Abstract
Sickle cell anemia is a common genetic disorder in African Americans. Opioid analgesics are traditionally the treatment for the severe pain associated with this disease. Here we reveal that the opioid antagonist naloxone possesses potent analgesic activity in two transgenic mouse models of sickle cell anemia (NY1DD and hBERK1) and not in their respective controls (ICR-CD1 and C57BL/6J) when administered by three parenteral routes [intracerebroventricular (i.c.v.), intrathecal, and subcutaneous]. In the NY1DD mice, naloxone (i.c.v.) possessed ≈300-fold greater potency than morphine (i.c.v.). Other opioid antagonists (naltrexone, norbinaltorphimine, and naltrindole) were substantially less effective in producing analgesia. Naloxone and morphine were synergistic in NY1DD mice, suggesting different receptor systems. Microarray analysis suggested naloxone-induced down-regulation of the CC chemokine receptor (CCR)5 in NY1DD mice but not in control mice. Pretreatment of control mice with CC chemokine ligand 5 [CCL5 (RANTES)] enabled naloxone to produce analgesia similar to that observed in NY1DD mice. Mu opioid receptor knockout mice treated similarly also displayed analgesia. That the effect of CCL5 was specifically related to CCR5 and/or CCR1 activation was demonstrated by antagonism of analgesia with the chemokine antagonist methionylated RANTES. Similar antagonism of naloxone-induced analgesia also was observed when NY1DD mice were pretreated with methionylated RANTES. These results indicate that CCR5/CCR1 receptors are directly or indirectly involved in analgesia produced by naloxone. The present study suggests that naloxone may be clinically useful in the treatment of pain associated with sickle cell disease and other disorders involving inflammation.
Keywords: chemokines, microarray, opioid antagonist
Sickle cell disease is the most common genetic disorder in African Americans, affecting 1 in 400, and 8% are carriers of the gene having the sickle cell trait (1). On a global scale, a quarter of a million babies are diagnosed with this illness every year. The pathophysiology of this disease is due to a point mutation in hemoglobin that gives rise to sickling of erythrocytes resulting from the polymerization of hemoglobin S. Sickling plays a significant role in promoting vasoocclusive events that lead to ischemia-induced inflammation and pain (2, 3). Sickle cell patients who have had surgery describe the pain associated with their disease as more severe than postoperative pain.
Sickle cell disease is characterized by increased inflammation and infection arising from severe microvascular occlusive crises. Chemokines and their receptors have been implicated in the development of vascular inflammatory disorders, and, in this regard, higher levels of proinflammatory cytokines and chemokines have been reported to be present in patients with sickle cell disease (4–6).
Opioid analgesics, most notably morphine, have been the mainstay for treatment of pain associated with this disease. However, adequate pain control has been problematic for several reasons, including tolerance, dependence, sedation, respiratory depression, nausea, constipation, and pruritus. Furthermore, because of the fear among many health professionals that patients will become addicted, many sickle cell patients in pain management are under-treated with opioids (1, 7, 8).
Because pain associated with sickle cell disease clearly presents a therapeutic challenge that has not been adequately addressed, a new approach to pharmacotherapy is desirable. Here we describe the results of studies in sickle cell mutant mice that suggest a counterintuitive approach to pain pharmacotherapy for sickle cell disease that may obviate the central and peripheral side effects associated with morphine.
Results and Discussion
Age-Related Increase in Pain Sensitivity in NY1DD Mice.
To evaluate the developmental progress of sickle cell disease, we measured tail-flick latency of NY1DD mice as a function of age. At 10 weeks of age NY1DD mice were found to have a tail-flick latency of 1.71 ± 0.06 sec (mean ± SE), which was substantially lower than 2.34 ± 0.05 sec, observed for C57BL/6J control mice (P < 0.0001). The tail-flick latency did not change significantly at 10 weeks or later. The 0.63-sec difference in reaction time reflects a significant increase in sensitivity to the thermal noxious stimulus that might be equated with hyperalgesia. In this regard, hyperalgesia appeared to be a developmental phenomenon, because NY1DD mice at 6 weeks of age displayed no statistical difference in tail-flick latency from C57BL/6J control mice (2.24 ± 0.06 vs. 2.38 ± 0.09, P = 0.25). In contrast, the control mice mean tail-flick latency remained constant (2.34 ± 0.08) over the same time period. The increased sensitivity to pain in NY1DD mice is most likely attributable to ischemia-induced inflammation as a consequence of the progression of the disease. In view of these results, all experiments were carried out with transgenic mice that were 10 weeks or older.
