Abstract
CD1d-restricted natural killer T (NKT) cells, expressing the invariant T cell antigen receptor (TCR) chain encoded by Vα14-Jα18 gene segments in mice and Vα24-Jα18 in humans [invariant NKT (iNKT) cells], contribute to immunoregulatory processes, such as tolerance, host defense, and tumor surveillance. iNKT cells are positively selected in the thymus by CD1d molecules expressed by CD4+/CD8+ cortical thymocytes. However, the identity of the endogenous lipid(s) responsible for positive selection of iNKT cells remains unclear. One candidate lipid proposed to play a role in positive selection is isoglobotrihexosylceramide (iGb3). However, no direct evidence for its physiological role has been provided. Therefore, to directly investigate the role of iGb3 in iNKT cell selection, we have generated mice deficient in iGb3 synthase [iGb3S, also known as α1–3galactosyltransferase 2 (A3galt2)]. These mice developed, grew, and reproduced normally and exhibited no overt behavioral abnormalities. Consistent with the notion that iGb3 is synthesized only by iGb3S, lack of iGb3 in the dorsal root ganglia of iGb3S-deficient mice (iGb3S−/−), as compared with iGb3S+/− mice, was confirmed. iGb3S−/− mice showed normal numbers of iNKT cells in the thymus, spleen, and liver with selected TCR Vβ chains identical to controls. Upon administration of α-galactosylceramide, activation of iNKT and dendritic cells was similar in iGb3S−/− and iGb3S+/− mice, as measured by up-regulation of CD69 as well as intracellular IL-4 and IFN-γ in iNKT cells, up-regulation of CD86 on dendritic cells, and rise in serum concentrations of IL-4, IL-6, IL-10, IL-12p70, IFN-γ, TNF-α, and Ccl2/MCP-1. Our results strongly suggest that iGb3 is unlikely to be an endogenous CD1d lipid ligand determining thymic iNKT selection.
Keywords: A3galt2, CD1d, glycosphingolipid, KRN7000, dendritic cell
Presentation of peptide antigens to T cells by MHC molecules constitutes a well established mechanism essential for development, priming, and reactivity of T cells to specific peptide antigens. CD1 molecules, which are evolutionarily related to MHC molecules (1), are nonpolymorphic transmembrane proteins that associate with β2-microglobulin and present various lipid antigens to T lymphocytes (2). In humans, the CD1 family encompasses five isoforms (CD1a to -e). Based on amino acid sequence homology, the five members of the human CD1 family have been assigned to either group 1, which comprises CD1a, -b, -c, and -e molecules, or group 2, which consists of the CD1d molecule (3). Group 1 CD1 molecules are not present in mice and rats, whereas CD1d molecules are highly conserved in all mammalian species studied thus far (3).
Natural killer T (NKT) cells represent a distinct lymphocyte population that coexpress NK surface markers such as NK1.1 (CD161) and T cell antigen receptor (TCR) (4, 5). A subset of NKT cells express an invariant TCR α-chain (Vα14-Jα18 in mouse and Vα24-Jα18 in human) with a restricted set of TCR β chains (Vβ2, Vβ7, and Vβ8.2 in mouse and Vβ11 in human) (4, 6, 7). These NKT cells are referred to as invariant NKT (iNKT) cells and, unlike MHC class I and II restricted T cells, they are restricted by CD1d molecules. α-Galactosylceramide (αGalCer), a glycosphingolipid (GSL) isolated from a marine sponge, was identified as a specific and strong ligand for this invariant TCR, causing a rapid release of large amounts of Th1 and Th2 type cytokines (8). Although iNKT cells constitute <1% of mouse lymphocytes, they seem to play an important role in defense against infections with bacteria (e.g., Mycobacterium and spirochete), fungi (e.g., Cryptococcus), and parasites (e.g., Trypanosoma), in tumor surveillance as well as in establishing peripheral tolerance (9, 10).
Development of iNKT cells follows a TCR instructive process, as for conventional MHC class I and II restricted T cells. However, iNKT cells are positively selected in the thymus by the presentation of endogenous lipid ligands by CD1d on double-positive (CD4+/CD8+) cortical thymocytes, whereas conventional T cells are selected by peptide-MHC class I or II expressed on cortical epithelial cells (11). Loading of CD1d molecules with lipid antigens occurs in a low pH endosomal/lysosomal compartment (12). Mice lacking CD1d or showing aberrant CD1d trafficking and antigen presentation, because of a mutation in the cytoplasmic tail of CD1d, display strongly diminished iNKT cell numbers as a consequence of a failure to undergo positive selection in the thymus (13, 14). Sphingolipid activator proteins, also known as saposins (15), and lysosomal proteases (16) are also required for normal iNKT cell selection in the thymus. The identity of the endogenous selecting ligand(s) is unknown, but evidence suggests it may be a GSL because of the inability of a glucosylceramide deficient cell line to stimulate iNKT cell hybridomas (17).
