Abstract
Mycobacterium tuberculosis parasitizes resting macrophages yet is killed by activated macrophages through both oxidative and nonoxidative mechanisms. Nonoxidative mechanisms are linked to the maturation of the bacteria-containing phagosome into an acidified, hydrolytically active compartment. We describe here a mechanism for killing Mycobacteria in the lysosomal compartment through the activity of peptides generated by the hydrolysis of ubiquitin. The induction of autophagy in infected macrophages enhanced the delivery of ubiquitin conjugates to the lysosome and increased the bactericidal capacity of the lysosomal soluble fraction. The accumulation of ubiquitinated proteins in the autophagolysosome provides one possible mechanism behind the antimicrobial activities observed for a range of pathogens in autophagous host cells.
Keywords: macrophage, phagosome, tuberculosis, lysosome
Mycobacterium tuberculosis parasitizes its host macrophages through arresting the normal maturation of its phagosome, and it resides in a compartment of relatively high pH (6.4) that fails to fuse with lysosomes (1–3). Much of the literature detailing the bactericidal activity of infected macrophages focuses on reactive oxygen and nitrogen intermediates produced by these cells upon activation by host cytokines (4). Although these mechanisms are undoubtedly key to the clearance of many pathogens, it is also clear that nonoxidative mechanisms dependent on the successful delivery of the pathogen to the acidic, hydrolytically active lysosomes also play an important role (5–7). Alternative pathways for bactericidal activity have recently come to the fore following the work of Gutierrez et al. (8). They demonstrated that the induction of autophagy in Mycobacterium-infected macrophages led to delivery of the bacterium to lysosomes and culminated in bacterial death. Similar lysosomal-mediated killing has also been reported for Streptococcus, Shigella, Legionella, and Salmonella (9–12), indicating that microbicidal activity through the induction of autophagy has broad implications for intracellular infections. In this work we describe a mechanism of mycobacterial killing in the lysosome by ubiquitin-derived peptides, which is enhanced by the induction of autophagy.
Results and Discussion
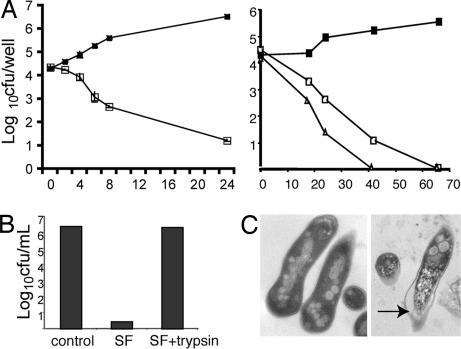
In preliminary studies designed to elucidate the nutrient source exploited by intracellular M. tuberculosis, we incubated bacteria in solubilized lysosomes isolated from resting bone-marrow derived macrophages (BMMΦ) and noted, to our surprise, that the lysosomal extract killed the bacteria. This soluble fraction (SF) was toxic to both M. tuberculosis and Mycobacterium smegmatis and, at 50 μg/ml, induced a 5 log reduction in bacterial number after 24 h (M. smegmatis) or 65 h (M. tuberculosis) of incubation (Fig. 1A). Separation by size-exclusion columns indicated that the bactericidal component of the SF was smaller than 10 kDa (data not shown), and activity of the lysate was abolished by trypsin treatment, implying that it is proteinaceous in nature (Fig. 1B). Electron microscopical analysis of SF-treated organisms indicated that the integrity of the bacterial cell wall was impaired and suggested that the bacterial cell wall was permeabilized (Fig. 1C). This ultrastructural change is similar to that observed upon treatment of bacteria with pore-forming cationic antimicrobial peptides or defensins, the proposed mechanism of which is to insert into the bacterial membrane and alter membrane permeability (13).
Fig. 1.
Bactericidal activity of lysosomal SF on M. smegmatis and M. tuberculosis. (A) Bacteria were incubated with buffer (filled squares) or with SF at 50 μg/ml (open squares) or 100 μg/ml (triangles). At the indicated time points, the viable bacteria were determined by plating cfu. (Left) M. smegmatis. (Right) M. tuberculosis. (B) The bactericidal activity of lysosomal SF is protease-sensitive. M. smegmatis was incubated overnight at 37°C with buffer, SF, or SF preincubated with trypsin, then the viable bacteria were determined. (C) SF-treated mycobacteria display membrane damage. M. smegmatis was incubated overnight with buffer or SF and then analyzed by electron microscopy. (Left) Control. (Right) SF-treated. An arrow indicates the compromised cell wall.