Effect of Opioid Antagonists and Morphine on NY1DD and hBERK1 Mice.
When NY1DD mice were treated with the opioid antagonist naloxone, we found that naloxone produced potent analgesia in the tail-flick assay by three parenteral routes of administration (Table 1). In this regard, naloxone behaved like a full agonist with a potency nearly 300-fold greater than that of morphine by the intracerebroventricular (i.c.v.) route of administration. At a dose of 25 pmol, the peak effect of naloxone was 10 min with a 60-min duration. Significantly, 3-week-old NY1DD mice exhibited only 12% analgesia, which increased to 70% at 9 weeks. These data parallel the tail-flick latency time, which reached a plateau at 10 weeks. In contrast, naloxone administered to either ICR-CD1 or C57BL/6J control mice exhibited no analgesic effect in the same dose range. To eliminate the possibility that this effect was an aberrant response or a strain-specific effect, a second transgenic sickle cell strain of mice, hBERK1, was tested. In this case, naloxone administered i.c.v. produced analgesia that was ≈130 times more potent than that of morphine.
Table 1.
The analgesic potency of naloxone and morphine in transgenic mouse models of sickle cell anemia
| Analgesic | ED50, 95% confidence interval |
||
|---|---|---|---|
| i.c.v., nmol/mouse | i.t., nmol/mouse | s.c., nmol/kg | |
| Naloxone | |||
| NY1DD | 0.014 (0.011–0.018) | 0.045 (0.035–0.057) | 2,830 (1,290–8,670) |
| hBERK1 | 0.030 (0.026–0.034) | ||
| Morphine | 3.95 (1.63–6.87) | 0.085 (0.039–0.149) | 2,360 (450–4,480) |
Naloxone and morphine tail-flick response was measured at the time of peak analgesic activity (10, 10, and 30 min, respectively). Values for morphine apply only to NY1DD mice.
Because the analgesic potency of naloxone differed only by a factor of three in the brain relative to the spinal cord of NY1DD mice (Table 1), it is possible that the same receptors mediate both the response in the cord and the brain. These data suggest that the potent analgesia elicited by naloxone is a feature associated with these transgenic mouse models of sickle cell anemia.
Naloxone is known to produce divergent effects as a function of dose. Thus, although hyperalgesia is known to occur at doses that effectively antagonize opioids, there are reports of paradoxical effects characterized by analgesia at low doses and hyperalgesia in the high-dose range (9–12). However, because we found that identical doses of naloxone produced potent analgesia in only the transgenic mouse, it would appear that the analgesic effect is modulated by the pathophysiology associated with the disease.
Binding studies of [3H]naloxone to brain membranes from NY1DD mice were carried out to determine whether there are substantial differences in Kd or Bmax for naloxone to account for its analgesic effect. The finding that the binding data for [3H]naloxone in C57BL/6J control vs. NY1DD mice exhibited no dramatic differences in Kd (20.8 vs. 10.5 nM) or Bmax (49.1 vs. 44.6 fmol per nanogram of protein) suggests the potent analgesic activity of naloxone in NY1DD mice is not related to a significant change in the affinity or number of target receptors relative to wild-type mice.
In contrast to i.c.v. administration, the intrathecal (i.t.) and s.c. analgesic potencies of morphine did not differ significantly from those of naloxone (Table 1). In this regard, the analgesic ED50 values of morphine in NY1DD mice were in the same range as the C57BL/6J controls when administered either by i.c.v., i.t., or s.c. routes (Table 2). The order of potency was i.t. > i.c.v. ≫ s.c., with the i.t. potency of morphine greater than that of i.c.v. by a factor of 25- to 100-fold. This differed from that of naloxone, which showed a 3-fold greater potency of i.c.v. over i.t. administration. The inverse i.t./i.c.v. potency ratios for naloxone vs. morphine may reflect their interaction with different receptor systems.
Table 2.
Morphine ED50 values in control and transgenic mouse models of sickle cell anemia
| Mouse model | ED50, 95% confidence interval |
||
|---|---|---|---|
| i.c.v., nmol/mouse | i.t., nmol/mouse | s.c., nmol/kg | |
| C57BL/6J | 2.70 (2.07–4.64) | 0.088 (0.06–0.122) | 6,470 (3,880–10,260) |
| NY1DD | 3.95 (1.63–6.87) | 0.085 (0.039–0.149) | 2,360 (450–4,480) |
The ED50 values were calculated by using a parallel line multiple regression assay. The individual values are based on the number of positive responses per total number of mice from three to four different doses of morphine.
