Abstract
The boronic acid moiety is a versatile functional group useful in carbohydrate recognition, glycoprotein pull-down, inhibition of hydrolytic enzymes and boron neutron capture therapy. The incorporation of the boronic-acid group into DNA could lead to molecules of various biological functions. We have successfully synthesized a boronic acid-labeled thymidine triphosphate (B-TTP) linked through a 14-atom tether and effectively incorporated it into DNA by enzymatic polymerization. The synthesis was achieved using the Huisgen cycloaddition as the key reaction. We have demonstrated that DNA polymerase can effectively recognize the boronic acid-labeled DNA as the template for DNA polymerization, that allows PCR amplification of boronic acid-labeled DNA. DNA polymerase recognitions of the B-TTP as a substrate and the boronic acid-labeled DNA as a template are critical issues for the development of DNA-based lectin mimics via in vitro selection.
INTRODUCTION
Boronic acid is a versatile functional group that has been explored for the development of various biologically important compounds (1–3), such as carbohydrate sensors and receptors (4–12), inhibitors of hydrolytic enzymes (1–3,13) and boron neutron capture therapy agents (14,15). We envision that the incorporation of the boronic acid moiety into nucleic acid has the potential to lead to the discovery of new aptamers (7,16,17) against carbohydrates, glycoproteins and glycolipids with specific focus on differentiating the carbohydrate portion. The design is built on earlier works demonstrating the feasibility of selecting modified DNA/RNA aptamers for various applications and the effect of DNA modification in general (18–28). Boronic acid-labeled aptamers with high specificity and affinity for certain carbohydrates could be used as tools for the rapid analysis of glycosylation patterns of proteins, peptides and lipids in the same way as lectin arrays are used (29–33). However, the number and specificity of carbohydrate aptamers could far surpass that of available natural lectins after the development of the appropriate platform technology. The key step in developing such boronic acid-labeled DNA aptamers is the successful demonstration of the design, synthesis and incorporation of boronic acid-labeled nucleotide into DNA by enzymatic polymerization. Herein, we describe our efforts in demonstrating this critical feasibility.
MATERIALS AND METHODS
Synthesis of the boronic acid-labeled thymidine triphosphate (B-TTP)
Chemicals were obtained from Aldrich, Frontier Scientific, Accros and Asymchem unless indicated otherwise. For all reactions analytical grade solvents were used. Anhydrous solvents were used for all moisture-sensitive reactions. NMR data was collected on a Varian Unity 300 MHz or a Bruker 400 MHz spectrometer. The chemical shifts are relative to TMS as an internal standard for 1H, the deuterated solvent used for 13C and 85% H3PO4 as an external reference for 31P. Mass spectra were recorded on a Waters Micromass LC-Q-TOF micro spectrometer at Georgia State University Mass Spectrometry Facilities.
8-Bromo-2-methylquinoline (2)
To the solution of 2-bromoaniline (5.0 g, 29.1 mmol) in 6N hydrochloric acid (15 ml) under reflux crotonaldehyde (2.2409 g, 32.0 mmol) was added dropwise. After refluxing for 8 h, the reaction mixture was cooled down and washed with 20 ml of ether, followed by the addition of zinc chloride (3.95 g). The reaction mixture was stirred for 30 min at room temperature and an additional 15 min at 0°C to yield a yellow precipitate. The solid was collected and washed with 3N cold hydrochloric acid, and then suspended in 2-propanol (20 ml) and stirred for 5 min at room temperature. The solid was filtered and washed with 2-propanol until the washing became colorless, and then washed with 20 ml of ether and dried in air. The solid was suspended in 15 ml of cold water followed by the addition of 5 ml of concentrated ammonium hydroxide. The mixture was vigorously shaken and then extracted with ether (3 × 20 ml). After drying over magnesium sulfate and concentration, a dark solid product was obtained, which was purified by chromatography (EtOAC:Hexanes 10:90) to give a white solid product (3.62 g, 56%) 1H NMR(400 MHz, CDCl3) δ 8.02 (2H, t, J = 8.4 Hz), 7.73 (1H, d, J = 8 Hz), 7.33 (2H, t, J = 8), 2.82 (3H, s); 13C NMR (75 MHz, CDCl3) δ 160.2, 144.7, 136.4, 132.8, 127.6, 127.3, 125.9, 124.0, 122.7, 25.6; EIMS, m/z 221/223 M/M + 2; Anal. Calcd for C10H8BrN: C, 54.08; H, 3.63; N, 6.31. Found: C, 54.25; H, 3.41; N, 5.89.