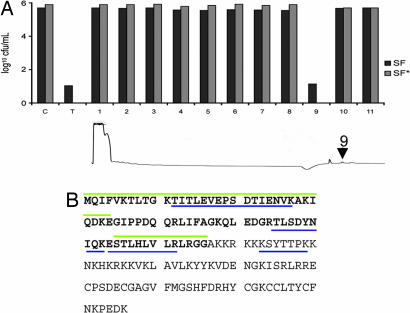
The lysosome contains a rich mixture of hydrolytic enzymes and proteins. To identify the component responsible for mycobacterial killing, SF was loaded onto a cation-exchange column and fractionated by HPLC. Each individual fraction was tested for bactericidal activity against M. smegmatis, and a single peak (peak 9) was found to retain the bactericidal activity (Fig. 2A). The HPLC-separated fraction was trypsinized and analyzed by nano liquid chromatography/tandem mass spectrometry, and, in two independent runs, four discrete peptides were obtained that corresponded to ubiquitin and the ribosomal subunit Rps27a, which is translated with the ubiquitin sequence as an N-terminal extension (Fig. 2B).
Fig. 2.
Identification of ubiquitin peptides by mass spectrometry. (A) HPLC purification of the fraction containing mycobactericidal compound. The bactericidal activity of the total (T) sample was compared with that of the buffer-treated control (C) and each fraction from SF and SF*. The arrow on the trace indicates the peak retaining bactericidal activity. (B) Amino acid sequence of ubiquitin as an N-terminal extension of ribosomal subunit Rps27A. Amino acids corresponding to the ubiquitin are in bold. The peptides identified by mass spectroscopy are underlined in blue, and the peptides Ub1 and Ub2 are overlined in green.
Ubiquitin is best characterized as a posttranslational addition to proteins that targets them for degradation through the proteosome (14). In addition to this “classic” route for protein breakdown, it is also known that ubiquitin regulates trafficking of proteins in the endocytic pathway. Monoubiquitinated proteins, most notably integral membrane proteins, are imported into multivesicular bodies (MVBs) and delivered to lysosomes for degradation (15, 16). However, lysosomally degraded proteins are thought to be deubiquitinated before import into the lysosome. Both datasets pose significant logistical problems to the proposal that bacterial killing by ubiquitin-derived peptides may be biologically significant. But it is also known that ubiquitination is a requirement for the formation MVBs. In previous experiments demonstrating the transference of cytosolic cargo into lysosomes in macrophages infected with Leishmania, we found that this transfer was enhanced upon the induction of autophagy and was routed through autophagosomes, which appeared comparable ultrastructurally with MVBs (17). Moreover, cells deficient in autophagy accumulate aggregates of ubiquitinated proteins in the cytosol (16, 18, 19).
We hypothesized that the formation of internal vesicular structures might facilitate transfer of membrane-associated ubiquitin or ubiquitinated protein aggregates to the lumen of the lysosome. This hypothesis is supported by the recent detection of ubiquitin in exosomes (20) and previous studies that documented ubiquitin within the lumen of MVBs and lysosomes (21, 22). Initial observations with immunofluorescence microscopy using antibodies to LAMP1 and to ubiquitin indicated that there was significant overlap between LAMP1-positive and ubiquitin-positive compartments in both resting BMMΦs as well as BMMΦs stimulated into autophagy by starvation [supporting information (SI) Fig. 6].
