Abstract
The proteasome maintains cellular homeostasis by degrading oxidized and damaged proteins, a function known to be impaired during aging. The proteasome also acts in a regulatory capacity through E3 ligases to mediate the spatially and temporally controlled breakdown of specific proteins that impact biological processes. We have identified components of a Skp1-Cul1-F-Box E3 ligase complex that are required for the extended lifespan of Caenorhabditis elegans insulin/insulin-like growth factor-1-signaling (IIS) mutants. The CUL-1 complex functions in postmitotic, adult somatic tissues of IIS mutants to enhance longevity. Reducing IIS function leads to the nuclear accumulation of the DAF-16/FOXO transcription factor, which extends lifespan by regulating downstream longevity genes. These CUL-1 complex genes act, at least in part, by promoting the transcriptional activity of DAF-16/FOXO. Together, our findings describe a role for an important cellular pathway, the proteasomal pathway, in the genetic determination of lifespan.
Keywords: aging, proteasome, ubiquitin, daf-2, insulin
The ability to maintain efficient protein turnover is a long-term challenge for cells and organisms that is amplified during aging (1). Improperly folded and defective proteins are degraded by the proteasome to maintain cellular homeostasis. These misfolded, oxidized, and damaged proteins accumulate with increasing age in extracts from the aging heart, lung, kidney, and liver in rats, and in different human tissues as well (1). The decline in the proteasomes' ability to function as a quality-control system is thought to contribute to, and to be aggravated by, the accumulation of damaged proteins. As a consequence, proteasomal inefficiency itself may contribute to the process of aging.
The proteasome also functions in a regulatory capacity by acting in a spatially and temporally controlled fashion to degrade specific proteins that govern biological processes. Regulated proteolysis influences many processes that require precise orchestration, including cell cycle progression, growth, and immunity (2). The substrates of regulated proteolysis receive a chain of ubiquitin (Ub) molecules that marks them for degradation. The attachment of Ub to a target protein involves its activation by an E1 (Ub-activating) enzyme and its subsequent transfer to an E2 (Ub-conjugating) enzyme. The E2 transfers Ub moieties to the substrate through its association with an E3 ligase (2).
E3 ligases play a key role in regulatory-proteasomal function. They physically recruit the ubiquitination target and thus determine the specificity and timing of degradation. A well known family of E3 ligases is the Skp1-Cul1-F-Box (SCF) family (3), which consists of scaffolding proteins called Cullins around which other components of the complex are organized. A Cullin simultaneously binds to an E2 enzyme carrying an activated Ub and to an adaptor. The adaptor, in turn, identifies and engages the substrate, thereby facilitating target ubiquitination (Fig. 1A). Cullins vary in their adaptor compatibility. For example, the Cul-1 complex employs two adaptor proteins: Cul-1 binds to a linker protein called Skp1, which in turn binds to a second adaptor protein containing an F-Box motif. The F-Box protein recognizes an individual substrate and recruits it to the Cul-1 complex for ubiquitination (Fig. 1A) (3). In general, multiple F-Box proteins, each with a unique substrate specificity, can bind the SCF complex, thus increasing the repertoire of cellular proteins whose degradation can be controlled. In this study, we have investigated the role of the SCF E3 ligase components in the regulation of aging.
Fig. 1.
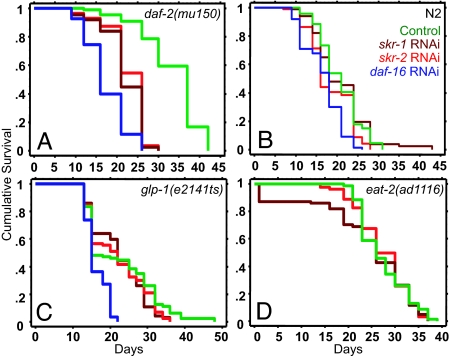
cul-1 RNAi shortens the extended lifespan of daf-2 mutants. (A) Schematic representation of the CUL-1 E3 ligase complex. (B–G) Lifespan curves of long-lived mutants and wild-type worms grown as adults on control empty vector (green lines), daf-16 RNAi (blue lines), and cul-1 RNAi (red lines). (B) daf-2(mu150). Control vector: mean = 33.4 ± 0.7, n = 89/90 (number of animals that died/total; see Materials and Methods); daf-16 RNAi: mean = 17.3 ± 0.5, n = 80/83, P < 0.0001 vs. control; cul-1 RNAi: mean = 25.2 ± 0.4, n = 80/85, P < 0.0001 vs. control. (C) daf-2(e1368). Control vector: mean = 32.7 ± 0.7, n = 58/105; daf-16 RNAi: mean = 22.1 ± 0.3, n = 76/105, P < 0.0001 vs. control; cul-1 RNAi: mean = 25.1 ± 0.4, n = 69/104, P < 0.0001 vs. control. (D) daf-2(e1370). Control vector: mean = 40.8 ± 1.0, n = 72/89; daf-16 RNAi: mean = 22.7 ± 0.5, n = 67/90, P < 0.0001 vs. control; cul-1 RNAi: mean = 31.1 ± 0.9, n = 77/83, P < 0.0001 vs. control. (E) N2. Control vector: mean = 20.7 ± 0.7, n = 67/88; daf-16 RNAi: mean = 16.6 ± 0.5, n = 67/87, P < 0.0001 vs. control; cul-1 RNAi: mean = 18.8 ± 0.6, n = 84/90, P = 0.3 vs. control. (F) glp-1(e2141ts). Control vector: mean = 23.2 ± 0.4, n = 86/87; daf-16 RNAi: mean = 17.8 ± 0.4, n = 79/84, P < 0.0001 vs. control; cul-1 RNAi: mean = 23.9 ± 0.3, n = 74/76, P = 0.4 vs. control. (G) eat-2(ad1116). Control vector: mean = 27.3 ± 0.6, n = 70/96; daf-16 RNAi: mean = 24.8 ± 0.6, n = 68/96, P = 0.005 vs. control; cul-1 RNAi: mean = 25.2 ± 0.5, n = 77/102, P = 0.2 vs. control.