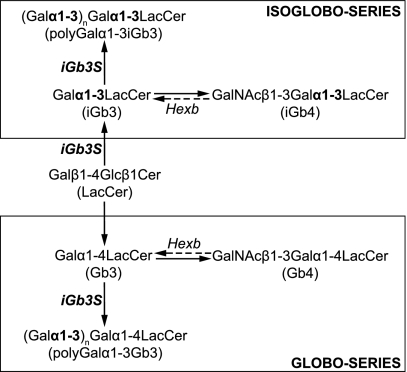
Zhou et al. (18) reported that in mice deficient in the β-subunit of β-hexosaminidase (Hexb−/−, a mouse model of the GSL lysosomal storage disorder; Sandhoff disease; Fig. 1), iNKT cells were present at greatly reduced frequency. It was reasoned that the lysosomal enzymes deficient in Sandhoff disease, β-hexosaminidase A and B, generate the natural lipid ligand in the thymus. Of the known degradation products resulting from the activity of these enzymes, only isoglobotrihexosylceramide (iGb3) showed in vitro stimulatory activity toward iNKT cells (18, 19). Therefore, it was concluded that iGb3 is the endogenous ligand mediating positive selection of iNKT cells in the thymus. A subsequent study using an iGb3-blocking lectin then implicated iGb3 in peripheral activation of iNKT cells by dendritic cells (DCs) (20).
Fig. 1.
Metabolic pathways of globo- and isoglobo-series of GSL. iGb3 and Gb3, the initial members of isoglobo- and globo-series, respectively, are synthesized from lactosylceramide (LacCer) by addition of a galactose (Gal) sugar moiety in α1–3 or α1–4 linkage, respectively. Through subsequent addition of N-acetylgalactosamine (GalNAc), iGb4 and Gb4 are formed. Lysosomal breakdown (dashed arrows) of iGb4 and Gb4 is performed by β-hexosaminidases A and B, which both require a functional β-subunit (Hexb). Furthermore, through the action of iGb3S on iGb3 and Gb3, polyGalα1–3iGb3 and polyGalα1–3Gb3 structures are formed. For sake of clarity, other enzymes and members of these GSL series have been omitted.
Doubts as to the relevance of iGb3 to iNKT cell selection in vivo has recently arisen based on the observation that iNKT cell numbers are decreased in other mouse models of GSL storage diseases, which would not primarily interfere with iGb3 formation (21, 22). These findings do not necessarily call into question the hypothesis of iGb3 being the iNKT-selecting endogenous ligand, because they are consistent with the alternative hypothesis that lipid accumulation per se could impede the loading of an endogenous ligand on CD1d, such as iGb3, resulting in the impaired positive selection of iNKT cells (21, 23).
One prerequisite for the involvement of iGb3 in iNKT cell selection is that this GSL must be present in the thymus. In an accompanying manuscript, Speak et al. (24) did not find any biochemical evidence for the existence of this lipid in the thymus. However, this could reflect very low levels of expression of this lipid, although that explanation was deemed unlikely, because no storage of iGb3 occurred in the thymus of a mouse model of Fabry disease (deficient in α-galactosidase), which would accumulate this lipid if it were present.
In this study, we have therefore taken a genetic approach to test whether iGb3 is a physiologically relevant selecting ligand in the mouse by generating mice deficient in iGb3 synthase [iGb3S, also known as α1–3galactosyltransgerase 2 (A3galt2)] using homologous recombination in murine embryonic stem cells. These mice show normal iNKT development, numbers, TCR Vβ usage, and function. In agreement with Speak et al. (24), we therefore conclude that iGb3 is not a physiologically relevant thymic iNKT-selecting ligand in vivo.
Results
The iGb3S-Deficient (iGb3S−/−) Mouse.
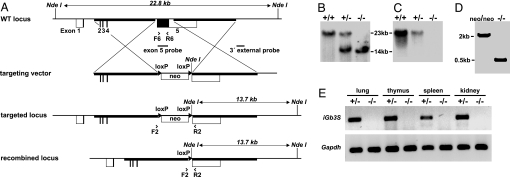
The coding sequence of the fifth exon of iGb3S, which is responsible for enzymatic activity (25, 26), was replaced by a loxP-flanked neomycin resistance cassette (Fig. 2A). Homologous recombination at the iGb3S locus was confirmed by Southern blot (Fig. 2 B and C). In selected tissues, iGb3S deficiency was also demonstrated at the mRNA level by RT-PCR (Fig. 2E). Mice were crossed with a transgenic mouse strain ubiquitously expressing Cre recombinase (27) to delete the loxP-flanked neomycin selection cassette, thereby excluding the possibility that the mutant phenotype is influenced by the neo cassette in an unpredictable way. The deletion of the selection cassette was verified by PCR (Fig. 2D). iGb3S−/− mice reproduced normally, and progeny were born at expected Mendelian ratios. They grew normally and exhibited no overt developmental or behavioral defects. Body and organ weights were normal (data not shown). Histological examination of bone marrow, thymus, lymph nodes, spleen, brain, eye, heart, lung, intestines, liver, pancreas, and kidney did not reveal differences between iGb3S−/− and iGb3S+/− littermates (data not shown).