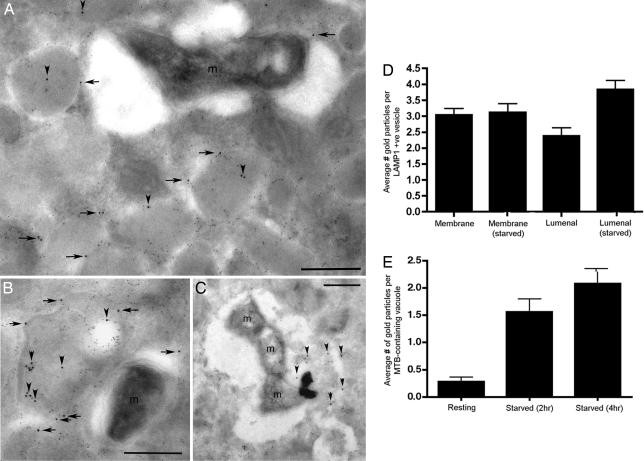
The localization of ubiquitin to the lysosome, in particular to the lumen of the vacuole where bacteria are delivered, is key to establishing a mechanism for ubiquitin-mediated killing. To determine whether the ubiquitin associated with the LAMP-positive vacuoles was membrane-associated or luminal, immunoelectron microscopy was performed. Immunoelectron microscopy of control and M. tuberculosis-infected BMMΦs demonstrated that the majority of conjugated mono- and polyubiquitin detected was either within LAMP1-positive vesicles or associated with the limiting membranes of these structures (Fig. 3 A and B). In cells treated by starvation for 2 h, M. tuberculosis can be seen in dense lysosomes that are also positive for ubiquitin. Within these lysosomal structures, the ubiquitin label was frequently found associated with internal, membrane-derived debris that was also LAMP1-positive (Fig. 3C and SI Fig. 7), which is consistent with delivery through invagination and the formation of MVBs. In untreated, M. tuberculosis-infected BMMΦ, there was little antiubiquitin signal in the bacteria-containing vacuoles (Fig. 3B); however, upon the up-regulation of autophagy by serum starvation, the delivery of ubiquitin to the bacteria-containing vacuoles was enhanced markedly (Fig. 3C). The shift in abundance of ubiquitin label into the lumen of LAMP1-positive vesicles and the lumen of bacteria-containing vacuoles was scored in Fig. 3 D and E, respectively. The increased ubiquitin signal in the lumen of lysosomes in serum-starved macrophages could be the result of enhanced delivery or decreased proteolysis (23).
Fig. 3.
Immunoelectron microscopy of M. tuberculosis-infected macrophages. Cells were probed with mouse antiubiquitin (12 nm gold) and rat anti-LAMP1 (6 nm gold). (A) Untreated, infected macrophage demonstrating that the bacteria-containing vacuoles have minimal ubiquitin signal. The ubiquitin signal is associated predominantly with the limiting membranes (horizontal arrows) or lumen (vertical arrowheads) of LAMP1-positive vesicles. (B) Untreated, infected macrophages where bacteria-containing vacuoles have minimal ubiquitin signal and where the ubiquitin signal is associated predominantly with internal, LAMP1-positive membranous material inside MVBs (vertical arrowheads). (C) In cells treated by starvation for 2 h, M. tuberculosis can be seen in vacuoles with flocculent lysosomal matrix that is also positive for ubiquitin (vertical arrowheads). (D) The distribution of ubiquitin signal associated with the limiting membrane or the lumen of LAMP1-positive vesicles was scored in resting versus 2-h starved macrophages and showed a significant increase in luminal label in the starved cells (n = 50 vacuoles). (E) The relative distribution of M. tuberculosis in vacuoles positive for luminal ubiquitin signal in untreated and starved BMMΦ is detailed ± SD (n = 50 vacuoles).
Our hypothesis hinges on ubiquitin accessing the lumen of the lysosome through the biogenesis of multivesicular bodies. We had demonstrated previously that comparable cytosol–lysosome transfer was enhanced in autophagous cells (17). To establish a connection, we examined the distribution of ubiquitin in macrophages from LC3-GFP mice. LC3, or Atg8, is an essential component of the endosomal sorting complex required for transport (ESCRT) machinery required for formation of MVBs and autophagosomes that is lipidated and associates with autophagosomes upon induction of autophagy (24). Furthermore, evidence indicates that LC3 binds to ubiquitin by means of a bridging protein, p62 (25). In resting and autophagic macrophages, LC3-GFP-positive autophagosomes colocalized with LAMP1 positive vacuoles (SI Fig. 8A). And although there was limited colocalization of ubiquitin and LC3-GFP in resting macrophages, upon induction of autophagy, the strongest ubiquitin-positive bodies in the cell were also positive for LC3 (SI Fig. 8B). These findings establish a connection between autophagy and the delivery of ubiquitin to the autophagosomes.