Other opioid antagonists produced analgesia in NY1DD mice, but these agonists were substantially less effective than naloxone and had a slower onset of action. Naltrexone, an antagonist known to be more potent than naloxone, gave a partial agonist response (33%, 8 nmol i.c.v.). The delta-selective opioid antagonist naltrindole (13) also produced partial agonism (10%, at 2.5 nmol per mouse). The selective kappa opioid antagonist norbinaltorphimine (14) had a 1-h peak effect and an ≈500-fold lower potency [ED50 = 1.55 (1.14–2.25) nmol per mouse i.c.v.] than that of naloxone. The 6-h peak effect of norbinaltorphimine [ED50 = 0.46 (0.46–1.66) nmol per mouse] by the i.t. route suggests that the primary site of action was not the spinal cord. Under the same conditions, these antagonists were inactive in control mice. The results of these experiments suggested that naloxone was not mediating significant analgesic activity via kappa or delta opioid receptors. Moreover, the greatly superior analgesic potency of naloxone over norbinaltorphimine, naltrindole, and naltrexone underscores its uniqueness among the opioid antagonists and suggests a possible nonopioid mechanism for its action.
In view of our results showing naloxone to possess high analgesic potency in NY1DD mice, we conducted studies to determine whether or not analgesic synergism exists between naloxone and morphine. The equieffective ratio for naloxone/morphine used was 1:300 (based on the morphine/naloxone ratio of ED50 values) to calculate and to determine the observed ED50 (15, 16). The observed ED50 for the combination was 0.75 nmol per mouse (0.70–0.81), whereas the theoretical ED50 based on an additive effect was 2.04 nmol per mouse (17). The results of this study revealed that naloxone and morphine combinations produce synergism of analgesia in the NY1DD mouse (Fig. 1). The synergism suggests that naloxone and morphine produce analgesia in the NY1DD mouse via different receptor systems (16). Among the numerous possibilities that could give rise to synergism are the existence of chemokine/opioid heterodimeric receptors (18, 19), which may selectively recognize naloxone in transgenic mice with sickle cell anemia, and the interaction of naloxone with different up-regulated neuronal pathways that modulate pain (20–22).
Fig. 1.
Synergism of naloxone and morphine in the NY1DD mice. Naloxone in the presence of morphine (▴) was 5.73 times (95% confidence interval, 4.62–7.11) more potent than naloxone alone (■), whereas morphine in the presence of naloxone (●) was 4.86 times (95% confidence interval, 2.04–9.12) more potent than morphine alone (▾).
Interaction of Naloxone with Chemokines.
To gain insight into the mode of action of naloxone as an analgesic we conducted cDNA microarray analyses of brains from NY1DD and C57BL/6J mice. Mice were pretreated with 25 pmol of naloxone or saline 10 min before their brains were harvested and flash-frozen in liquid nitrogen. The mouse BMAP cDNA microarrays (University of Minnesota) contain 11,592 unique expressed-sequence tags representing 11,532 genes. The images were processed by using GENEPIX PRO (Axon Laboratories, Union City, CA), and normalizations and other analyses were performed by using GENEPRING 7.0 (Agilent Technologies, Palo Alto, CA). The results were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.nih.gov/projects/geo).
Significantly, the CC chemokine receptor (CCR)5 chemokine receptor mRNA was down-regulated by naloxone in NY1DD mice but not in C57BL/6J mice. It has been previously shown that morphine up-regulates CCR5 expression (23), whereas CCR5 receptor agonists down-regulate its expression (24, 25). The down-regulation of the CCR5, which could be a consequence of naloxone-induced activation, presents several mechanistic scenarios that include the interaction of naloxone with opioid/chemokine heterodimeric receptors (18) or direct interaction of naloxone with the CCR5 receptor.
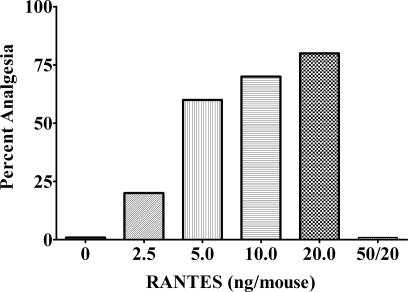
Given that the microarray studies implicated the possible involvement of naloxone with the chemokine receptors in NY1DD mice, we pretreated control mice (ICR-CD1 and C57BL/6J) i.c.v. with the chemokine receptor agonist murine RANTES [CC chemokine ligand (CCL)5] for 20 min before administration of 25 pmol of naloxone (i.c.v.) to peak at the same time. RANTES is known to interact mainly with CCR1 and CCR5 (26, 27). Under these conditions, naloxone was transformed to an analgesic with potency comparable with that observed in NY1DD mice (Fig. 2). The analgesia was dose-dependent, because increasing doses of CCL5 produced corresponding increases in the analgesic effect of naloxone.