Fig. 2.
Generation of iGb3S-deficient mouse. (A) Targeting strategy showing murine iGb3S WT locus, targeting vector and the targeted locus before and after Cre-mediated recombination together with relevant restriction sites and probes for Southern blot analysis. Coding and noncoding regions of exon 1–5 are marked by filled and nonfilled boxes, respectively. Homology arms are depicted as bold lines. Arrowheads show the position and orientation of PCR primers. Triangles flanking the PGK-gb2-neomycin selection cassette (neo) symbolize loxP sequences. (B) Southern blot analysis using 3′ external probe, as indicated in A, shows shortening of the diagnostic 22.8-kb-long NdeI fragment down to 13.7 kb after homologous recombination. (C) A replica of the Southern blot filter as used in B was hybridized with a probe specifically recognizing the coding part of exon 5 to demonstrate its deletion in iGb3S−/− mice. (D) Cre-mediated deletion of the neomycin selection cassette was confirmed by PCR by using primers F2 and R2 as a shortening of the 2,044-bp-long product to 494 bp. neo/neo and −/−, targeted locus before and after Cre-mediated deletion of neomycin selection cassette, respectively. (E) Detection of iGb3S-specific mRNA by RT-PCR by using total RNA from lung, thymus, spleen, and kidney of iGb3S+/− and iGb3S−/− animals; GAPDH was used as a positive control.
GSL Composition.
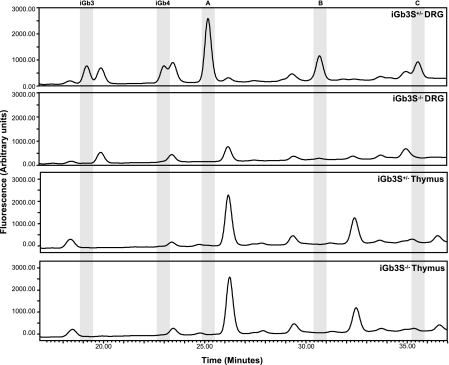
The only tissue in the mouse with proven expression of iGb3 is the dorsal root ganglion (DRG) [Speak et al. (24)]. Thus HPLC analysis of DRG was performed to confirm the absence of isoglobo-series in iGb3S−/− mice. Both iGb3S+/+ (not shown) and iGb3S+/− mice (Fig. 3) showed a GSL profile identical to that reported by Speak et al. (24) on DRG from C57BL/6 mice. As would be expected, iGb3 and consequently other members of the isoglobo-series [isoglobotetrahexosylceramide (iGb4) and isoglobopentahexosylceramide] were not detected in iGb3S−/− mice (Fig. 3). Additionally, no poly Galα1–3 (i)Gb3 was detected in the iGb3S−/− DRG (Fig. 3). Thymus, spleen, liver, and bone marrow GSLs were studied from iGb3S+/+, iGb3S+/−, and iGb3S−/− mice, and there was no difference in the GSL profiles (Fig. 3 and data not shown). This is in agreement with the accompanying manuscript of Speak et al. (24), because there is no evidence for isoglobo-series GSLs in these organs from control mice. There was no evidence for any compensatory changes in other GSLs as a consequence of iGb3S deficiency (Fig. 3 and data not shown).
Fig. 3.
HPLC analysis of GSL composition of DRG and thymus. Highly sensitive HPLC analysis [see Speak et al. (24) for details] demonstrates the absence of iGb3 in DRG of iGb3S−/− mice compared with controls. iGb4 is the biosynthetic derivative of iGb3 in the isoglobo-series and is consequently also absent. An additional three peaks are absent in iGb3S−/− DRG (labeled A–C) compared with controls. These peaks are sensitive to digestion with α1–3,6-galactosidase and are therefore most likely poly-Galα1–3 (i)Gb3 GSLs. There is no evidence for any of these species being present in iGb3S+/− thymus, and no difference is seen between iGb3S+/− and iGb3S−/− thymus. Profiles are representative of three independent observations on three different animal pairs with equivalent protein quantities processed for the analysis.
iNKT Cell Number and Usage of TCR Vβ Chains in iGb3S−/− Mice.
