti-tubulin labels different stages of spermatogenesis (arrowhead) within testes of control animals (C) but not in RNAi animals (D). This antibody also labels ciliated excretory ducts (arrow in C). (E and F) Anti-H3-S10P labels individual mitotic figures (arrows) and germ cell cysts (arrowhead) in the testes of control animals (E), but germ cell cysts were not observed in RNAi animals (F). (G) Overlay of C and E. and (H) Overlay of D and F. (Scale bars, 100 μm.) (I–N) In situ hybridization for gonad markers. Animals were fixed 2 months after amputation. (I and J) T-plastin is expressed in the testes of controls (n = 10) but not in RNAi animals (n = 10). (K and L) germinal H4 is expressed in testes of controls (n = 12) but not in RNAi animals (n = 12). (Scale bars, 1 mm.) (M and N) germinal H4 is expressed in the ovaries (arrowheads) of control animal (M); expression is absent from RNAi animal (N). (Scale bars, 500 μm.)
As additional markers to examine testes regeneration, we analyzed T-plastin and germinal H4 expression in control and dsRNA-treated worms. Control animals had robust expression of T-plastin (Fig. 4I) and germinal H4 (Fig. 4K) in testes, providing further evidence of proper testes regeneration. In contrast, T-plastin expression was not detected in nanos RNAi worms (Fig. 4J), and testes expression of germinal H4 was not observed (Fig. 4L). germinal H4 is also expressed in ovaries and somatic neoblasts (33). In control planarians, this marker enabled us to visualize the regenerated ovaries (Fig. 4M). In nanos RNAi animals, ovarian expression of germinal H4 was not observed (Fig. 4N). Together, these results demonstrate that nanos function is required for proper regeneration of the planarian gonads.
The above analysis was performed on animals after several months of RNAi treatment; thus, we could not distinguish between early and late effects on the regeneration process. To analyze further the progression of the nanos RNAi phenotype, we conducted similar RNAi experiments, except that we fixed animals 2 weeks after amputation (animals were starved from amputation until fixation). FISH to detect germinal H4 mRNA revealed that control animals regenerated testes primordia, visible as germinal H4-positive cell clusters (n = 10; Fig. 5 A–D). In contrast, such clusters were not observed in nanos RNAi animals; only neoblast staining was observed (n = 11; Fig. 5 E and F). We conclude that the nanos RNAi phenotype during regeneration is the result of failure to form or maintain testes primordia during early stages of testes regeneration rather than loss of mature testes after normal regeneration. Intriguingly, sexually mature animals fed nanos dsRNA every 4–5 days over the course of 1 month lost their gonads (10 of 10; data not shown), suggesting that Smed-nanos is also required to maintain germ cell-derived structures in adult planarians.
Fig. 5.
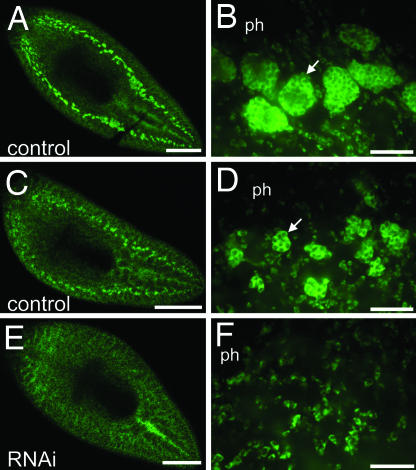
Smed-nanos RNAi knockdown animals do not regenerate testes primordia. Animals were fixed 14 days after amputation posterior to the ovaries and processed to detect germinal H4 mRNA by FISH. (A–D) Control animals. (E and F) nanos RNAi animals. (B, D, and F) Confocal images corresponding to the postpharyngeal regions of the planarians shown in A, C, and E. Control animals regenerated germinal H4-positive testes primordia (n = 10). (A and B) Well developed testes lobes (arrow in B) were observed in the largest of these specimens. (C and D) The remaining animals developed smaller clusters of germinal H4-positive testes primordia (arrow in D). (E and F) germinal H4-positive dorsal clusters were not detected in nanos RNAi animals (n = 11); only somatic neoblasts were observed. ph, pharynx. [Scale bars, 500 μm (A, C, and E); 50 μm (B, D, and F).]