Fig. 2.
The analgesic effect of naloxone in ICR-CD1 control mice. In the ICR-CD1 mice, naloxone had an increasing analgesic effect when pretreated for 20 min with increasing doses of RANTES. The analgesic effect of naloxone that was induced by RANTES was inhibited by a 2-h pretreatment of 50 ng of Met-RANTES per mouse before the RANTES injection. At these doses and in the time period used, RANTES did not elicit an analgesic effect when tested by itself. All injections were given i.c.v. The C57BL/6J mice were tested with 20 ng of RANTES per mouse and with 50 ng of Met-RANTES per mouse, with the same results.
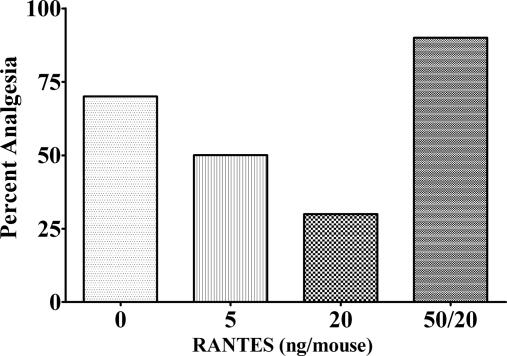
When C57BL/6J mice were pretreated under identical conditions with CCL5 and then administered morphine, decreased analgesia as a function of increasing dose of CCL5 was observed (Fig. 3). This observation was presumably attributable to cross-desensitization between CCR5 and mu opioid receptors (18, 28, 29). To further test the involvement of the chemokine receptors, the chemokine antagonist methionylated RANTES (Met-RANTES), selective for CCR1 and CCR5, was administered (50 ng per mouse) 2 h before naloxone in NY1DD mice. This dose reduced analgesia in NY1DD mice from 80% to 9%. Met-RANTES also blocked the effect of CCL5 in suppressing morphine analgesia. These results suggested a fundamental difference between the receptor systems activated by morphine and naloxone in producing analgesia.
Fig. 3.
The analgesic effect of morphine on C57BL/6J control mice. A 20-min pretreatment with RANTES at two different doses inhibited the analgesic effect of morphine on the C57BL/6J mice. The antagonist ED50 of RANTES was 5.50 ng per mouse (95% confidence interval, 2.58–9.81). This antagonist effect of RANTES on morphine was reversed by a 2-h pretreatment of 50 ng of Met-RANTES per mouse before the RANTES injection. All injections were i.c.v.
To determine whether the mu opioid receptors that mediate morphine analgesia were involved in the analgesic effect of naloxone, mu receptor knockout mice were used (30). These knockout mice were pretreated with CCL5 and then administered naloxone in a manner identical to that used with C57BL/6J mice. Under these conditions, the knockout mice displayed naloxone-induced analgesia that was indistinguishable from C57BL/6J mice. In view of these results, it appears that mu receptors are not involved in mediating naloxone analgesia in NY1DD and CCL5-pretreated mice. These results support our synergism studies, which suggest that naloxone and morphine activate different receptor systems. Moreover, given the inability of the delta and kappa opioid antagonists naltrindole and norbinaltorphimine to produce potent analgesia in either NY1DD mice or CCL5-pretreated control mice, it is possible that neither delta nor kappa opioid receptors are involved in the analgesic action of naloxone.
The data suggest that naloxone and CCL5 target a receptor system that contains a CCR, possibly CCR5 or CCR1. The obligate participation of both naloxone and CCL5 for potent analgesia and the microarray data that revealed naloxone-induced down-regulation of CCR5 in NY1DD mouse brain suggest that naloxone functions as an analgesic by interacting with an unknown target (a “naloxone receptor”) that is heterodimerized with CCR5 or CCR1 and that both the naloxone receptor and the chemokine receptor in this complex must both be occupied to effect a potent analgesic response. The reports of heterodimerization of CCRs and the dimerization induced by CCL5 (27) are in keeping with this model. Further investigation is required to determine the identity of such a receptor.