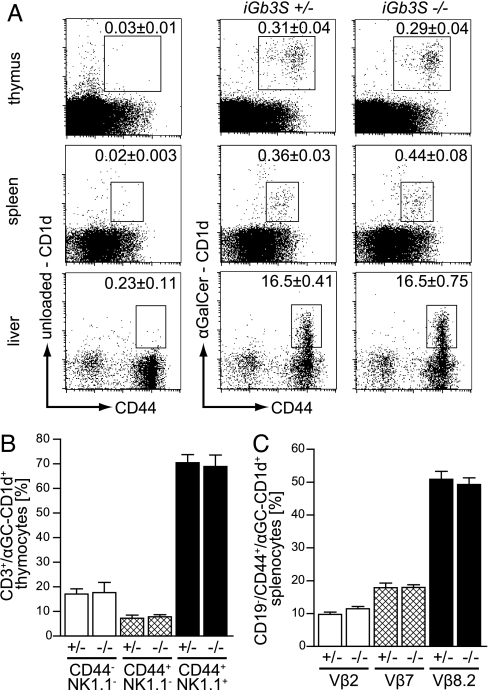
Previously, iGb3 was indirectly implicated as the endogenous ligand responsible for positive iNKT selection in the thymus (18). To test its role directly, we determined iNKT frequencies in thymus, spleen, and liver of iGb3S−/− animals, using αGalCer-loaded CD1d tetramers. The iNKT cell population was present in the thymus, spleen, and liver of iGb3S−/− mice at the same frequency as in iGb3S+/− animals (Fig. 4A). There was no difference in CD1d expression in thymocytes and splenic DCs between iGb3S−/− and iGb3S+/− mice (data not shown). During the course of thymic maturation, iNKT cells up-regulate surface expression of NK1.1 and CD44. Using these markers, three discrete developmental stages can be identified: immature, CD44−/NK1.1−; semimature, CD44+/NK1.1−; and mature, CD44+/NK1.1+ (28). No differences were observed in any of these developmental stages between iGb3S−/− mice and heterozygous littermates when investigating 5- (data not shown) as well as 8-week-old animals (Fig. 4B).
Fig. 4.
iNKT population shows no deviations in number, development, and TCRVβ usage in iGb3S−/− mice. (A) Thymocytes, splenocytes, and hepatic leukocytes from 8- to 10-week-old iGb3S+/− and iGb3S−/− mice were stained with anti-CD19 and -CD44 antibodies and with either αGalCer-loaded CD1d-tetramers to visualize iNKT population or unloaded CD1d-tetramers to define the amount of nonspecific binding. Plots were gated on lymphocytes. In case of spleen, CD19+ cells have been gated out. Numbers indicate the percentage of CD1d-tetramer+/CD44+ cells in the lymphocyte gate ± SEM; n = 5. (B) Thymocytes from 8-week-old iGb3S+/− and iGb3S−/− mice were stained with anti-NK1.1, -CD3, and -CD44 antibodies and αGalCer-loaded CD1d-tetramers to determine the amount of cells in immature (CD44−/NK1.1−), semimature (CD44+/NK1.1−), and mature (CD44+/NK1.1+) stage. Analysis was gated on CD3+/αGalCer-CD1d+ cells, and bars represent the percentage of cells with the depicted phenotype ± SEM; n = 4. (C) Splenocytes from 8- to 10-week-old iGb3S+/− and iGb3S−/− mice were stained with αGalCer-loaded CD1d-tetramers, anti-CD19 and -CD44, and either TCRVβ2, -7, or -8.2 antibodies. Analysis was gated on CD19−/αGalCer-CD1d+/CD44+ lymphocytes, and the bars represent the percentage of cells positive for the corresponding TCRVβ-chain ± SEM; n = 6. No significant differences could be seen between iGb3S+/− and iGb3S−/− mice in A–C.
During thymic positive selection of iNKTs, the invariant Vα14-chain pairs almost exclusively with Vβ2, -7, or -8.2, possibly as a consequence of the endogenous ligand(s) (11). Therefore, the proportion of these TCR Vβ-chains in the iNKT population was investigated in iGb3S−/− mice. The TCR Vβ-chain usage was identical in the iGb3S−/− and control animals (Fig. 4C).
Response to αGalCer Injection in iGb3S−/− Mice.
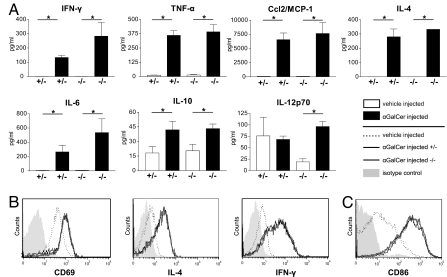
Upon stimulation with αGalCer, iNKT cells promptly secrete cytokines and exert a stimulatory effect on DCs (14, 29–32). To compare the in vivo function of iNKT cells of iGb3S−/− and iGb3S+/− mice, αGalCer or vehicle was injected intravenously. No significant differences in the response of iGb3S−/− and iGb3S+/− animals could be seen with respect to any of the cytokines measured in sera (Fig. 5A). Similarly, in the same experimental setting, flow cytometry did not show any difference in the up-regulation of surface CD69 and in the increase of intracellular IL-4 and IFN-γ in the iNKT cell populations of iGb3S−/− and iGb3S+/− mice (Fig. 5B). Furthermore, no difference was revealed in the maturation of splenic DCs, as defined by surface CD86 expression (Fig. 5C).