The induction of autophagy in Mycobacterium bovis bacillus Calmette–Guérin- and M. tuberculosis-infected macrophages through either serum starvation or treatment with the drug rapamycin was reported to lead to significant killing of intracellular bacteria by Gutierrez et al. (8). However, the induction of autophagy by cholesterol depletion of Mycobacterium avium-infected macrophages failed to impact bacterial survival (26). Despite this disparity, we were able to reproduce bacterial killing in M. tuberculosis-infected BMMΦ (SI Fig. 9). Furthermore, we demonstrate that this enhanced killing was due not only to the increased delivery of bacteria to the lysosome but also to the increased mycobactericidal capacity of the lysosomal fraction isolated from autophagic BMMΦ (SF*) compared with SF in bactericidal assays with M. smegmatis and M. tuberculosis (Fig. 4A). HPLC fractionation of SF* and in vitro assays determined that bactericidal activity was present exclusively in a single peak that eluted from the column identically to that of SF (Fig. 2A). The amount of ubiquitin present in SF and SF* was determined by immunoblot analysis to be 30 μM and 40 μM, respectively (Fig. 4C). Therefore, the increase in bactericidal activity of SF* correlates with an enrichment of ubiquitin in lysosomal compartments determined both biochemically and by immunoelectron microscopy.
Fig. 4.
Bacterial killing by BMMΦ upon induction of autophagy and activity of SF, SF*, and SF IFN against Mycobacteria. (A) Bactericidal activity of SF and SF*. M. smegmatis (Left) and M. tuberculosis (Right) were incubated in the presence of 50 μg/ml SF or SF* purified from control and autophagic BMMΦ, and bacterial viability was determined. Statistical significance between SF and SF* treatment was determined by Student's t test: ∗, P < 0.5; ∗∗, P < 0.05; ∗∗∗, P < 0.01; ∗∗∗∗, P < 0.005. (B) Bactericidal activity against M. smegmatis of 50 μg/ml SF purified from control, SF*, and SF IFN was determined. Statistical significance between SF and SF* treatment was determined by Student's t test: ∗, P < 0.5; ∗∗, P < 0.05. (A and B) The average ± SD of three independent experiments is shown. (C) (Left) Immunoblot analysis of SF, SF*, and SF IFN compared with ubiquitin standards. (Right) Signals quantitated by densitometry.
These experiments used serum starvation or rapamycin treatment to induce autophagy; however, it has been shown that IFN-γ activation, which is more physiologically relevant to M. tuberculosis infection, is also an inducer of autophagy. Consistent with our hypothesis, SF was isolated from IFN-γ-activated macrophages (SF IFN) and found to be more bactericidal than SF from control macrophages (Fig. 4B). As shown in Fig. 4C, this increase was mirrored by an increase in the concentration of ubiquitin to 51 μM, as determined by immunoblot analysis. Comparative blot analysis of the SFs revealed a concomitant increase in level of LC3-GFP, linking the increased ubiquitin concentration to the autophagous process (SI Fig. 10).
Interestingly, there is a body of literature that ascribes antimicrobial activity to ubiquitin. Ubiquitin has been identified as a constituent of several extracts that have antibacterial activity, including medium conditioned by insect cells, the mucosal layers of the colon and cervix, and the secretory granules of chromaffin cells (27–30). To demonstrate formally that ubiquitin was capable of killing Mycobacterium, we incubated purified ubiquitin with bacteria in vitro and found that intact ubiquitin did not display bactericidal activity. But it is anticipated that lysosomal ubiquitin would undergo cleavage by the diverse proteinases present, generating ubiquitin-derived peptides. In support of this hypothesis, bacterial killing was observed upon preincubation of full-length ubiquitin with the lysosomal proteinases cathepsins B, D, or L (Fig. 5A). No killing was seen in the presence of cathepsins B, D, or L in the absence of ubiquitin. Consistent with these data, the mycobactericidal activity of the lysosomal fraction could be enhanced by the addition of intact ubiquitin to the lysosomal extract (Fig. 5B). Combined, these data suggest strongly that ubiquitin-derived peptides are responsible for the bactericidal activity.
Fig. 5.
Bactericidal activity of ubiquitin and ubiquitin-derived peptides. (A) Bactericidal activity against M. smegmatis of purified ubiquitin and cathepsin-treated ubiquitin. Bacteria were incubated with 25 μM purified ubiquitin or 25 μM preincubated with cathepsin B, D, or L overnight. (B) Bactericidal activity against M. smegmatis of purified ubiquitin and SF-treated ubiquitin. Bacteria were incubated overnight with 100 μM purified ubiquitin or 25 μM preincubated with 25 μg/ml SF (SF25). Statistical significance between SF25 and SF25+Ub100 treatment was determined by Student's t test (∗∗∗∗, P < 0.005). (C) M. tuberculosis viability in the presence of varying concentrations of Ub2 peptide over the indicated time course was determined by plating cfu.