To examine whether nanos was also required for normal postembryonic development of the planarian reproductive system, we performed RNAi experiments on newly hatched worms. D1 hatchlings were fed nanos dsRNA every 4–5 days for a period of 2–3 months (it takes ≈2 months to reach reproductive maturity). Although these planarians grew normally, they did not develop ovaries (n = 8) or testes (n = 15) (as assayed by the molecular markers described above) or gonopores (n = 15); whereas control animals had ovaries (n = 8), fully developed testes (n = 14), and gonopores (n = 14; data not shown). Thus, nanos is required for the postembryonic development of the planarian reproductive system.
Smed-nanos Expression Suggests That Asexual Planarians Also Specify Germ Cells.
Surprisingly, Northern blot analysis revealed that nanos mRNA was also expressed in asexual planarians, at levels comparable with those observed in sexual animals (SI Fig. 7). Therefore, we performed whole-mount in situ hybridization to examine nanos expression in asexual worms. In asexual planarians, nanos mRNA was detected in cells with a distribution similar to that of the presumptive testes primordia of sexual worms (Fig. 6A and SI Fig. 10A); similar expression of Djnos was also reported in asexual D. japonica (32). This observation prompted us to test additional markers of the reproductive structures in asexual planarians (33). Of 14 genes examined that label the mature gonads in sexual planarians, 13 did not produce staining comparable with nanos in asexual worms (data not shown). However, germinal H4 (33) was also expressed in asexual planarians, labeling dorsolateral clusters of cells in a pattern similar to nanos (Fig. 6C and SI Fig. 10B) as well as somatic neoblasts, as indicated by double staining with anti-SMEDWI-1 antibodies (SI Fig. 10 C–E) (35).
Fig. 6.
Smed-nanos expression in the asexual strain of S. mediterranea and the effect of Smed-nanos RNAi in asexual planarians. (A) Whole-mount in situ hybridization to nanos labels clusters of dorsolateral cells reminiscent of the testes pattern in sexual planarians (n = 34). (B) Cells observed in A are undetectable 3 days after γ-irradiation (30 Gy) (n = 22). (C) germinal H4 is expressed in the neoblasts and clusters of cells similar to nanos-positive cells (n = 18). (D) γ-irradiation (30 Gy) eliminates germinal H4 expression (n = 15). (E and F) Control and RNAi animals fixed 1 month after amputation. (E) Animals fed control bacteria have germinal H4 expression similar to untreated planarians (n = 8 heads and 12 trunk pieces). (F) nanos RNAi planarians lose expression of germinal H4 expression from the dorsal cell clusters, whereas neoblast staining is unaffected (n = 9 heads and 11 trunk pieces). All animals were imaged dorsally by using differential interference contrast microscopy. (Scale bars, 200 μm.) (Insets) Higher magnification views of the boxed areas. (Inset scale bars, 100 μm.)
Previous work suggested that in asexual planarians, the only proliferating cells are neoblasts, the stem cells responsible for the animal's regenerative abilities, whereas differentiated cell types are postmitotic. After a lethal dose of γ-irradiation, neoblasts are eliminated, whereas differentiated cells are unaffected (35, 42, 43). Three days after γ-irradiation (30 Gy) the expression of nanos and germinal H4 mRNAs was eliminated from asexual planarians (Fig. 6 B and D). Thus, these genes may be expressed in a subset of the proliferating cell population, or neoblasts may give rise to short-lived germ cells in asexual planarians. Further experiments will be needed to clarify the basis of the radiation sensitivity of the presumptive germ cells.
Although nanos mRNA is detected in a specific cell population in asexual planarians, one possible explanation for the failure of these animals to develop mature reproductive organs is that nanos is not functional. To address this issue, we performed nanos RNAi experiments on asexual planarians (Fig. 6 E and F). Animals were fed twice with nanos dsRNA and then amputated 7 days after the second feeding; dsRNA feeding was resumed after 1 week, and animals were fixed 1 month after amputation. nanos RNAi knockdown animals regenerated and grew normally, but they lacked the dorsolateral population of germinal H4-expressing cells, whereas neoblast labeling was unaffected (Fig. 6F). A similar experiment was also performed on intact, asexual planarians. Animals were fed nanos dsRNA every 4–5 days for 1 month. All of the nanos RNAi animals (n = 15) lacked the dorsolateral population of germinal H4-positive cells observed in controls (n = 15; data not shown). Thus, nanos function is required for maintaining expression of germinal H4 in presumptive germ cells in intact and regenerating asexual planarians.