Fig. 5.
In vivo response to αGalCer injection is unaffected in iGb3S−/− mice. iGb3S+/− and iGb3S−/− mice (12 week old) were injected intravenously with 1 μg of αGalCer or vehicle. (A) Serum concentrations of indicated cytokines were measured 3 h after injection by cytometric bead array technology. iGb3S−/− mice did not significantly differ in the cytokine response from controls. Depicted are means ± SEM; n = 5; ∗, P < 0.05. (B) Hepatic leukocytes were isolated 2 h after injection and stained with anti-CD3, anti-CD44, or αGalCer-loaded CD1d-tetramers and either CD69 or intracellularly with anti-IL-4 or anti-IFN-γ. Histograms were gated on CD3+/αGalCer-CD1d+/CD44+ lymphocytes. Filled curve, isotype control binding in the gate; dotted line, vehicle-injected control; bold gray line, iGb3S−/−; thin black line, iGb3S+/−. (C) Surface expression of CD86 was investigated in splenic DCs 12 h after injection. Histograms were gated on CD11c+/MHCII+ cells. Legend as in B. One representative image from three independent analyses is shown.
Discussion
Based on investigations in Hexb−/− mice, iGb3 was proposed to be the endogenous ligand essential for the positive selection of mouse iNKT cells in the thymus (18). Although iGb3 can stimulate iNKT cells in vitro (18, 19) [Speak et al. (24)], there is currently no direct evidence supporting the hypothesis that iGb3 plays a role as an endogenous selecting ligand in vivo. Although iGb3S mRNA has been detected in multiple murine tissues, including the thymus (33) (and our data, Fig. 2E), iGb3 is undetectable in mouse or human thymus [Speak et al. (24)]. To rule out the possibility that trace amounts of iGb3 could be present below the detection limits of the assay but still above the necessary physiological threshold to select iNKT cells, we have generated an iGb3S−/− mouse.
The iGb3S−/− mouse with a deletion of the catalytic region of the iGb3S gene had no detectable isoglobo-series of GSL (iGb3, -4, and -5) or other GSL generated by iGb3S [e.g., Poly Galα1–3 (i)Gb3] in the DRG (Fig. 3), the sole location in the mouse where these GSLs have been identified [Speak et al. (24)]. α1,3galactosyltransferase, which also synthesizes the Galα1–3Gal epitopes in nonprimate tissues, was unable to compensate for the absence of iGb3S activity, in agreement with previous reports of the substrate specificities of these two enzymes. α1,3galactosyltransferase is specific for an N-acetyllactosamine substrate on glycoproteins or GSLs, whereas iGb3S is specific for GSL substrates (33, 34).
We have found that iNKT cells in the thymus, spleen, and liver of iGb3S−/− mice did not differ in number, TCR Vβ usage, or development or function from iGb3S+/− littermates (Figs. 4 and 5). These data strongly suggest that iGb3 is not an endogenous ligand responsible for thymic iNKT selection in vivo. An alternative possibility to account for the presence of normal numbers of iNKT cells in iGb3S−/− mice is that in the absence of iGb3, other GSLs take on the role of iGb3 and are responsible for the positive selection of iNKT cells. Although we cannot rule out this possibility, we think it is unlikely because of the lack of compensatory changes in other GSLs (Fig. 3, thymus) and the lack of alteration in TCR Vβ usage or iNKT cell development (Fig. 4 B and C), which may occur if the selecting ligand had changed. On the basis of these findings, we propose that iGb3 is not the physiological selecting ligand in vivo. This is further supported by the fact that iNKT cell loss is not specific to models that affect isoglobo-series catabolism (e.g., Hexb−/−) and is instead observed in multiple storage disorder mouse models (21, 22). Thus the lack of iNKT cells in all these models probably results from lipid storage per se rather than the specific absence of iGb3. Furthermore, the absence of an iNKT phenotype in the iGb3S−/− mouse is in agreement with Speak et al., who could not detect iGb3 in mouse or human thymus using a highly sensitive HPLC assay (24).
In conclusion, the results in this and the accompanying manuscript [Speak et al. (24)] strongly suggest that iGb3 is not involved in the thymic selection of iNKT cells. Other lipid ligands must now be sought.