Previous work by Kieffer et al. reported that peptides derived from ubiquitin were capable of microbicidal activity against a range of organisms (28). They identified two endoproteinase-derived peptide sequences Ub1 and Ub2 (Fig. 2B) that exhibited differential activity against fungal and bacterial pathogens. We synthesized peptides corresponding to these sequences and found, consistent with the published data (28), that no activity was observed with Ub1, but Ub2 was capable of mediating significant antibacterial activity against both M. smegmatis and M. tuberculosis. Using a growth inhibition assay on M. tuberculosis, we determined that the Ub2 peptide exhibited an minimum inhibitory concentration of 5 μM, indicating potent antimycobacterial activity that is consistent with the concentrations of ubiquitin found in the SFs (Fig. 5C).
Conclusions
This work details how ubiquitin-derived peptides contribute to the bactericidal capacity of the lysosomal milieu. The lysosomal compartment contains a complex mixture of hydrolytic enzymes, including proteases and lipases, and it is likely that ubiquitin-derived peptides in the lysosomal lumen would act synergistically with these other compounds to promote bacterial killing. The antibacterial properties of ubiquitin-derived peptides, compared with the relatively inert full-length ubiquitin, are similar to those reported for several other naturally occurring peptide fragments of larger proteins. Domains of lactoferrin and hemoglobin, termed lactoferricin and hemocidin, respectively, are bactericidal against Gram-negative and Gram-positive bacteria at micromolar concentrations (31, 32). Antibacterial properties are also ascribed to peptides that correspond to fragments of histones and ribosomal proteins L30, S30, and S19 (27, 30, 33). A 6.6-kDa antimicrobial peptide fragment of the S30 ribosomal protein was isolated from the cytosol of a macrophage-like cell line by Hiemstra et al. (34), who designated it ubiquicidin because it shares 38% overall identity with the full-length ubiquitin molecule. The bactericidal ubiquitin peptides used here are distinct from ubiquicidin, which was not identified as a component in the lysosomal soluble fraction.
Our observations also expand our understanding of the antibacterial capacity of macrophages and reveal an unappreciated role for the lysosomal degradation of ubiquitinated proteins. Immunoelectron microscopy revealed an unexpectedly strong lysosomal localization for ubiquitin-conjugated proteins, indicating that macrophages, in particular those undergoing autophagy, appear to traffic a large proportion of ubiquitinated proteins to the lysosome. The accumulation of ubiquitinated proteins in the autophagic vacuole and the autophagolysosome may provide a regulatable source for bactericidal, ubiquitin-derived peptides. Our data demonstrate a functional correlation between the concentration of lysosome-associated ubiquitin and the bactericidal activity of the organelle. This finding provides a potential killing mechanism behind the previous reports of enhanced bactericidal activity in autophagous cells infected with bacterial pathogens (8–12).
Intriguingly, genes encoding two different E3 ubiquitin ligases have been implicated in susceptibility to infection by M. tuberculosis and Mycobacterium leprae. Mira et al. (35) reported that mutations in the regulatory region of the Parkinson's disease gene PARKIN or PARK2 correlated strongly with susceptibility to leprosy. Parkin is a HECT E3 ligase that mediates polyubiquitination at lysine-48, targeting its substrate to the proteosome. However, it has also been shown to ubiquitinate proteins at lysine-63, and this reaction leads to aggregation of ubiquitinated proteins (36). Intracellular aggregates of ubiquitinated proteins are known to accumulate in the absence of autophagy, suggesting that this pathway may lead to delivery of ubiquitin to the lysosomal network (18, 19). The other locus identified as correlating with susceptibility to M. tuberculosis encodes UBE3A, another HECT E3 ligase, but it has not been characterized to the same degree as PARKIN (37). The data are provocative, however, given the pleotropic roles of E3 ubiquitin ligases in immune regulation, the suggestion of a direct effect remains speculative (38, 39).
Materials and Methods
Maintenance of Bacterial Cultures and Cells.