Our results provide evidence for conservation of nanos function in epigenetic germ cell specification in a representative of a basal protostome lineage. The functional genomic resources available for studying S. mediterranea will facilitate the identification and functional characterization of genes required for epigenetic germ cell specification and development of the reproductive system. Such studies should provide additional insight into the extent to which the mechanisms of germ cell specification have been conserved between diverse phylogenetic lineages.
Methods
Planarian Culture.
Clonal lines of hermaphroditic (33) and asexual (51) S. mediterranea were used for all experiments. Sexual planarians were maintained in 0.75× Montjuïc salts at 18°C and asexuals in 1× Montjuïc salts at 21°C (41). Animals were fed weekly with organic calf liver and starved 1 week before use.
In Situ Hybridization.
Whole-mount in situ hybridization was performed and imaged as described for sexual (33) and asexual animals (41). Within a given experiment, samples were developed with substrate for the same amount of time. For details on FISH, see SI Methods. For γ-irradiation experiments, planarians were exposed to 10 or 30 Gy as described previously (35).
RNAi.
RNAi feedings were performed as described previously (42). Control animals were fed bacteria containing pPR242 plasmid vector alone. For nanos RNAi, the corresponding cDNA was subcloned into pPR242 at the ApaI/NotI sites and confirmed by DNA sequencing.
Immunofluorescence.
Planarians were killed with 2% HCl, fixed in modified Schaudin's fixative (35), and stained as described previously (35, 41). Images were taken with a SteREO Lumar microscope (Zeiss, Thornwood, NY). Confocal images were obtained with a CARV confocal microscope (BD Biosciences, Rockville, MD) as described previously (35).
Supplementary Material
Acknowledgments
We thank Cristiana Hentea for help with cloning Smed-nanos, David Forsthoefel for helpful comments on the manuscript, Peter Reddien (Whitehead Institute, Massachusetts Institute of Technology) for providing pPR242, Howard Ducoff for use of the gamma source, and the anonymous reviewers whose constructive criticisms greatly improved this work. The Washington University Genome Sequencing Center generated the planarian genomic sequence data used here. R.M.Z. is a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research. This work was supported by National Science Foundation CAREER Award IBN-0237825 and National Institutes of Health Grant R01 HD043403 (to P.A.N.). P.A.N. was a Damon Runyon Scholar supported by Damon Runyon Cancer Research Foundation Grant DRS 33-03.
Abbreviation
- D
day.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Smed-nanos sequence reported in this paper has been deposited in the GenBank database (accession no. EF035555).
This article contains supporting information online at www.pnas.org/cgi/content/full/0609708104/DC1.
References
- 1.Seydoux G, Braun RE. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Nieuwkoop PD, Sutasurya LA. Primordial Germ Cells in the Chordates. Cambridge, UK: Cambridge Univ Press; 1979. [Google Scholar]
- 3.Nieuwkoop PD, Sutasurya LA. Primordial Germ Cells in the Invertebrates: From Epigenesis to Preformation. Cambridge, UK: Cambridge Univ Press; 1981. [Google Scholar]
- 4.Extavour CG, Akam M. Development (Cambridge, UK) 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- 5.Johnson AD, Drum M, Bachvarova RF, Masi T, White ME, Crother BI. Evol Dev. 2003;5:414–431. doi: 10.1046/j.1525-142x.2003.03048.x. [DOI] [PubMed] [Google Scholar]
- 6.Santos AC, Lehmann R. Curr Biol. 2004;14:R578–R589. doi: 10.1016/j.cub.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Seydoux G, Schedl T. Int Rev Cytol. 2001;203:139–185. doi: 10.1016/s0074-7696(01)03006-6. [DOI] [PubMed] [Google Scholar]
- 8.McLaren A. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 9.Matsui Y, Okamura D. Bioessays. 2005;27:136–143. doi: 10.1002/bies.20178. [DOI] [PubMed] [Google Scholar]
- 10.Newmark PA, Sánchez Alvarado A. Nat Rev Genet. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- 11.Reddien PW, Sánchez Alvarado A. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 12.Curtis WC. Proc Boston Soc Nat Hist. 1902;30:515–559. [Google Scholar]
- 13.Gremigni V. Boll Zool. 1974;41:359–377. [Google Scholar]
- 14.Morgan TH. Arch Ent Mech Org. 1902;13:179–212. [Google Scholar]