Studies in worms, yeast, flies, and mice have identified several pathways that influence aging (4). In Caenorhabditis elegans, these include lifespan-extending processes activated by inhibition of the insulin/insulin-like growth factor-1 (IGF-1)-signaling (IIS) pathway (5), caloric restriction (6), and inhibition of mitochondrial respiration (7). In addition, a pathway regulated by the reproductive system that partially overlaps with the IIS pathway also affects lifespan (8). Reduction-of-function IIS-pathway mutations delay the anatomical and behavioral signs of aging and double lifespan (5). In the wild type, the IIS pathway is activated through DAF-2, the insulin/IGF-1 receptor, which activates a conserved PI-3 kinase/PDK/AKT, SGK cascade that results in phosphorylation of the FOXO transcription factor DAF-16 (4). When the IIS pathway is inhibited, this phosphorylation of DAF-16 is prevented, causing DAF-16 to accumulate in the nucleus (9–11), where it activates (or represses) individual antioxidant, antimicrobial, metabolic, and other genes whose combined activities produce large changes in lifespan (12, 13).
In this study, we demonstrate a role for proteasomal E3 ligases in the genetic control of organismal aging. We show that components of a putative SCF CUL-1 complex function in the postmitotic adult somatic tissues of daf-2/IIS-receptor mutants to promote longevity. These genes are required for DAF-16/FOXO to activate its target gene sod-3 in daf-2 mutants. This CUL-1 complex is not required for the longevity of wild-type animals or other long-lived mutants. Nor is it required for DAF-16/FOXO to initiate an alternative developmental state, dauer formation, when insulin/IGF-1 signaling is inhibited during development, or for DAF-16 to extend lifespan in response to germ-line removal. Thus this complex may be required to link the lifespan-extending activity of DAF-16 specifically to the insulin/IGF-1 pathway.
Results
Impairing General Proteasome Function Shortens C. elegans Lifespan Indiscriminately.
To assess the importance of general proteasomal function, we systematically depleted various structural and enzymatic subunits of the proteasome. We initiated RNAi during adulthood to circumvent the requirement for proteasomal function during development. Proteasomal-RNAi treatments elicited a dramatic shortening of lifespan in wild-type animals as well as in long-lived daf-2/IIS-receptor mutants, and a glp-1(e2141ts) mutant, whose lifespan is extended by germ cell loss (14) [supporting information (SI) Table 1]. This suggests that general proteasomal function is essential for viability of adult animals. In contrast, inactivating regulatory proteasomal function by RNAi-knockdown of the six predicted worm Cullins, CUL-1 to -6 (15), had distinct effects on the extended lifespan of daf-2(mu150) mutants. Some knockdowns suppressed lifespan extension (cul-1, cul-3), some had less significant effects (cul-4), and others had no effect at all (cul-5) (SI Table 1). However, unlike the general proteasomal subunits, RNAi-knockdown of Cullins did not shorten the lifespan of glp-1(e2141ts) mutants or wild-type worms (SI Table 1). [In fact, some RNAi treatments further extended the glp-1-mutant lifespan (SI Table 1).] These findings suggest that reducing Cullin function does not generally shorten lifespan, but instead that Cullin complexes might selectively influence the longevity of particular mutants. We chose to focus on cul-1 because cul-1 RNAi caused the most pronounced effect on daf-2 mutants' lifespan.
cul-1 Function Is Required Specifically for the Extended Lifespan of daf-2 Mutants.
In multiple experiments, cul-1 RNAi significantly reduced the extended lifespan of daf-2(mu150) mutants (Fig. 1B and SI Table 2) as well as daf-2(e1368) and daf-2(e1370) mutants (Fig. 1 C and D and SI Table 2). cul-1 RNAi also shortened the extended lifespans of animals carrying mutations in other IIS-pathway genes (data not shown). In contrast, the lifespan of wild-type worms was not affected (Fig. 1E and SI Table 2). To investigate whether cul-1 was required for lifespan extension by other means, we asked whether cul-1 RNAi shortened the long lifespan of the germ-line-defective glp-1(e2141ts) mutants. We found that it did not (Fig. 1F and SI Table 2). Caloric restriction increases lifespan in many organisms, including C. elegans. eat-2 mutants are used as a model for caloric restriction in worms (6). We found that cul-1 RNAi had no effect on the lifespan extension of eat-2(ad1116) mutants (Fig. 1G). Thus cul-1 function appears to be required specifically for the extended lifespan of IIS-pathway mutants.
cul-1 Acts in Postmitotic Adult Somatic Tissues to Regulate Lifespan.
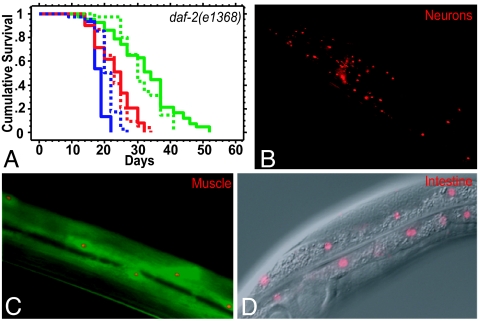
As in many other organisms, C. elegans cul-1 functions as a negative regulator of the cell cycle (15). In our experiments, cul-1 RNAi was initiated from the first day of adulthood, when all of the tissues are postmitotic except for the proliferating germ line. To investigate whether cul-1 RNAi might shorten the lifespan of daf-2 mutants by increasing germ-line proliferation (15), we removed the reproductive system by killing the gonad precursor cells Z1 and Z4 with a laser microbeam. cul-1 RNAi still shortened the lifespan of daf-2(e1368) mutants (Fig. 2A and SI Table 3), as well as that of gonad-ablated daf-2(mu150) mutants (data not shown). To determine whether cul-1 was expressed in somatic tissues of adult worms, we generated transgenic animals expressing a Pcul-1::rfp transcriptional reporter construct. We found that the transgene was expressed in the adult neurons, muscles, and intestine (Fig. 2 B–D). Together these experiments demonstrate that cul-1 functions in the somatic tissues of adult daf-2 mutants to influence lifespan.