Conclusions
The unusual ability of the pure opioid antagonist naloxone to function as a potent analgesic in transgenic mouse models of sickle cell anemia and in control mice treated with CCL5 implicates chemokine receptors in mediating this effect. The finding that the CCR1/CCR5 antagonist Met-RANTES was capable of antagonizing naloxone-induced analgesia in both NY1DD and CCL5-pretreated control mice supported the involvement of chemokine receptors. Because the analgesic effect of naloxone in CCL5-pretreated mu opioid receptor knockout mice was not significantly different from pretreated control mice, it appears that the mu opioid system is not involved.
The key role of the chemokine system in the CNS has only recently been appreciated, and, in this connection, chemokines have been suggested to be endogenous regulators of opioid analgesia and tolerance (28, 31). In view of the involvement of chemokines in a variety of inflammatory diseases, the results of the present study suggest that naloxone may be clinically useful as an analgesic in the treatment of pain associated with sickle cell disease and other diseases involving chronic inflammation (11, 12, 32). The absence of morphine-like central side effects of naloxone may offer a superior approach for the treatment of pain in such conditions.
Materials and Methods
Mice.
All experimental animals were housed in groups of 4–10 in a temperature- and humidity-controlled environment. Animals were maintained on a 12-h light/dark cycle and had unlimited access to food and water. The six types of male mice studied were ICR-CD1 (Harlan Sprague, Madison, WI); wild-type controls: C57BL/6J (The Jackson Laboratory, Bar Harbor, ME); and the transgenic mouse models of sickle cell anemia: NY1DD, hBERK1, and HbA-BERK controls (Robert Hebbel, University of Minnesota) (33–35) and MORKO (gift from Sabita Roy, University of Minnesota) (30). Briefly, NY1DD mice having a C57BL/6J background are homozygous for deletion of the murine beta major globin and carry a single copy of linked transgenes for human alpha and beta S globins; C57BL/6J wild-type mice were used as controls. hBERK1 sickle mice have a mixed genetic background, are homozygous for knockout of murine alpha globin, heterozygous for knockout of murine beta globin, and carry a single copy of linked transgenes for human alpha and beta S globins; controls are HbA-BERK mice, which have the same mixed genetic background but express normal human hemoglobin rather than sickle cell hemoglobin. Animals were studied at 10–12 weeks of age. In the time-course study, the mice were 6–10 weeks of age. All experiments were approved by the Institutional of Animal Care and Use Committee of the University of Minnesota.
Analgesic Testing.
Antinociception was measured by using the modified radiant heat tail-flick test (36). Briefly, a radiant heat source was applied to the dorsal side of the tail, and the latency to flick away from the heat source was recorded. The average latency period for the tail to flick in the control mice is 2–3 sec. The data were made quantal by designating a positive antinociceptive response of an animal as those that increased their latency to tail flick (after drug treatment) by at least three standard deviations above the mean of the baseline latency of the whole group (37). The test is stopped manually at a maximum of 4 sec, which is normally greater than the three standard deviations required for positive antinociception. At least three groups of 8–10 mice were used for each drug paradigm, and each mouse was used only once. ED50 values and 95% confidence intervals were calculated by using the parallel line assay (38). When ED50 values were compared, all of the data were analyzed together, and values with separate 95% confidence intervals at P < 0.05 were considered significantly different.
Drugs and Administration.
Morphine sulfate, naloxone, and naltrexone HCl were supplied by Mallinckrodt (St. Louis, MO); norbinaltorphimine and naltrindole were obtained by synthesis in the laboratory of P.S.P. Recombinant murine RANTES (CCL5) was purchased in a 20-μg volume from PEPROTECH, Inc. (Rocky Hill, NJ). Recombinant CCL5/Met-RANTES was purchased in 25-μg volume from R & D Systems (Minneapolis, MN). All drugs administered s.c. were given in a volume of 10 ml/kg; i.c.v. and i.t. injections were given in a volume of 5 μl per mouse (39, 40). All drug injections were timed so that they all reached their peak effect at the final endpoint.
Synergism.
The doses for the synergism studies were calculated with the assistance of Carolyn Fairbanks (15). Briefly, a prestock solution of naloxone and morphine was made based on the ED50 values (naloxone, 0.014 nmol; morphine, 3.95 nmol). From the prestock solution, the other dilutions were made as follows (naloxone/morphine): 0.75:225; 1.5:450; 3:900; and 6:1,800. The naloxone portion plotted separately is labeled as naloxone in the presence of morphine, and the morphine portion plotted separately is labeled as morphine in the presence of naloxone. An interaction was considered synergistic if the observed combined ED50 value was significantly less (95% confidence intervals did not overlap) than the calculated theoretical additive combined ED50 value (16, 17).
Binding Studies.