8-Bromo-2-bromomethylquinoline (3)
To a solution of 2 (2.5477 g, 11.47 mmol) in carbon tetrachloride (40 ml) NBS (2.2461 g, 12.62 mmol) and 20 mg of AIBN were added. The mixture was refluxed overnight under regular light, and then filtered to remove the solid. Evaporation of the solvent gave a yellow solid product that was purified by chromatography (hexanes:DCM 80:20) to yield a white solid (1.33 g, 39%). 1H NMR (400 MHz, CDCl3) δ 8.16 (1H, d, J = 8.4 Hz), 8.05 (1H, d, J = 7.2 Hz), 7.78 (1H, d, J = 7.6 Hz), 7.65 (1H, d, J = 8.4 Hz), 7.41 (1H, t, J = 7.6 Hz), 4.78 (3H, s); 13C NMR (75 MHz, CDCl3) δ 158.3, 144.7, 138.0, 133.9, 128.9, 127.7, 127.6, 125.1, 122.4, 34.6; EIMS, m/z 299/300/301 (M/M + 1/M + 2); Anal. Calcd for C10H7Br2N: C, 39.91; H, 2.34; N, 4.65. Found: C, 40.13; H, 2.281; N, 4.34.
(8-Bromo-quinolin-2-ylmethyl)-methylamine (4)
To a solution of 3 (1 g, 3.32 mmol) in THF (5 ml) methylamine (10.5 ml, 40% aqueous solution) was added. The solution was stirred for 30 min and then extracted with EtOAc (30 ml). The organic phase was washed with DI water (2 × 20 ml), dried over anhydrous magnesium sulfate and concentrated to give a red oily product, that was purified by column chromatography (MeOH:DCM 1:99) to yield a yellow solid (0.8 g, 96%). 1H NMR (400 MHz CDCl3) δ 8.09 (1H, d, J = 8.4 Hz), 8.02 (1H, d, J = 7.2 Hz), 7.77 (1H, d, J = 8 Hz), 7.49 (1H, d, J = 8.4 Hz), 7.36 (1H, t, J = 8.0 Hz), 4.12 (2H, s), 2.58 (3H, s); FABMS, m/z 251/253 (M + H/M + 2 + H); Anal. Calcd for C11H11BrN2: C, 52.61; H, 4.42; N, 11.16. Found: C, 52.17; H, 4.46; N, 11.10.
(8-Bromo-quinolin-2-ylmethyl)-methylcarbamic acid tert-butyl ester (5)
To a solution of 4 (0.7501 g, 2.99 mmol) in methanol (Boc)2O (1.4992 g, 6.87 mmol) and triethylamine (2.1 ml, 14.9 mmol) were added. The mixture was stirred at room temperature for 2 h, and then concentrated in vacuo to remove all the solvent. The residue was dissolved in DCM (20 ml) and then washed with DI water (2 × 10 ml) and brine (10 ml). The organic solution was dried over MgSO4 and concentrated to give yellow oil. Purification by chromatography (hexanes:EtOAc 10:90) yielded a light yellow oily product. 1H NMR (400 MHz CDCl3) δ 8.16 (1H, t, J = 8.4 Hz), 8.06 (1H, d, J = 6.9 Hz), 7.80 (1H, d, J = 7.8 Hz), 7.41 (2H, m), 4.81 (2H, s), 3.03 (3H, d, J = 11.7 Hz), 1.4–1.6 (9H); 13C NMR (100 MHz, CDCl3) δ 160.0, 144.9, 137.5, 133.4, 128.7, 127.7, 126.9, 124.9, 120.7, 119.8, 80.1, 55.5, 35.2, 28.7; ESIMS, m/z 351/353 (M/(M + 2),100); Anal. Calcd for C16H19BrN2O2: C, 54.71; H, 5.45; N, 7.98. Found: C, 54.97; H, 5.62; N, 7.75.