Fig. 2.
cul-1 acts in postmitotic adult somatic tissues to regulate the lifespan of daf-2 mutants. (A) cul-1 RNAi shortens the extended lifespan of gonad-ablated daf-2 mutants. Lifespans of gonad precursor (Z1 and Z4) ablated daf-2(e1368) mutants (solid lines) and unablated control worms (dotted lines) grown as adults on bacteria expressing dsRNA for cul-1 (red), daf-16 (blue), and control vector (green) are shown. Z1,Z4(−) control vector (green solid line): mean = 32.6 ± 1.4, n = 43/43; Z1,Z4(−) cul-1 RNAi (red solid line): mean = 23.0 ± 0.6, n = 54/54, P < 0.0001 vs. control; Z1,Z4(−) daf-16 RNAi (blue solid line): mean = 18.2 ± 0.36, n = 30/30, P < 0.0001 vs. control; unablated control vector (green dotted line): mean = 31.4 ± 1.0, n = 35/43; unablated cul-1 RNAi (red dotted line): mean = 23.1 ± 0.9, n = 39/45, P < 0.0001 vs. control; unablated daf-16 RNAi (blue dotted line): mean = 20.8 ± 0.5, n = 40/45, P < 0.0001 vs. control. Similar results were obtained with additional repetitions of this experiment (SI Table 3). (B–D) cul-1 expression in adult somatic tissues. A Pcul-1::rfp construct reveals expression in adult neurons (head neurons are shown in B), muscles (C; colocalization with Pmyo-3::gfp coinjection marker), and intestinal cells (D; composite fluorescence-differential interference contrast microscopy image). The red fluorescent protein signal appears nuclear because of the presence of a nuclear localization signal in the construct for ease of cell identification.
skr-1 and skr-2 Are Required for the Longevity of daf-2 Mutants.
We sought to identify other components of the CUL-1 complex that influences the lifespan of daf-2 mutants. Skp1 in worms is represented by a group of 21 Skp1-related (SKR) proteins (16, 17). We subjected daf-2(mu150) mutants to RNAi for 15 of the 21 skr genes for which RNAi bacteria were available, and we examined the effects on longevity. We found that RNAi depletion of skr-1 and skr-2 shortened the lifespan of daf-2(mu150) mutants by up to 44% (Fig. 3A and SI Table 2). Some of the other skr RNAi clones also shortened the extended lifespan of daf-2(mu150) mutants, but not to the same extent (SI Table 4). skr-1 and skr-2 RNAi also shortened the lifespan extensions of daf-2(e1368) and daf-2(e1370) mutants substantially (SI Table 4). As with cul-1 RNAi, we did not observe any effect on the lifespan of wild-type animals (Fig. 3B) or the extended lifespans of glp-1(e2141ts) or eat-2(ad1116) mutants (Fig. 3 C and D). SKR-1 and SKR-2 have been shown to interact physically with C. elegans CUL-1 (16, 17). The two genes share 83% nucleotide identity and are predicted to produce cross-RNAi effects. Therefore, we refer to these genes as skr-1/2. These findings indicate that skr-1/2, like cul-1, are required for the extended lifespan of daf-2 mutants.
Fig. 3.
skr-1/2 are required for the extended lifespan of daf-2 mutants but not other long-lived mutants or wild-type worms. Lifespan experiments of long-lived mutants and wild-type worms grown as adults on control empty vector (green lines), daf-16 RNAi (blue lines), skr-1 RNAi (deep red lines), and skr-2 RNAi (bright red lines). (A) daf-2(mu150). Control vector: mean = 33.4 ± 0.7, n = 89/90; daf-16 RNAi: mean = 17.3 ± 0.5, n = 80/83, P < 0.0001 vs. control; skr-1 RNAi: mean = 22.0 ± 0.5, n = 88/91, P < 0.0001 vs. control; skr-2 RNAi: mean = 22.8 ± 0.4, n = 92/92, P < 0.0001 vs. control. (B) N2. Control vector: mean = 20.7 ± 0.7, n = 67/88; daf-16 RNAi: mean = 16.6 ± 0.5, n = 67/87, P < 0.0001 vs. control; skr-1 RNAi: mean = 20.7 ± 0.7, n = 82/86, P = 0.71 vs. control; skr-2 RNAi: mean = 21.4 ± 0.9, n = 67/81, P = 0.19 vs. control. (C) glp-1(e2141ts). Control vector: mean = 22.4 ± 1.0, n = 82/85; daf-16 RNAi: mean = 16.1 ± 0.3, n = 88/88, P < 0.0001 vs. control; skr-1 RNAi: mean = 21.8 ± 0.8, n = 88/91, P = 0.19 vs. control; skr-2 RNAi: mean = 22.1 ± 0.7, n = 92/92, P = 0.1 vs. control. (D) eat-2(ad1116). Control vector: mean = 27.3 ± 0.6, n = 70/96; skr-1 RNAi: mean = 23.8 ± 1.1, n = 66/94, P = 0.2 vs. control; skr-2 RNAi: mean = 27.3 ± 0.6, n = 67/93, P = 0.8 vs. control.
F-Box Adaptors That Influence the Lifespan of daf-2 Mutants.