- 15.Baguñà J, Saló E, Auladell C. Development (Cambridge, UK) 1989;107:77–86. [Google Scholar]
- 16.Baguñà J, Carranza S, Pala M, Ribera C, Giribet G, Arnedo MA, Ribas M, Riutort M. Ital J Zool. 1999;66:207–214. [Google Scholar]
- 17.Weisblat DA. Curr Biol. 2006;16:R453–R455. doi: 10.1016/j.cub.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann R, Nusslein-Volhard C. Development (Cambridge, UK) 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, Lehmann R. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- 20.Forbes A, Lehmann R. Development (Cambridge, UK) 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi S, Yamada M, Asaoka M, Kitamura T. Nature. 1996;380:708–711. doi: 10.1038/380708a0. [DOI] [PubMed] [Google Scholar]
- 22.Deshpande G, Calhoun G, Yanowitz JL, Schedl PD. Cell. 1999;99:271–281. doi: 10.1016/s0092-8674(00)81658-x. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi Y, Hayashi M, Kobayashi S. Proc Natl Acad Sci USA. 2004;101:10338–10342. doi: 10.1073/pnas.0401647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramaniam K, Seydoux G. Development (Cambridge, UK) 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- 25.Koprunner M, Thisse C, Thisse B, Raz E. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki A, Tsuda M, Saga Y. Development (Cambridge, UK) 2007;134:77–83. doi: 10.1242/dev.02697. [DOI] [PubMed] [Google Scholar]
- 28.Jaruzelska J, Kotecki M, Kusz K, Spik A, Firpo M, Reijo Pera RA. Dev Genes Evol. 2003;213:120–126. doi: 10.1007/s00427-003-0303-2. [DOI] [PubMed] [Google Scholar]
- 29.Extavour CG, Pang K, Matus DQ, Martindale MQ. Evol Dev. 2005;7:201–215. doi: 10.1111/j.1525-142X.2005.05023.x. [DOI] [PubMed] [Google Scholar]
- 30.Chang CC, Lee WC, Cook CE, Lin GW, Chang T. Intl J Dev Biol. 2006;50:413–421. doi: 10.1387/ijdb.052100cc. [DOI] [PubMed] [Google Scholar]
- 31.Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Dev Biol. 2006;300:406–415. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 32.Sato K, Shibata N, Orii H, Amikura R, Sakurai T, Agata K, Kobayashi S, Watanabe K. Dev Growth Differ. 2006;48:615–628. doi: 10.1111/j.1440-169X.2006.00897.x. [DOI] [PubMed] [Google Scholar]
- 33.Zayas RM, Hernandez A, Habermann B, Wang Y, Stary JM, Newmark PA. Proc Natl Acad Sci USA. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cebrià F, Newmark PA. Development (Cambridge, UK) 2007;134:833–837. doi: 10.1242/dev.02794. [DOI] [PubMed] [Google Scholar]
- 35.Guo T, Peters AH, Newmark PA. Dev Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Ghirardelli E. In: Regeneration in Animals and Related Problems. Kiortsis V, Trampusch HAL, editors. Amsterdam: North–Holland; 1965. pp. 177–184. [Google Scholar]
- 37.Fedecka-Bruner B. Bull Biol Fr Belg. 1967;101:255–319. [PubMed] [Google Scholar]
- 38.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 39.Sánchez Alvarado A, Newmark PA. Proc Natl Acad Sci USA. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cebrià F, Kobayashi C, Umesono Y, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Itoh M, Taira M, Sanchez Alvarado A, et al. Nature. 2002;419:620–624. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]
- 41.Cebrià F, Newmark PA. Development (Cambridge, UK) 2005;132:3691–3703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- 42.Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A. Dev Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sanchez Alvarado A. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 44.Salvetti A, Rossi L, Lena A, Batistoni R, Deri P, Rainaldi G, Locci MT, Evangelista M, Gremigni V. Development (Cambridge, UK) 2005;132:1863–1874. doi: 10.1242/dev.01785. [DOI] [PubMed] [Google Scholar]
- 45.Newmark PA, Reddien PW, Cebria F, Sanchez Alvarado A. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11861–11865. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franquinet R, Lender T. Z Mikrosk Anat Forsch. 1972;86:481–495. [PubMed] [Google Scholar]
- 47.Silveira M, Porter KR. Protoplasma. 1964;59:240–265. [Google Scholar]
- 48.Farnesi RM, Marinelli M, Tei S, Vagnetti D. Riv Biol. 1977;70:113–136. [PubMed] [Google Scholar]
- 49.Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 50.Newmark PA, Sánchez Alvarado A. Dev Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- 51.Sánchez Alvarado A, Newmark PA, Robb SM, Juste R. Development (Cambridge, UK) 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.