M. tuberculosis wild-type strain CDC 1551 was provided by Ian Orme (Colorado State University, Fort Collins, CO). M. smegmatis mc2155 was obtained from American Type Culture Collection (Manassas, VA). Mycobacterial strains were maintained in Middlebrook 7H9 liquid medium (BD Biosciences, San Jose, CA) or on Middlebrook 7H11 agar (BD Biosciences) plates supplemented with oleic acid, albumin, dextrose, catalase (OADC; BD Biosciences). BMMΦ were isolated from BALB/c mice or from LC3-GFP transgenic mice (LC3-GFP 54) and maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FCS (Invitrogen)/5% horse serum (Invitrogen)/1.5 g/liter sodium pyruvate (Invitrogen)/20% L cell-conditioned medium.
Isolation of Bactericidal Soluble Fraction.
Lysosomes were isolated from BALB/c BMMΦ as described previously (40), and SF was obtained as follows. Confluent T150 were incubated for 2 h with 20 mg/ml iron dextran and then chased in culture medium. Macrophages were harvested by scraping into homogenation buffer (250 mM sucrose/20 mM Hepes/0.5 mM EGTA/0.1% gelatin, pH 5.5) and lysed by multiple passages through a tuberculin syringe with a 25-gauge needle. Cell debris and unbroken cells were removed by centrifugation at 500 × g for 10 min. The supernatant was applied to a MiniMACS column (Miltenyi Biotech, Auburn, CA) placed on a magnetic stand to retain iron-loaded lysosomes. After two washes with homogenation buffer, the column was removed from the magnetic stand and the lysosomes eluted in 1 ml of homogenation buffer. To prepare SF, the lysosomes were centrifuged 12,000 × g for 5 min. The pellets from 2.5 × 108 cells were then resuspended in 1 ml of 20 mM sodium acetate buffer (pH 5.5) containing 1% Tween 20. This lysate was then centrifuged at 100,000 × g, 4°C, for 50 min. The resulting supernatant (SF) was stored at −20°C. Total protein content was estimated by using the detergent-compatible Bradford protein assay. To prepare SF*, autophagy was induced by amino acid and serum starvation. Macrophages were incubated for 2 h at 37°C in Earle's balanced salt solution (starvation medium) and then incubated in 20 mg/ml iron dextran for 2 h and chased for 2 h in starvation medium. Macrophages were harvested, and SF was purified as described above. To prepare SF IFN, macrophages were activated overnight by using 100 units/ml IFN-γ, then the standard protocol was followed.
The standard assay to determine antimycobacterial activity of soluble fraction was as follows: 1 × 105 cfu of log-phase M. tuberculosis or M. smegmatis was incubated in medium containing 50 μg/ml or 100 μg/ml of SF in sodium acetate buffer or an equivalent amount of sodium acetate/Tween 20 buffer (control) for the indicated time. After treatment, serial dilutions were plated to determine the colony-forming units per milliliter of SF-treated bacteria compared with the control. In all bactericidal assays, three independent experiments were performed.
Purification of Antimycobacterial Component by HPLC.
To purify the bactericidal compound further, SF was applied to a cation-exchange column (Partisil 5 SCX; Whatman, Florham Park, NJ) equilibrated in 20 mM sodium acetate buffer at a flow rate of 1 ml/min. After a 30-min wash, fractions were eluted over 20 min in a gradient of 0–1.0 M NaCl by using a System Gold HPLC (Beckman, Fullerton, CA). After desalting, the bactericidal activity of each fraction was determined as described above. The bactericidal fraction was analyzed by nano liquid chromatography/tandem mass spectrometry by the Cornell Biotechnology Resource Center Proteomics and Mass Spectroscopy Facility.
Sample Preparation for Electron Microscopy and Immunoelectron Microscopy.
Macrophages were infected at a multiplicity of infection of 10:1, then incubated at 37°C in standard culture medium or medium containing 50 μg/ml rapamycin (Sigma, St. Louis, MO) or Earle's basic salt solution (starvation medium) to induce autophagy. At 2 and 4 h after infection, the medium was removed, and the cells were fixed in cold 200 mM Pipes containing 4% paraformaldehyde. Parallel infections were performed, and the bacterial viability in control macrophages (at 4 h after infection) and macrophages undergoing autophagy (at 2 and 4 h after infection) was determined by cfu assay. After processing as detailed previously (41), sections were probed with mouse monoclonal antibody (FK2) against conjugated ubiquitin (Biomol, Plymouth Meeting, PA) and rat ID4B against LAMP1. Antibody binding was visualized with 12 nm of gold-conjugated goat anti-mouse IgG and 6 nm of gold-conjugated goat anti-rat IgG (Jackson ImmunoResearch, West Grove, PA). SF-treated M. smegmatis were fixed in 2% glutaraldehyde for 1 h at 4°C and processed into Spurr's resin.