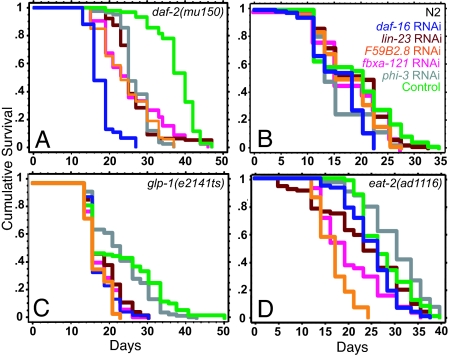
To identify F-Box adaptor proteins that influence the lifespan of daf-2 mutants, we performed a targeted F-Box RNAi screen to isolate suppressors of daf-2(mu150) extended lifespan. Based on the annotated gene information on the family of >400 genes encoding F-Box proteins predicted by Kipreos and Pagano (18) [a recent study has increased the predicted number to 520 (19)], we made a selective sublibrary of RNAi clones from a whole-genome feeding RNAi library (20). In a screen of this F-Box sublibrary, we found several clones that significantly shortened the extended lifespan of daf-2(mu150) mutants (data not shown). Of these, four clones produced a substantial reduction in lifespan when the RNAi treatment was performed only in adulthood (Fig. 4A). One of these clones corresponded to the gene lin-23, which encodes an F-Box protein known to function in the C. elegans CUL-1 complex for the regulation of larval cell cycles (21). The other clones corresponded to the genes fbxa-121 (Y18D10A.18), phi-3 (Y46G5A.6), and an unknown F-Box gene, F59B2.8 (Fig. 4A). We found either no effect or only a small reduction in the lifespan of wild-type worms in response to knockdown of any of these genes (Fig. 4B). Of the four genes, only phi-3 RNAi did not shorten the lifespan of any other long-lived mutants tested (Fig. 4 A–D). Loss of function of lin-23, fbxa-121, and F59B2.8 shortened the extended lifespans of glp-1(e2141ts) and eat-2(ad1116) mutants, although to different degrees (Fig. 4 C and D). This heterogeneity suggests that phi-3 may selectively promote the lifespan of IIS-pathway mutants, whereas other adaptors might influence different longevity pathways as well.
Fig. 4.
Effects of RNAi inactivation of F-Box adaptors on lifespan. Lifespan experiments with long-lived mutants and wild-type worms subjected as adults to feeding RNAi for control vector (green lines), daf-16 (blue lines), and genes encoding the following F-Box adaptor proteins: lin-23 (red lines), F59B2.8 (orange lines), fbxa-121 (pink lines), and phi-3 (gray lines). (A) daf-2(mu150). Control vector: mean = 37.8 ± 0.5, n = 80/91; daf-16 RNAi: mean = 17.8 ± 0.3, n = 92/92, P < 0.0001 vs. control; lin-23 RNAi: mean = 26.7 ± 0.3, n = 78/84, P < 0.0001 vs. control; F59B2.8 RNAi: mean = 24.4 ± 0.7, n = 86/87, P < 0.0001 vs. control; fbxa-121 RNAi: mean = 25.8 ± 0.9, n = 81/86, P < 0.0001 vs. control; phi-3 RNAi: mean = 27.0 ± 0.4, n = 84/84, P < 0.0001 vs. control. (B) N2. Control vector: mean = 18.6 ± 0.8, n = 70/88; daf-16 RNAi: mean = 15.6 ± 0.4, n = 86/90, P < 0.0001 vs. control; lin-23 RNAi: mean = 18.6 ± 0.6, n = 82/94, P = 0.68 vs. control, P < 0.0001 vs. daf-16 RNAi; F59B2.8 RNAi: mean = 17.5 ± 0.5, n = 74/89, P = 0.07 vs. control, P = .06 vs. daf-16 RNAi; fbxa-121 RNAi: mean = 18.3 ± 0.7, n = 59/86, P = 0.64 vs. control, P = 0.02 vs. daf-16 RNAi; phi-3 RNAi: mean = 15.7 ± 0.5, n = 80/91, P = .007 vs. control, P = 0.31 vs. daf-16 RNAi. (C) glp-1(e2141ts). Control vector: mean = 22.4 ± 1.0, n = 82/85; daf-16 RNAi: mean = 16.1 ± 0.3, n = 88/88, P < 0.0001 vs. control; lin-23 RNAi: mean = 17.9 ± 0.4, n = 85/90, P < 0.0001 vs. control; F59B2.8 RNAi: mean = 16.8 ± 0.3, n = 85/88, P < 0.0001 vs. control; fbxa-121 RNAi: mean = 17.0 ± 0.4, n = 90/90, P < 0.0001 vs. control; phi-3 RNAi: mean = 22.2 ± 0.7, n = 92/94, P = 0.2 vs. control. (D) eat-2(ad1116). Control vector: mean = 27.3 ± 0.6, n = 70/96; lin-23 RNAi: mean = 23.0 ± 0.9, n = 85/90, P < 0.02 vs. control; F59B2.8 RNAi: mean = 16.6 ± 0.4, n = 64/96, P < 0.0001 vs. control; fbxa-121 RNAi: mean = 20.2 ± 0.9, n = 90/90, P < 0.0001 vs. control; phi-3 RNAi: mean = 29.5 ± 0.7, n = 74/94, P = 0.008 vs. control.
The CUL-1 Complex Genes Affect DAF-16/FOXO Activity.