Brains from three control or sickle-cell-affected mice were removed, and the cerebellum was dissected away and discarded. The brains from each group were pooled and homogenized (Polytron homogenizer at setting 4 for 20 sec; Brinkmann, Westbury, NY) in 10× (wt/vol) ice-cold Hepes buffer (0.25 mM, pH 7.4). The homogenate was centrifuged at 27,000 × g for 15 min (4°C). The supernatant discarded, and the pellet was resuspended in 20× (wt/vol) ice-cold buffer and put on ice for 90 min to exhaust endogenous opiate sources. The suspension was then centrifuged as above, and the supernatant was discarded, and the pellet was resuspended in 10× (wt/vol) ice-cold buffer. This procedure was repeated three times. The final pellet was resuspended in enough buffer to make a 2% (wt/vol) solution, which was used in the experiments. For binding experiments, [3H]naloxone was added in various concentrations; Hepes buffer was added to make a final volume of 0.100 ml. To this mixture, 0.400 ml of the homogenate suspension was added. All concentrations were performed in triplicate. Nonspecific binding was measured by using 10 μM naloxone. Tubes were then incubated at room temperature for 90 min and filtered by using a harvestor (Brandel, Gaithersburg, MD) and GF/C filter paper (Whatman, Florham Park, NJ) presoaked in 0.25% polyethylenimine in water. Filter papers were then placed in scintillation vials, and 4.0 ml of scintillation mixture (Econo-Safe; RPI, Inc., Mt. Prospect, IL) was added. Radioactivity was counted in a LS 6200 scintillation counter (Beckman, Fullerton, CA). Binding data were analyzed by using PRISM software (GraphPad, San Diego, CA); Kd and Bmax values were calculated by using the global fitting of total and nonspecific binding curves model. The protein concentration was determined by using the Lowry method. Each experiment was repeated two or three times, and the mean and SEM were calculated (41, 42).
RNA Isolation and Microarray Hybridizations.
C57BL/6J wild-type and NY1DD transgenic mouse model(s) of sickle cell anemia were administered doses of 25 pmol of naloxone per mouse or 0.9% saline (5 μl); after 10 min the mice were killed, and their brains were removed and frozen in liquid N2 before being transferred to an −80°C freezer. These frozen brains were then used for RNA isolation and further analyses.
RNA was isolated by using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the recommended protocol of the manufacturer. Reverse transcription and cDNA labeling was performed by using the SuperScript Direct cDNA labeling system (Invitrogen) and 20 μg of total RNA. A total of three replications were performed. These replications were true biological replications because, for each new experiment, RNA isolated from a different tissue sample was used. Mouse BMAP cDNA microarrays spotted with 11,592 unique expressed sequence tags (University of Minnesota) were used. To conduct the experiments, a reference design was used in which RNA isolated from the naloxone treatments were labeled with Cy5-dCTP (GE-Healthcare Biosciences Corp., Piscataway, NJ) and compared with the RNA isolated from the saline treatment for the respective mouse strain. The control RNA was labeled with Cy3-dCTP (GE-Healthcare Biosciences Corp.). Final hybridizations were performed with 50 μl of the hybridization mixture, which contained 30 μl of formamide, 12 μl of 20× SSC (1× standard saline citrate = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), 2 μl of 5% SDS, 3 μl of 10 mg/ml ssDNA, and 3 μl of 10 mg/ml poly(A)-RNA, and labeled cDNA at 42°C for 16 h.
Microarray and RT-PCR Analyses.
Image processing was performed (GENEPIX PRO 6.0; Axon Instruments, Union City, CA), and genes with obvious printing and hybridization errors were discarded. Lowess normalization was performed by using GENESPRING 7.0 (Agilent Technologies) on all of the experiments; then, multiple criteria were used to identify significant genes. To identify genes that were differentially expressed in each treatment, replicate experiments were pooled together, and genes with ≥1.5-fold difference over all of the replications were considered to be significant. The different data sets, results, and raw data can be accessed through the National Center for Biotechnology Information Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/projects/geo).
For validating the microarray results, quantitative RT-PCR was performed on five randomly selected genes. The primers were designed by using MacVector software (Accelrys, San Diego), and care was taken to ensure that the primers spanned exon–intron splice sites for negating nonspecific amplification. The RNA isolated was treated with DNase I (Invitrogen), and then a quantitative PCR was performed in an iQ5Cycler (Bio-Rad, Hercules, CA) by using iQ SYBR Green Supermix (Bio-Rad), and the data were analyzed by using the 2−ΔΔCT method.