Bactericidal Activity of Ubiquitin.
Purified ubiquitin that corresponds to the N-terminal ubiquitin sequence of Rps27a was purchased from Sigma. The bactericidal activity of ubiquitin was determined by incubating 1 × 105 cfu of M. smegmatis with 25 μM ubiquitin or buffer-only control overnight at 37°C. The number of surviving bacteria was determined by plating serial dilutions. Preincubation of 25 μM ubiquitin with 0.5 unit of cathepsin B, cathepsin D, or cathepsin L (Sigma) was performed at 37°C overnight before performing the standard bactericidal assay on M. smegmatis.
Quantification of Ubiquitin in SF.
To quantitate the relative amount of ubiquitin present in SF, an immunoblot was performed on equivalent protein amounts from SF and SF* compared with a known amount of purified ubiquitin standard. The immunoblot was probed with monoclonal antibody against ubiquitin and a horseradish peroxidase anti-mouse secondary antibody. Signals were quantitated by using ImageQuant software (National Institutes of Health, Bethesda, MD).
Bactericidal Activity of Ubiquitin Peptides.
Ubiquitin peptides Ub1–34 (MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKE) and Ub2 (STLHLVLRLRGG) were synthesized by GenScript Corp. (Piscataway, NJ). To determine bactericidal activity against M. smegmatis, the standard bactericidal assay described above was performed. To determine bactericidal activity against M. tuberculosis, 5 × 105 cfu/ml was incubated over 4 days with the indicated concentrations of Ub2, and the number of surviving bacteria was determined by plating cfu.
Supplementary Material
Acknowledgments
We thank Dr. Sheng Zhang, Associate Director for Proteomic and Mass Spectrometry, for expert help. This work was supported by National Institutes of Health Awards AI057086 and AI067027 (to D.G.R.). G.E.P. was supported by the Heiser Program for Research in Leprosy and Tuberculosis. The LC3-GFP mice were a gift from Drs. T. Yoshimori (CREST, Japan Science and Technology Agency, Kawaguchi-Saitama, Japan) and C. Munz (The Rockefeller University, New York, NY).
Abbreviations
- BMMΦ
bone marrow-derived macrophages
- MVBs
multivesicular bodies
- SF
soluble fraction from resting BMMΦ
- SF*
soluble fraction from starved BMMΦ
- SF IFN
IFN-γ-activated macrophages.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700036104/DC1.
References
- 1.Deretic V, Singh S, Master S, Harris J, Roberts E, Kyei G, Davis A, de Haro S, Naylor J, Lee HH, Vergne I. Cell Microbiol. 2006;8:719–727. doi: 10.1111/j.1462-5822.2006.00705.x. [DOI] [PubMed] [Google Scholar]
- 2.Russell DG. Nat Rev Mol Cell Biol. 2001;2:569–577. doi: 10.1038/35085034. [DOI] [PubMed] [Google Scholar]
- 3.Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 4.Nathan C, Shiloh MU. Proc Natl Acad Sci USA. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacMicking JD, Taylor GA, McKinney JD. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 6.Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. J Immunol. 1998;160:1290–1296. [PubMed] [Google Scholar]
- 7.Via LE, Fratti RA, McFalone M, Pagan-Ramos E, Deretic D, Deretic V. J Cell Sci. 1998;111:897–905. doi: 10.1242/jcs.111.7.897. [DOI] [PubMed] [Google Scholar]
- 8.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 9.Amer AO, Swanson MS. Cell Microbiol. 2005;7:765–778. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 13.Brogden KA. Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 14.Ciechanover A. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 15.Hicke L, Schubert HL, Hill CP. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 16.Shirk AJ, Anderson SK, Hashemi SH, Chance PF, Bennett CL. J Neurosci Res. 2005;82:43–50. doi: 10.1002/jnr.20628. [DOI] [PubMed] [Google Scholar]