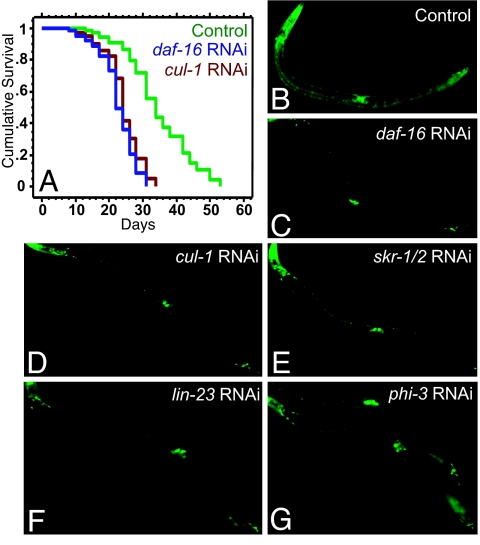
The extended lifespan of daf-2 mutants is completely dependent on the DAF-16/FOXO transcription factor (5). In daf-2 mutants, DAF-16 accumulates in the nucleus, where it regulates the expression of genes that influence longevity (9–13). This led us to investigate whether the CUL-1 complex might influence lifespan by affecting DAF-16. In principle, CUL-1 complex-RNAi could reduce (i) the level of DAF-16 protein, (ii) nuclear localization of DAF-16, and/or (iii) transcriptional activity of DAF-16. In daf-2 mutants subjected to RNAi-depletion of cul-1, skr-1/2, and the four F-Box genes, we found no noticeable reduction in either the level of DAF-16-GFP or its nuclear accumulation (22) (data not shown). Similarly, cul-1 RNAi still produced a strong suppression of the extended lifespans of daf-16; daf-2 mutants expressed a functional, constitutively nuclear form of DAF-16 (DAF-16AM-GFP) (23) (Fig. 5A; see SI Table 5 for background information).
Fig. 5.
RNAi of cul-1 complex genes affects DAF-16 transcriptional activity. (A) daf-16(mu86); daf-2(e1370); Pdaf-16::daf-16AM::gfp worms were subjected to RNAi treatment from day 1 of adulthood. Empty vector control (green curve): mean = 34.6 ± 0.6, n = 94/102; daf-16 RNAi (blue curve): mean = 22.7 ± 0.7; n = 84/90, P < 0.0001 vs. control; cul-1 RNAi (red curve): mean = 24.3 ± 0.3, n = 95/104, P < 0.0001 vs. control. (B–G) daf-2(e1370); Psod-3::gfp worms subjected to RNAi of cul-1 complex genes from the L4 stage through day 2 of adulthood. sod-3 expression is increased in daf-2 mutants (B) in a daf-16-dependent manner (C). This increased expression is significantly reduced in RNAi of cul-1 (D), skr-1/2 (E), and lin-23 (F). A milder, variable effect was observed on phi-3 RNAi (G). Data from multiple trials are shown in SI Table 5. Bar graph representation is found in SI Fig. 7.
To ask whether DAF-16 transcriptional activity was modified, we monitored Psod-3::gfp, a direct transcriptional reporter of DAF-16 activity that is up-regulated in daf-2 mutants in a daf-16 dependent fashion (12, 22, 24). We found that RNAi inhibition of cul-1, skr-1/2 and lin-23 in daf-2(e1370) mutants also produced a significant reduction in Psod-3::gfp expression (Fig. 5 B–F, SI Table 5, and SI Fig. 7). phi-3 and F59B2.8 RNAi treatments produced a more variable and milder reduction in GFP levels (Fig. 5G, SI Table 5, and SI Fig. 7). There was no significant effect following fbxa-121 RNAi (SI Table 5 and SI Fig. 7). These experiments suggested that CUL-1 complexes containing the F-Box protein LIN-23 (and possibly PHI-3) may influence the ability of DAF-16 to regulate the expression of its target genes.
A Mutation in the PAPP Domain of lin-23 Shortens the Extended Lifespan of daf-2 Mutants.
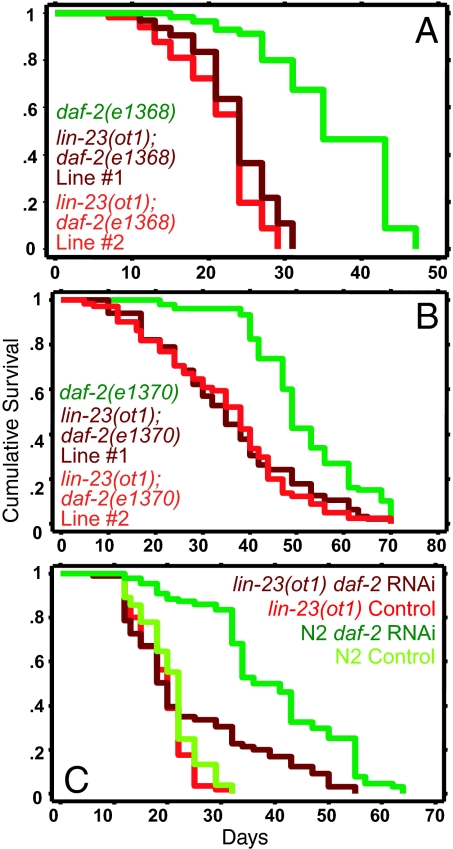
LIN-23 contains two protein–protein interaction domains, shown in other organisms to bind substrates. One domain, containing seven WD repeats, is required for LIN-23's cell cycle function (21, 25, 26). A second domain, the PAPP domain, is predicted to bind SH3- and WW-domain-containing proteins (SI Fig. 8) (25). lin-23(ot1) mutants contain a single amino acid substitution in the PAPP domain. They display overextended axons in some neurons but no cell cycle defects (25). We asked whether this mutant could recapitulate the RNAi effect of lin-23 reduction of function on the lifespan of daf-2 mutants. We found that lin-23(ot1) reduced the lifespan extension of daf-2 mutants (Fig. 6 A and B and SI Table 6) and also attenuated the lifespan extension induced by daf-2 RNAi (Fig. 6C and SI Table 6). These data showed that lin-23 is required for the extended lifespan of daf-2 mutants for a function that is independent of its developmental role in cell cycle regulation.
Fig. 6.