The generation of the iGb3S−/− mouse may help to unravel the significance of the highly restricted distribution of iGb3 in the mouse, where to date it has been found only in DRG. Studies are in progress to see whether the iGb3S−/− mouse has phenotypic abnormalities in the peripheral nervous system that may provide insight into the functional role of this enigmatic GSL.
Methods
Generation of iGb3S−/− Mouse.
Clone MPMGc121G13146Q8 from the 129/Ola mouse genomic cosmid library number 121 from the German Resource Center for Genome Research (Berlin, Germany) containing the iGb3S locus was used to construct a targeting vector by means of Red/ET homologous recombination technology (Gene Bridges, Dresden, Germany) (35). First, a 13.5-kb fragment corresponding to the region from 128.262.983 to 128.276.500 on chromosome 4 (NCBIm36 assembly) and bearing the iGb3S gene was subcloned into the plasmid vector pBluescript M13(+) (Stratagene, Heidelberg, Germany). Then the coding sequence of the exon 5 was replaced by a loxP-flanked neomycin selection cassette (loxP-PGK-gb2-neo-loxP cassette; Gene Bridges) in an extent corresponding to the region 128.269.096–128.270.244 (Fig. 2A). The targeting vector was electroporated into E14 embryonic stem (ES) cells, and 384 G418-resistant clones were picked, expanded, and characterized by Southern blot analysis by using the 3′ external probe (Fig. 2A). Of eight positive ES cell clones, six were microinjected into C57BL/6 blastocysts. In iGb3S−/− mice, homologous recombination at the iGb3S-locus and deletion of the iGb3S exon 5 were confirmed by Southern blot analysis (Fig. 2 B and C). For Cre-mediated deletion of the loxP-flanked neomycin selection cassette, iGb3S-deficient mice were crossed with a Cre deleter strain (27). Deletion of the selection cassette was proven by PCR by using the primers F2 5-CCC AGA TAA CCC TGA CTT GG-3 and R2 5-ATT GTC AAG GTG TGG GAA CC-3 (2,044 bp before and 494 bp after deletion; Fig. 2D). Mice were kept under specific pathogen-free conditions and backcrossed for four generations to C57BL/6 (Charles River, Wiga, Sulzfeld, Germany) before analysis.
Southern Blot Analysis.
Genomic DNA was digested by NdeI (New England Biolabs, Frankfurt am Main, Germany), blotted onto positively charged nylon membranes (Roche, Mannheim, Germany), and hybridized with specific probes, which were detected by using the digoxigenin (DIG) luminescent detection kit (Roche) according to the manufacturer's instructions. DIG-dUTP-labeled probes were synthesized by the PCR DIG probe synthesis kit (Roche) and primers as follows: 3′ external probe (660 bp): 5′-GAG GGG GAA CAA AAA GAA AAC-3′ and 5′-TAG GTT GGA ATG GCG AGC-3′; exon 5 probe (571 bp): F6: 5′-GAA CGT GGT GTA CTA TGT GTT TAC G-3′ and R6: 5′-GAT CTC TCT CCT CCA GAT AAT TTC C-3′.
RNA Isolation and RT-PCR.
After phenol/chloroform extraction (36) and digestion by RNase-free DNaseI (turbo DNA free; Ambion, Huntingdon, U.K.), 3 μg of total RNA was reverse-transcribed in 20 μl of total volume by using SuperscriptII (Invitrogen, Karlsruhe, Germany), according to the manufacturer's instructions. RT-PCR was performed with 1 μl of cDNA and the primers F6 and R6, as above.
Organ Preparation and Flow Cytometry.
Thymi and spleens were mechanically disrupted in six-well plates and digested for 15 min in 5 ml of digestion solution of RPMI medium 1640/10 mM Hepes/0.1% BSA/0.5 mg/ml collagenase IA/4.5 k units/ml DNase I (all Sigma, Schnelldorf, Germany) at 37°C and 5% CO2. After repetitive pipetting, the organ homogenates were sieved through a 100-μm filter (BD, Heidelberg, Germany) and washed with FACS buffer (PBS containing 0.1% BSA). Liver preparation was performed as described in ref. 37. For lipid loading, PE-labeled CD1d tetramers (Proimmune, Oxford, U.K.) were incubated overnight at room temperature with 12-fold molar excess of αGalCer (Axxora, Lörrach, Germany) dissolved at 0.2 mg/ml in PBS plus 0.5% Tween 20 or were left unloaded and incubated solely with PBS plus 0.5% Tween 20 (30). Cells (106) were resuspended in 80 μl of FACS buffer and stained with 1 pmol of either αGalCer-loaded or unloaded CD1d tetramer for 30 min at room temperature. Cells were washed with 2 ml of FACS buffer and incubated with anti-CD16/32 (Fcγ RIII/II) antibody (BD) at 4°C for 5 min followed by staining with monoclonal antibodies in combinations as indicated in Results: anti-TCR-Vβ2 (B20.6), anti-TCR-Vβ7 (TR310), anti-TCR-Vβ8.1 and -8.2 (MR5–2), anti-CD1d (CD1.1; 1B1), anti-CD3ε (145–2C11), anti-CD69 (H1.2F3), armenian hamster IgG1 (G235–2356), rat IgG2bκ (A95–1; all BD), anti-MHCII (M5/114.15.2), anti-NK1.1 (PK136), anti-CD11c (N418), anti-CD19 (MB19–1), anti-CD44 (IM7), and anti-CD86 (PO3.1; all eBioscience, San Diego, CA). Intracellular cytokine staining was performed by using Cytofix/Cytoperm, Perm/Wash Buffer, anti-IL-4 (11B11), and anti-IFN-γ (XMG1.2) (all BD) according to the manufacturer's instructions. Samples were analyzed on a FACSCalibur flow cytometer (BD) after gating on lymphocytes in forward- and side-scatter plots.