- 17.Schaible UE, Schlesinger PH, Steinberg TH, Mangel WF, Kobayashi T, Russell DG. J Cell Sci. 1999;112:681–693. doi: 10.1242/jcs.112.5.681. [DOI] [PubMed] [Google Scholar]
- 18.Kamimoto T, Shoji S, Hidvegi T, Mizushima N, Umebayashi K, Perlmutter DH, Yoshimori T. J Biol Chem. 2006;281:4467–4476. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buschow SI, Liefhebber JM, Wubbolts R, Stoorvogel W. Blood Cells Mol Dis. 2005;35:398–403. doi: 10.1016/j.bcmd.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Low P, Doherty FJ, Fellinger E, Sass M, Mayer RJ, Laszlo L. FEBS Lett. 1995;368:125–131. doi: 10.1016/0014-5793(95)00624-i. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz AL, Ciechanover A, Brandt RA, Geuze HJ. EMBO J. 1988;7:2961–2966. doi: 10.1002/j.1460-2075.1988.tb03158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yates RM, Hermetter A, Taylor GA, Russell DG. Traffic. 2007;8:241–250. doi: 10.1111/j.1600-0854.2006.00528.x. [DOI] [PubMed] [Google Scholar]
- 24.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjorkoy G, Lamark T, Johansen T. Autophagy. 2006;2:138–139. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- 26.de Chastellier C, Thilo L. Cell Microbiol. 2006;8:242–256. doi: 10.1111/j.1462-5822.2005.00617.x. [DOI] [PubMed] [Google Scholar]
- 27.Howell SJ, Wilk D, Yadav SP, Bevins CL. Peptides. 2003;24:1763–1770. doi: 10.1016/j.peptides.2003.07.028. [DOI] [PubMed] [Google Scholar]
- 28.Kieffer AE, Goumon Y, Ruh O, Chasserot-Golaz S, Nullans G, Gasnier C, Aunis D, Metz-Boutigue MH. FASEB J. 2003;17:776–778. doi: 10.1096/fj.02-0699fje. [DOI] [PubMed] [Google Scholar]
- 29.Svensson I, Calles K, Lindskog E, Henriksson H, Eriksson U, Haggstrom L. Appl Microbiol Biotechnol. 2005;69:92–98. doi: 10.1007/s00253-005-1958-6. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Griffiths WJ, Jornvall H, Agerberth B, Johansson J. Eur J Biochem. 2002;269:512–518. doi: 10.1046/j.0014-2956.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- 31.Kuwata H, Yip TT, Yip CL, Tomita M, Hutchens TW. Biochem Biophys Res Commun. 1998;245:764–773. doi: 10.1006/bbrc.1998.8466. [DOI] [PubMed] [Google Scholar]
- 32.Liepke C, Baxmann S, Heine C, Breithaupt N, Standker L, Forssmann WG. J Chromatogr. 2003;791:345–356. doi: 10.1016/s1570-0232(03)00245-9. [DOI] [PubMed] [Google Scholar]
- 33.Kim HS, Park CB, Kim MS, Kim SC. Biochem Biophys Res Commun. 1996;229:381–387. doi: 10.1006/bbrc.1996.1814. [DOI] [PubMed] [Google Scholar]
- 34.Hiemstra PS, van den Barselaar MT, Roest M, Nibbering PH, van Furth R. J Leukocyte Biol. 1999;66:423–428. doi: 10.1002/jlb.66.3.423. [DOI] [PubMed] [Google Scholar]
- 35.Mira MT, Alcais A, Nguyen VT, Moraes MO, Di Flumeri C, Vu HT, Mai CP, Nguyen TH, Nguyen NB, Pham XK, et al. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 36.Lim KL, Dawson VL, Dawson TM. Neurobiol Aging. 2006;27:524–529. doi: 10.1016/j.neurobiolaging.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 37.Cervino AC, Lakiss S, Sow O, Bellamy R, Beyers N, Hoal-van Helden E, van Helden P, McAdam KP, Hill AV. Hum Mol Genet. 2002;11:1599–1603. doi: 10.1093/hmg/11.14.1599. [DOI] [PubMed] [Google Scholar]
- 38.Liu YC. Annu Rev Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 39.Mueller DL. Nat Immunol. 2004;5:883–890. doi: 10.1038/ni1106. [DOI] [PubMed] [Google Scholar]
- 40.Pethe K, Swenson DL, Alonso S, Anderson J, Wang C, Russell DG. Proc Natl Acad Sci USA. 2004;101:13642–13647. doi: 10.1073/pnas.0401657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beatty W, Russell DG. Methods Mol Med. 2001;54:281–293. doi: 10.1385/1-59259-147-7:281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.