A mutation in the PAPP domain of lin-23 shortens the extended lifespan of daf-2 mutants. (A and B) The lin-23(ot1) mutation shortens the extended lifespan of daf-2 mutants. (A) daf-2(e1368) (green curve): mean = 36.3 ± 1.1, n = 43/72; lin-23(ot1); daf-2(e1368) line #2 (bright red curve): mean = 21.6 ± 0.7, n = 46/68, P < 0.0001 vs. control; lin-23(ot1); daf-2(e1368) line #1 (deep red curve): mean = 23.7 ± 0.6, n = 56/86, P < 0.0001 vs. control. (B) daf-2(e1370) (green curve): mean = 55.2 ± 2.0, n = 58/86; lin-23(ot1); daf-2(e1370) line #1 (deep red curve): mean = 37.4 ± 2.2, n = 41/79, P < 0.0001 vs. control; lin-23(ot1); daf-2(e1370) line #2 (bright red curve): mean = 38.8 ± 1.6, n = 57/90, P < 0.0001 vs. control. In the above experiments, worms were grown at 20°C on normal food (OP50) for their entire life. (C) lin-23(ot1) mutants display significantly reduced lifespan extension in response to daf-2 RNAi as compared with wild-type worms. lin-23(ot1) mutants grown at 20°C on normal OP50 bacteria from hatching until L4 and then shifted to feeding RNAi bacteria expressing dsRNA for the control empty vector (bright red curve, mean = 19.1 ± 0.5, n = 62/106) and daf-2 (deep red curve, mean = 24.3 ± 1.5, n = 68/105, P < 0.0001 vs. control). N2 worms grown under similar conditions on control empty vector (bright green curve, mean = 20.7 ± 0.6, n = 58/111) and daf-2 RNAi (deep green curve, mean = 39.7 ± 1.4, n = 74/106, P < 0.0001 vs. control). Similar results were obtained when the worms were grown at 20°C during development and transferred to 25°C as adults, except with daf-2(e1370). lin-23(ot1) extended the lifespan of daf-2(e1370) mutants at 25°C (SI Table 6).
Discussion
In this study, we have shown that individual proteins known to function together in a CUL-1 E3 ligase complex are required for the extended lifespan of C. elegans IIS-pathway mutants. Our findings suggest that this complex extends lifespan, at least in part, by promoting the transcriptional activity of the FOXO transcription factor DAF-16. This is previously unrecognized evidence that E3 ligase genes can promote longevity.
General Proteasome Function Is Required for the Viability of Adult Worms.
The proteasome performs housekeeping functions (such as degradation of misfolded, oxidized, and damaged proteins) as well as regulatory functions (spatially and temporally controlled breakdown of proteins that impact specific biological processes). We found that in both wild-type animals and long-lived mutants, inactivating general proteasome function shortened lifespan dramatically. The apparent uniformity of these effects across different genotypes suggests that the housekeeping function of the proteasome is required in the adult to prevent the accumulation of misfolded or oxidized proteins and/or to perform other essential functions. It is possible that general proteasomal genes also have specific functions in lifespan regulation, but their requirement for cell viability precluded the identification of such function(s).
The Mechanism of Regulation of Aging by the CUL-1 Complex.
To our knowledge, there have been no attempts to ask whether proteasomal E3 ligases influence the rate of aging in any organism. We found that RNAi depletion of different E3 ligase subunits that are known to determine the specificity of proteasomal function had distinct effects on the lifespan of different long-lived mutants and wild-type worms. One set, comprising cul-1, skr-1/2, and four F-box genes, seemed particularly interesting to us because its members were required for the longevity of IIS mutants but not that of wild-type worms. Orthologues of CUL-1, SKR-1/2, and one of these F-box proteins, LIN-23, are known to form complexes that target specific proteins for degradation (27). Thus we propose that these proteins also form a complex that affects lifespan.
There are at least two possible mechanisms that explain the effect of inhibiting the CUL-1 complex on lifespan. The first possibility is that cul-1 and skr-1/2 affect lifespan as part of the housekeeping activity of the proteasome. In this model, animals with reduced CUL-1 complex activity are healthier than animals with reduced levels of general proteasomal subunits simply because fewer, or nonessential, housekeeping functions are affected. Alternatively, a CUL-1 complex could ubiquitinate and degrade one or more proteins that actively regulate the rate of aging. These two possibilities are not mutually exclusive. However, if the CUL-1 complex performed a general housekeeping function, then reducing cul-1 or skr-1/2 activity would be expected to shorten the lifespans of all animals, including wild-type animals and multiple long-lived mutants. Instead, cul-1 and skr-1/2 RNAi only affected the lifespan of IIS-pathway mutants. In addition, RNAi of cul-1, skr-1/2, and some of the F-box proteins we analyzed prevented daf-2 mutants from up-regulating sod-3, a direct DAF-16 target gene. Thus the simplest interpretation of our findings is that the CUL-1 complex performs a regulatory role within the IIS pathway.
Based on our results, a CUL-1 complex composed of CUL-1, SKR-1/2 and one or more of the F-Box proteins we identified seems likely to influence the lifespan of IIS mutants by ubiquitination, and probably degradation, of proteins that regulate IIS. In principle, the target of this complex could be a member of the IIS-kinase cascade. If so, then the levels of this kinase should be lower in daf-2 mutants than in wild-type animals. However, we found that daf-2 mutants do not display reduced levels of AKT-1, AKT-2, or PDK-1 (SI Fig. 9, SI Movies 1 and 2). The levels of one IIS-pathway kinase, SGK-1, have been reported to be reduced in daf-2(e1370) mutants grown at 25°C (28), but we observed no change at 20°C, where CUL-1-complex RNAi strongly affects lifespan. Moreover, at 25°C, there was no change in SGK-1 levels upon RNAi of the CUL-1 complex genes (data not shown). Thus the known IIS kinases are probably not targeted by the proteins identified in our study. Consistent with this interpretation, DAF-16 nuclear localization, which is regulated by this kinase cascade, was not affected by CUL-1 complex RNAi, and CUL-1 function is still required in animals carrying the constitutively nuclear DAF-16AM protein (Fig. 5A).