Acknowledgments
We thank Dr. Carolyn Fairbanks, Dr. Sabita Roy, Michael Powers, Stephana Choong, Fuad Abdulla, and Dr. Wayne Xu for advice and assistance. This work was supported by National Institutes of Health Grants DA01533 (to P.S.P.) and HL55552 (to R.P.H.).
Abbreviations
- CCL
CC chemokine ligand
- CCR
CC chemokine receptor
- i.c.v.
intracerebroventricular(ly)
- i.t.
intrathecal(ly)
- Met
methionylated.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE 6578).
References
- 1.Platt A, Eckman JR, Beasley J, Miller G. J Emerg Nurs. 2002;28:297–303. doi: 10.1067/men.2002.125268. [DOI] [PubMed] [Google Scholar]
- 2.Hebbel RP, Osarogiagbon R, Kaul DK. Microcirculation. 2004;11:129–153. [PubMed] [Google Scholar]
- 3.Dampier C, Shapiro BS. In: Pain in Infants, Children and Adolescents. Schechter NC, Berde CB, Yaster M, editors. Philadelphia: Lippincott Williams & Wilkins; 2003. pp. 489–516. [Google Scholar]
- 4.Pathare A, Kindi SA, Daar S, Dennison D. Hematology. 2003;8:329–337. doi: 10.1080/10245330310001604719. [DOI] [PubMed] [Google Scholar]
- 5.Hibbert JM, Hsu LL, Bhathena SJ, Irune I, Sarfo B, Creary MS, Gee BE, Mohamed AI, Buchanan ID, Al-Mahmoud A, et al. Exp Biol Med. 2005;230:68–74. doi: 10.1177/153537020523000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murdoch C, Finn A. Blood. 2000;95:3032–3043. [PubMed] [Google Scholar]
- 7.Shapiro BS, Benjamin LJ, Payne R, Hedrich G. J Pain Symptom Manage. 1997;14:168–174. doi: 10.1016/S0885-3924(97)00019-5. [DOI] [PubMed] [Google Scholar]
- 8.Marlowe KF, Chicella MF. Pharmacotherapy. 2002;22:484–491. doi: 10.1592/phco.22.7.484.33675. [DOI] [PubMed] [Google Scholar]
- 9.Levine JD, Gordon NC, Fields HL. Nature. 1979;278:740–741. doi: 10.1038/278740a0. [DOI] [PubMed] [Google Scholar]
- 10.Carmody JJ, Carroll PR, Morgans D. Life Sci. 1979;24:1149–1152. doi: 10.1016/0024-3205(79)90050-x. [DOI] [PubMed] [Google Scholar]
- 11.Woolf CJ. Brain Res. 1980;189:593–597. doi: 10.1016/0006-8993(80)90375-3. [DOI] [PubMed] [Google Scholar]
- 12.Kayser V, Guilbaud G. Brain Res. 1981;226:344–348. doi: 10.1016/0006-8993(81)91110-0. [DOI] [PubMed] [Google Scholar]
- 13.Portoghese PS, Sultana M, Takemori AE. Eur J Pharmacol. 1988;146:185–186. doi: 10.1016/0014-2999(88)90502-x. [DOI] [PubMed] [Google Scholar]
- 14.Takemori AE, Ho BY, Naeseth JS, Portoghese PS. J Pharmacol Exp Ther. 1988;246:255–258. [PubMed] [Google Scholar]
- 15.Fairbanks CA, Stone LS, Kitto KF, Nguyen HO, Posthumus IJ, Wilcox GL. J Pharmacol Exp Ther. 2002;300:282–290. doi: 10.1124/jpet.300.1.282. [DOI] [PubMed] [Google Scholar]
- 16.Tallarida RJ. Drug Synergism and Dose-Effect Data Analysis. Boca Raton, FL: Chapman & Hall/CRC; 2000. pp. 57–75. [Google Scholar]
- 17.Voorsuij AJZ, Nass CAG. Arch Int Pharmacodyn. 1957;109:211–228. [PubMed] [Google Scholar]
- 18.Chen C, Li J, Bot G, Szabo I, Rogers TJ, Lu-Chen LY. Eur J Pharm. 2004;483:175–186. doi: 10.1016/j.ejphar.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki S, Chuang LF, Yau P, Doi RH, Chuang RY. Exp Cell Biol. 2002;280:192–200. doi: 10.1006/excr.2002.5638. [DOI] [PubMed] [Google Scholar]
- 20.Gillman MA, Lichtigfeld FJ. Neurolog Res. 1985;7:106–119. doi: 10.1080/01616412.1985.11739709. [DOI] [PubMed] [Google Scholar]