Cytokine Profiles.
Female mice were injected intravenously with 1 μg of αGalCer (Axxora) dissolved in 100 μl of vehicle solution of 0.5% Tween 20 in PBS or with vehicle solution as a control. Blood samples were collected after 3 h, and serum concentrations of IL-6, IL-10, IL-12p70, IFN-γ, TNF- α, Ccl2/MCP-1, and IL- 4 were measured by cytometric bead array technology by using a mouse inflammation kit and IL-4 flex set, respectively, FACSCalibur (all BD) and FCAP array software (Soft Flow, Pécs, Hungary).
HPLC.
Liver and DRG were homogenized in water by using a tight-fitting dounce, and 1 volume of homogenate was mixed with 4 volumes of chloroform:methanol, 1:2 (vol/vol). Splenocytes, bone marrow, and thymocytes were resuspended in water, and 1 volume of the cell suspension was mixed with 4 volumes of chloroform:methanol, 1:2 (vol/vol). The samples were left to mix gently overnight, and the remainder of the GSL extraction, purification, and HPLC was performed as described in Speak et al. (24).
Statistical Analysis.
The Mann–Whitney test was performed to compare data sets; in the case of serum cytokine elevation upon αGalCer administration, the one-tailed test was used; in all other cases, the two-tailed test was used. Differences were considered significant if P < 0.05. Numbers of independent observations per group are indicated with every result.
Acknowledgments
We thank R. Jennemann for the genomic iGb3 clone, F. van der Hoeven and U. Kloz for blastocyst microinjection, the group of Prof. G. Schütz for the Cre-deleter mouse strain, and Mahnaz Bonrouhi for excellent technical assistance (all at the German Cancer Research Center). This work was supported by Deutsche Forschungsgemeinschaft Grants SFB 405:B10 (to H.-J.G.) and LU 612/4–1 (to B.L.). A.O.S. is supported by a scholarship from the Glycobiology Institute, University of Oxford.
Abbreviations
- αGalCer
α-galactosylceramide
- DC
dendritic cell
- DRG
dorsal root ganglion
- GSL
glycosphingolipid
- iGb3
isoglobotrihexosylceramide
- iGb3S
iGb3 synthase
- iGb3S−/−
iGb3S-deficient
- iGb4
isoglobotetrahexosylceramide
- NKT
natural killer T
- iNKT
invariant NKT
- TCR
T cell antigen receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
See Commentary on page 5713.