Our previous experiments, and those of others, have demonstrated that the DAF-2 pathway has outputs in addition to the phosphorylation of DAF-16 on consensus AKT phosphorylation sites (11, 28, 29). For instance, in a daf-16; daf-2 mutant background, DAF-16AM extends lifespan substantially (≈80%), but in a daf-16(−); daf-2(+) background, DAF-16AM extends lifespan by only ≈30% (11). These and other findings have suggested that in long-lived daf-2 mutants, the nuclear translocation of DAF-16 may be accompanied by the modification of one or more cofactors. Genome-wide RNAi screens in worms have identified a number of new genes whose RNAi depletion extends lifespan and that are predicted to function in the IIS pathway (30, 31). The CUL-1 complex may target one or more of these proteins.
In animals with low IIS, DAF-16 not only acts in adults to extend lifespan, it also acts during development to promote dauer formation (32). We did not observe any noticeable effects on dauer formation in daf-2 mutants on RNAi of CUL-1-complex genes (data not shown). DAF-16 also acts during adulthood to increase lifespan in response to germ-line removal (8, 11, 14), and the CUL-1 complex was not required for this process either. Thus the CUL-1 complex may impart specificity to DAF-16 activity by linking DAF-16 activity to reduced IIS.
Proteasomal Regulation of IIS Pathways in Vertebrates.
In vertebrates, a CUL-1 complex has been shown to inhibit the activity of FOXO proteins by catalyzing their degradation (33–35). Likewise, the E3 ligase NEDD4–1 catalyzes the degradation of another conserved component of the IIS cascade, the phosphatase PTEN, which potentiates FOXO activity (36). This contrasts with our findings that in C. elegans, a CUL-1 complex is required for the activity of DAF-16/FOXO under conditions of low IIS. Given the many similarities between IIS pathways in worms and vertebrates, it will be interesting to learn whether specific CUL-1 complexes may promote FOXO activity under conditions of reduced IIS activity in vertebrates as well as in worms, and thus potentially enhance their longevity.
Materials and Methods
Strains.
The following strains were used in this study: CF1844 fer-15(b26) II; daf-2(mu150) III; fem-1(hc17) IV; CF1041 daf-2(e1370) III; DR1572 daf-2(e1368) III; CB4037 glp-1(e2141ts) III; CF1908 eat-2(ad1116); CF1579 daf-2(e1370) III; muIs74 [pAD76 (Psod-3::gfp, rol-6)], CF1380 daf-16(mu86) I; daf-2(e1370) III; muEx158(Pdaf-16::daf-16AM::gfp, Psur-5::gfp). The lin-23(ot1) mutant (OH1476; lin-23(ot1) II; oxIs12 [Punc-47::gfp] X) was outcrossed thrice to our laboratory N2 stock (N2A) to generate the strain CF2279 lin-23(ot1) II; oxIs12 (Punc-47::gfp) X. CF2279 was crossed to CF1041 daf-2(e1370) and DR1572 daf-2(e1368) to generate double mutants strains (SI Table 6).
Lifespan Analysis.
All lifespans were performed as described (30). For RNAi experiments, worms were grown on OP50 bacteria until the L4 stage (day 0) or first day of adulthood (day 1), as applicable, and then transferred to plates containing RNAi bacteria for the duration of adult life. daf-2(mu150) mutants were grown at 20°C for ≈24 h and then shifted to 25°C for sterility. For glp-1(e2141ts) lifespan assays, animals were raised at 25°C to eliminate germ cells, then shifted to 20°C at day 1 for the rest of the assay. Lifespans of all other mutants and wild-type worms were performed at 20°C. The n in lifespan figures is the number of animals that died/total. The total number of observations equals the number of animals that died plus the number of censored animals that crawled off the plate, exploded, bagged, or became contaminated.
RNAi Experiments and F-Box Screen.
RNAi by feeding was generally performed as described (30). For the F-Box screen, a sublibrary of C. elegans RNAi clones was isolated from the library described in ref. 20. In some instances RNAi clones described elsewhere were used (37). Approximately 100 eggs of the daf-2(mu150) mutant strain CF1844 were transferred to RNAi plates and allowed to grow at 20°C for ≈24 h before transferring to 25°C. Negative (empty vector) control [pAD12 (7)] and positive control daf-16 RNAi bacteria [pAD43 (7)] were seeded on multiple plates and monitored throughout the screen. Plates were screened for suppressors of lifespan extension on approximately day 16 (≈50% worms dead on pAD43 control plates; >80% alive on pAD12 plates) and approximately day 25 (≈100% worms dead on pAD43; >50% alive on pAD12). The identity of all RNAi clones isolated from the library was verified by sequencing of inserts with an M13-forward primer and, upon start of every lifespan analysis, by PCR with T7 primers. All positive hits from the screen were retested by using standard lifespan assays described above.
Molecular Biology and Sequencing.
A Pcul-1::rfp promoter was generated as described (38). Approximately 5 kb of cul-1 upstream promoter region was amplified by using the primers: forward A: 5′-GAG AGC AAA GTC GCC CAC AAT CAC ATC-3′ and reverse B: 5′-ACG CTT CTT CTT TGG CAT GGT GGT CCA AAC CAC CTC GGA ATC ACA CGT-3′. Pmyo-3::gfp with a nuclear-localization signal (NLS) was amplified from the plasmid pPZ024 by using the primers: forward PM034: 5′-ACC ACC ATG CCA AAG AAG AAG CGT-3′ and reverse PM031: 5′-AAG GGC CCG TAC GGC CGA CTA GTA GG-3′. PCR products from these two reactions were pooled and amplified with the following nested primers to obtain the transcriptional fusion: forward A*: 5′-GAG TTG CCA AAG ATG AGC GGT GCT C-3′ and reverse PM075: GGA AAC AGT TAT GTT TGG TAT ATT GGG AAT GTA TTC TG-3′.