2-[(tert-Butoxycarbonyl-methyl-amino)-methyl]-quinoline-8-boronic acid (6)
To a flask charged with 5 (0.4440 g, 1.26 mmol), bis(neopentyl glycolato)diboron (0.3427 g, 1.52 mmol), Pd(dppf)Cl2 (0.0310 g, 0.038 mmol) and KOAc (0.3722 g, 3.79 mmol) in a nitrogen atmosphere anhydrous DMSO (10 ml) was added. The mixture was stirred at 80°C overnight. After cooling down, the reaction mixture was poured into DCM (20 ml) and washed with DI water (4 × 30 ml). The organic solution was dried over MgSO4 and concentrated to give dark oil. Purification by column chromatography (MeOH/DCM, 1:99) yielded a yellow oily product (0.3313 g, 82%). 1H NMR (400 MHz CDCl3) δ 8.45 (1H, d, J = 5.4 Hz), 8.14 (1H, d, J = 6.6 Hz), 7.97 (1H, d, J = 8.1 Hz), 7.623 (1H, t, J = 7.5 Hz), 7.49 (1H, d, J = 8.4 Hz), 4.80 (2H, d, J = 6.0 Hz), 3.09 (3H, d, J = 4.2 Hz), 1.3–1.5 (9H); 13C NMR (100 MHz, CDCl3) δ 157.3, 156.8, 156.2, 150.3, 139.4, 137.4, 129.7, 127.2, 126.6, 119.1, 118.9, 80.5, 74.7, 34.7, 27.4, 23.9; ESIMS, m/z 315, M − 1.
Compound 7
To a solution of 6 (0.226 g, 0.72 mmol) in DCM (20 ml) TFA (5 ml) was added. The solution was stirred for 1 h, and then concentrated in vacuo to give yellow oil that was then dissolved in dry THF (10 ml). To this mixture azido acetic acid (79 mg, 0.79 mmol), N,N′-carbonyldiimidazole (CDI) (174 mg, 1.07 mmol) and i-PrNEt (0.25 ml, 1.43 mmol) at 0°C were added. The mixture was stirred overnight at room temperature and then concentrated to almost dryness. Purification by silica gel column (DCM:MeOH, 100:1) yielded a yellow solid (0.160 g, 68%). 1H NMR (400 MHz CDCl3) δ 8.45 (1H, d, J = 5.4 Hz), 8.14 (1H, d, J = 6.6 Hz), 7.97 (1H, d, J = 8.1 Hz), 7.623 (1H, t, J = 7.5 Hz), 7.49 (1H, d, J = 8.4 Hz), 4.80 (2H, d, J = 6.0 Hz), 4.30 (2H, d, J = 5.5 Hz), 3.09 (3H, d). ESIMS, m/z 300, M + 1.
5-[3-(Trifluoroacetamido)-propynyl]-2′-deoxyuridine 9
5-Iodo-2′-deoxyuridine (0.35 g, 1.0 mmol) was dissolved in degassed anhydrous DMF (10 ml). Copper (I) iodide (0.038 g, 0.2 mmol) was added and the reaction mixture was stirred under nitrogen in the dark by wrapping the reaction flask with aluminum foil for 30 min. Triethylamine (0.3 ml, 2.0 mmol) was added to the reaction mixture, followed by N-propynyltrifluoroacetamide (0.45 g, 2.97 mmol) and tetrakis(triphenylphosphine) palladium (0) (0.11 g, 0.10 mmol). The reaction mixture was stirred overnight with an aluminum foil wrap at room temperature. Then solvent was removed and the residue was purified with a silica gel column (methanol:DCM 1:20) to give a light yellow solid (0.25 g, 67%). 1H NMR (300 MHz, CD3OD) δ 8.4 (1H, s), 6.22 (1H, t), 4.39 (1H, m), 4.26 (2H, s), 3.82 (1H, m), 3.74 (2H, m), 2.38–2.20 (2H, m). ESIMS (m/z): 378 (M + 1).