- 21.Kim DH, Fields HL, Barbaro NM. Brain Res. 1990;516:37–40. doi: 10.1016/0006-8993(90)90894-h. [DOI] [PubMed] [Google Scholar]
- 22.Crain SM, Shen KF. Trends Pharmacol Sci. 1990;11:77–81. doi: 10.1016/0165-6147(90)90322-y. [DOI] [PubMed] [Google Scholar]
- 23.Proudfoot AEI, Power CA, Hoogewerf AJ, Montjovent M-O, Borlat F, Offord RE, Wells NC. J Biol Chem. 1996;271:2599–2603. doi: 10.1074/jbc.271.5.2599. [DOI] [PubMed] [Google Scholar]
- 24.Steele AD, Szabo I, Bednar F, Rogers TJ. Cytokine Growth Factor Rev. 2002;13:209–222. doi: 10.1016/s1359-6101(02)00007-2. [DOI] [PubMed] [Google Scholar]
- 25.Fabry ME, Nagel RL, Pachnis A, Suzuka SM, Costantini F. Proc Natl Acad Sci USA. 1992;89:12150–12154. doi: 10.1073/pnas.89.24.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez-Frade JM, Vila-Coro AJ, Martin A, Nieto M, Sanchez-Madrid F, Proudfoot AE, Wells TN, Martinez-A C, Mellado M. J Cell Biol. 1999;144:755–765. doi: 10.1083/jcb.144.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rollins BJ. Blood. 1997;90:909–928. [PubMed] [Google Scholar]
- 28.Szabo I, Chen X-H, Xin L, Adler MW, Howard OMZ, Oppenheim JJ, Rogers TJ. Proc Natl Acad Sci USA. 2002;99:10276–10281. doi: 10.1073/pnas.102327699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang N, Rogers TJ, Caterina M, Oppenheim JJ. J Immunol. 2004;173:594–599. doi: 10.4049/jimmunol.173.1.594. [DOI] [PubMed] [Google Scholar]
- 30.Loh HH, Liu HC, Cavalli A, Yang W, Chen YF, Wei LN. Mol Brain Res. 1998;54:321–326. doi: 10.1016/s0169-328x(97)00353-7. [DOI] [PubMed] [Google Scholar]
- 31.Adler MW, Geller EB, Chen X, Rogers TJ. Am Assoc Pharm Sci. 2006;7:E865–E869. doi: 10.1208/aapsj070484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kayser V, Besson JM, Guilbaud G. Prog Brain Res. 1988;77:301–312. doi: 10.1016/s0079-6123(08)62796-x. [DOI] [PubMed] [Google Scholar]
- 33.Solovey A, Kollander R, Shet A, Milbauer LC, Choong S, Panoskaltsis-Mortani A, Blazar BR, Kelm RJ, Jr, Hebbel RP. Blood. 2004;104:840–846. doi: 10.1182/blood-2003-10-3719. [DOI] [PubMed] [Google Scholar]
- 34.Fabry ME, Nagel RL, Pachnis A, Suzuka SS, Constantine F. Proc Natl Acad Sci USA. 1992;89:12150–12154. doi: 10.1073/pnas.89.24.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabry ME, Sengupta SM, Suzuka SM, Constantine F, Rubin EM, Hofrichter G, Manci E, Culberson D, Factor SM, Nagel RL. Blood. 1995;86:2419–2428. [PubMed] [Google Scholar]
- 36.Tulunay FC, Takemori AE. J Pharmacol Exp Ther. 1974;190:395–400. [PubMed] [Google Scholar]
- 37.Tallarida RJ. Drug Synergism and Dose–Effect Data Analysis. Boca Raton, FL: Chapman and Hall/CRC; 2000. pp. 91–116. [Google Scholar]
- 38.Finney DJ. Statistical Methods in Biological Assay. 2nd Ed. New York: Hafner; 1964. [Google Scholar]
- 39.Haley TJ, McCormick WG. Brit J Pharmacol. 1957;12:12–15. doi: 10.1111/j.1476-5381.1957.tb01354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hylden JL, Wilcox GL. Eur J Pharmacol. 1980;67:313–316. doi: 10.1016/0014-2999(80)90515-4. [DOI] [PubMed] [Google Scholar]
- 41.Oliver H, Lowry NJ, Rosebrough A, Lewis F, Rose J, Randall J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 42.Werling LL, Zarr GD, Brown SR, Cox BM. J Pharmacol Exp Ther. 1985;233:722–728. [PubMed] [Google Scholar]