Transgenic Strains.
The Pcul-1::rfp transcriptional reporter construct was injected (40 ng/μl), along with the coinjection marker Pmyo-3::gfp (75 ng/μl) into N2 to generate independent transgenic lines (indicated by muEx designation). muEx345–muEx349 were established into strains CF2283a–CF2283e, respectively.
Microscopy.
Psod-3::gfp and Ppdk-1::pdk-1::gfp pictures were captured by using a Retiga EXi Fast1394 CCD digital camera (QImaging, Burnaby, BC, Canada) attached to a Zeiss Axioplan 2 compound microscope (Zeiss Corporation, Jena, Germany). Openlab 4.0.2 software (Improvision, Coventry, U.K.) was used for image acquisition. GFP assays were conducted on either a Zeiss M2Bio or Leica MZ16F (Wetzlar, Germany) dissecting microscope with fluoresence attachment. Photographs of Pakt-1::akt-1::gfp and Pakt-2::akt-2::gfp were taken by using a Leica DFC340 black and white camera.
Supplementary Material
Acknowledgments
We are grateful to Oliver Hobert, Gary Ruvkun, Maren Hertweck, Ralf Baumeister, Thimo Kurz, and Bruce Bowerman for strains and reagents; M. Gaglia, M. Hansen, A. Kao, S. J. Lee, and L. Mitic for helpful comments on the manuscript; P. Zhang for Pmyo-3::gfp reagents; other C.K. laboratory members for support; and J. Alcedo and J. R. Berman for their comments on this article. A.G. gratefully acknowledges the continuous support from Cori Bargmann and Andrew Fire in the critical review and discussion of this work. C.K. is an American Cancer Society Professor and a founder of Elixir Pharmaceuticals. A.G. was supported by a Glen/American Federation for Aging Research (AFAR) junior postdoctoral fellowship and the Larry Hillblom Foundation for Aging Research postdoctoral grant. S.H.-K. was supported by a Human Frontier Science Program fellowship. This work was supported by National Institutes of Health Grant R01 AG011816 (to C.K.).
Abbreviations
- SCF
Skp1-Cul1-F-Box
- IGF-1
insulin-like growth factor-1
- Ub
ubiquitin
- IIS
insulin/IGF-1 signaling.
Note Added in Proof.
While this study was in press, Li et al. (39) published results identifying an E3 ligase that affects aging in an opposite fashion.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700638104/DC1.
References
- 1.Gaczynska M, Osmulski PA, Ward WF. Mech Ageing Dev. 2001;122:235–254. doi: 10.1016/s0047-6374(00)00246-3. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Willems AR, Schwab M, Tyers M. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 4.Kenyon C. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 6.Lakowski B, Hekimi S. Proc Natl Acad Sci USA. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 8.Hsin H, Kenyon C. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 9.Henderson ST, Johnson TE. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee RY, Hench J, Ruvkun G. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 11.Lin K, Hsin H, Libina N, Kenyon C. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 12.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 13.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 14.Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 15.Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 16.Nayak S, Santiago FE, Jin H, Lin D, Schedl T, Kipreos ET. Curr Biol. 2002;12:277–287. doi: 10.1016/s0960-9822(02)00682-6. [DOI] [PubMed] [Google Scholar]
- 17.Yamanaka A, Yada M, Imaki H, Koga M, Ohshima Y, Nakayama K. Curr Biol. 2002;12:267–275. doi: 10.1016/s0960-9822(02)00657-7. [DOI] [PubMed] [Google Scholar]
- 18.Kipreos ET, Pagano M. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-5-reviews3002. REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas JH. Genome Res. 2006;16:1017–1030. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 21.Kipreos ET, Gohel SP, Hedgecock EM. Development (Cambridge, UK) 2000;127:5071–5082. doi: 10.1242/dev.127.23.5071. [DOI] [PubMed] [Google Scholar]
- 22.Libina N, Berman JR, Kenyon C. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 23.Lin K, Dorman JB, Rodan A, Kenyon C. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 24.Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 25.Mehta N, Loria PM, Hobert O. Genetics. 2004;166:1253–1267. doi: 10.1534/genetics.166.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dreier L, Burbea M, Kaplan JM. Neuron. 2005;46:51–64. doi: 10.1016/j.neuron.2004.12.058. [DOI] [PubMed] [Google Scholar]
- 27.Latres E, Chiaur DS, Pagano M. Oncogene. 1999;18:849–854. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- 28.Hertweck M, Gobel C, Baumeister R. Dev Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- 29.Paradis S, Ruvkun G. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen M, Hsu AL, Dillin A, Kenyon C. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riddle DL. C. elegans II. Woodbury, NY: Cold Spring Harbor Lab Press; 1997. [Google Scholar]
- 33.Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, Tindall DJ. Proc Natl Acad Sci USA. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plas DR, Thompson CB. J Biol Chem. 2003;278:12361–12366. doi: 10.1074/jbc.M213069200. [DOI] [PubMed] [Google Scholar]
- 35.Aoki M, Jiang H, Vogt PK. Proc Natl Acad Sci USA. 2004;101:13613–13617. doi: 10.1073/pnas.0405454101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, et al. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamesch P, Milstein S, Hao T, Rosenberg J, Li N, Sequerra R, Bosak S, Doucette-Stamm L, Vandenhaute J, Hill DE, Vidal M. Genome Res. 2004;14:2064–2069. doi: 10.1101/gr.2496804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hobert O. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- 39.Li W, Gao B, Lee SM, Bennett K, Fang D. Dev Cell. 2007;12:235–246. doi: 10.1016/j.devcel.2006.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.