References
- 1.Kasahara M, Nakaya J, Satta Y, Takahata N. Trends Genet. 1997;13:90–92. doi: 10.1016/s0168-9525(97)01065-2. [DOI] [PubMed] [Google Scholar]
- 2.Brigl M, Brenner MB. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 3.Porcelli SA, Modlin RL. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 4.Kronenberg M, Gapin L. Nat Rev Immunol. 2002;2:557–568. doi: 10.1038/nri854. [DOI] [PubMed] [Google Scholar]
- 5.Makino Y, Kanno R, Ito T, Higashino K, Taniguchi M. Int Immunol. 1995;7:1157–1161. doi: 10.1093/intimm/7.7.1157. [DOI] [PubMed] [Google Scholar]
- 6.Arase H, Arase N, Ogasawara K, Good RA, Onoe K. Proc Natl Acad Sci USA. 1992;89:6506–6510. doi: 10.1073/pnas.89.14.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lantz O, Bendelac A. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, et al. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 9.Hansen DS, Schofield L. Int J Parasitol. 2004;34:15–25. doi: 10.1016/j.ijpara.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Taniguchi M, Harada M, Kojo S, Nakayama T, Wakao H. Annu Rev Immunol. 2003;21:483–513. doi: 10.1146/annurev.immunol.21.120601.141057. [DOI] [PubMed] [Google Scholar]
- 11.Wei DG, Curran SA, Savage PB, Teyton L, Bendelac A. J Exp Med. 2006;203:1197–1207. doi: 10.1084/jem.20060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts TJ, Sriram V, Spence PM, Gui M, Hayakawa K, Bacik I, Bennink JR, Yewdell JW, Brutkiewicz RR. J Immunol. 2002;168:5409–5414. doi: 10.4049/jimmunol.168.11.5409. [DOI] [PubMed] [Google Scholar]
- 13.Chiu YH, Park SH, Benlagha K, Forestier C, Jayawardena-Wolf J, Savage PB, Teyton L, Bendelac A. Nat Immunol. 2002;3:55–60. doi: 10.1038/ni740. [DOI] [PubMed] [Google Scholar]
- 14.Smiley ST, Kaplan MH, Grusby MJ. Science. 1997;275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 15.Zhou D, Cantu C, III, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, et al. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honey K, Benlagha K, Beers C, Forbush K, Teyton L, Kleijmeer MJ, Rudensky AY, Bendelac A. Nat Immunol. 2002;3:1069–1074. doi: 10.1038/ni844. [DOI] [PubMed] [Google Scholar]
- 17.Stanic AK, De Silva AD, Park JJ, Sriram V, Ichikawa S, Hirabyashi Y, Hayakawa K, Van Kaer L, Brutkiewicz RR, Joyce S. Proc Natl Acad Sci USA. 2003;100:1849–1854. doi: 10.1073/pnas.0430327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou D, Mattner J, Cantu C, III, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, et al. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 19.Xia C, Yao Q, Schumann J, Rossy E, Chen W, Zhu L, Zhang W, De Libero G, Wang PG. Bioorg Med Chem Lett. 2006;16:2195–2199. doi: 10.1016/j.bmcl.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 20.Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, III, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, et al. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 21.Gadola SD, Silk JD, Jeans A, Illarionov PA, Salio M, Besra GS, Dwek R, Butters TD, Platt FM, Cerundolo V. J Exp Med. 2006;203:2293–2303. doi: 10.1084/jem.20060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sagiv Y, Hudspeth K, Mattner J, Schrantz N, Stern RK, Zhou D, Savage PB, Teyton L, Bendelac A. J Immunol. 2006;177:26–30. doi: 10.4049/jimmunol.177.1.26. [DOI] [PubMed] [Google Scholar]
- 23.Godfrey DI, McConville MJ, Pellicci DG. J Exp Med. 2006;203:2229–2232. doi: 10.1084/jem.20061787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Speak AO, Salio M, Neville DCA, Fontaine J, Priestman DA, Platt N, Heare T, Butters TD, Dwek RA, Trottein F, et al. Proc Natl Acad Sci USA. 2007;104:5971–5976. doi: 10.1073/pnas.0607285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keusch JJ, Manzella SM, Nyame KA, Cummings RD, Baenziger JU. J Biol Chem. 2000;275:25308–25314. doi: 10.1074/jbc.M002629200. [DOI] [PubMed] [Google Scholar]
- 26.Wiggins CA, Munro S. Proc Natl Acad Sci USA. 1998;95:7945–7950. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwenk F, Baron U, Rajewsky K. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 29.Kitamura H, Iwakabe K, Yahata T, Nishimura S, Ohta A, Ohmi Y, Sato M, Takeda K, Okumura K, Van Kaer L, et al. J Exp Med. 1999;189:1121–1128. doi: 10.1084/jem.189.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomura M, Yu WG, Ahn HJ, Yamashita M, Yang YF, Ono S, Hamaoka T, Kawano T, Taniguchi M, Koezuka Y, et al. J Immunol. 1999;163:93–101. [PubMed] [Google Scholar]
- 32.Vincent MS, Leslie DS, Gumperz JE, Xiong X, Grant EP, Brenner MB. Nat Immunol. 2002;3:1163–1168. doi: 10.1038/ni851. [DOI] [PubMed] [Google Scholar]
- 33.Milland J, Christiansen D, Lazarus BD, Taylor SG, Xing PX, Sandrin MS. J Immunol. 2006;176:2448–2454. doi: 10.4049/jimmunol.176.4.2448. [DOI] [PubMed] [Google Scholar]
- 34.Taylor SG, McKenzie IF, Sandrin MS. Glycobiology. 2003;13:327–337. doi: 10.1093/glycob/cwg030. [DOI] [PubMed] [Google Scholar]
- 35.Muyrers JP, Zhang Y, Stewart AF. Trends Biochem Sci. 2001;26:325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 36.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 37.Korten S, Anderson RJ, Hannan CM, Sheu EG, Sinden R, Gadola S, Taniguchi M, Hill AV. Infect Immun. 2005;73:849–858. doi: 10.1128/IAI.73.2.849-858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]