Compound 10
To compound 9 (0.25 g, 0.66 mmol) dissolved in methanol was added ammonium hydroxide. The mixture was stirred overnight followed by solvent removal. The residue was dried under vacuum and then dissolved in DMF. Pentynoic acid (68 mg, 0.69 mmol) and benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBop) (0.99 mmol) were added under ice-bath cooling. The reaction mixture was stirred overnight at room temperature. Then solvent was evaporated and the residue was purified by silica gel chromatography (methanol:DCM 1:15) to give the product (155 mg, 65%). 1H NMR (300 MHz CD3OD): δ 8.29 (1H, s), 6.22 (1H, t), 4.39 (1H, m), 4.14 (2H, s), 3.90 (1H, m), 3.74 (2H, m), 2.41 (4H, m), 2.26 (3H, m). 13C NMR (100 MHz, CD3OD) δ 170.8, 162.0, 148.2, 142.7, 97.2, 87.2, 86.5, 84.3, 80.9, 72.5, 69.4, 67.7, 59.9, 39.0, 33.1, 27.7, 12.8. ESIMS (m/z): 362 (M + 1).
Compound 11
Compound 10 (0.15 g, 0.4 mmol) was dried in vacuo over P2O5 overnight and then dissolved in anhydrous trimethylphosphate (0.6 ml) under nitrogen. Proton sponge (also dried overnight over P2O5) (0.102 g, 0.48 mmol) was added to the solution in one portion. Then the reaction mixture was cooled in an ice bath and POCl3 was added drop wise via a syringe with stirring. The reaction mixture was stirred on ice for 2 h and then a mixture of 0.98 g of bis-tri-n-butylammonium pyrophosphate (dissolved in DMF 1.6 ml) and 0.6 ml tri-n-butylamine was added in one portion. The mixture was stirred at room temperature for 10 min and then triethylammonium bicarbonate solution (0.1 M, pH 8, 10 ml) was added. The reaction mixture was stirred at room temperature for an additional hour and purified with a DEAE-Sephadex A-25 column using a linear gradient of ammonium bicarbonate (0–0.6 M) followed by freeze drying to give the final product as a white powder (84 mg, 35%). 1H NMR (300 MHz, D2O): δ 8.21 (1H, s), 6.33 (1H, t), 4.68 (1H, m), 4.26 (5H, m), 2.57 (4H, m), 2.45 (3H, m). 13C NMR (100 MHz, D2O) δ 174.1, 164.2, 150.1, 144.5, 98.7, 89.4, 85.5, 85.2, 83.0, 73.1, 70.1, 69.7, 65.0, 38.5, 33.9, 29.2, 14.0; 31P NMR (161 MHz, D2O): δ−10.04 (γP, d), −11.30 (αP, d), −22.96 (βP, t). ESIMS (m/z): 601 (M), 521 (M−80).
Compound 12
The azide compound 7 (0.014 g, 0.046 mmol) and triphosphate compound 11 (0.009 g, 0.015 mmol) were suspended in 150 μl of a mixture of EtOH:H2O:t-butyl alcohol (3:2:5). To this mixture were added 5 μl of 1.12 M sodium ascorbate aqueous solution and 5 μl of 0.54 M CuSO4 aqueous solution. The mixture was stirred at room temperature overnight and then filtered to remove the unreacted azide compound. The filtrate was purified by a DEAE-sephadex A-25 column. Fractions were collected by monitoring the UV absorbance at 289 nm. The combined fractions were lyophilized to yield a white powder product (3 mg, 20%). 1H NMR (300 MHz CD3OD): δ 8.52 (1H, m), 8.11 (1H, m), 7.88 (1H, m), 7.68 (1H, s), 7.54 (2H, m), 7.15 (1H, m), 5.94 (1H, dt), 5.60 (2H, d), 5.01 (2H, d), 4.43 (1H, m), 4.06 (3H, m), 3.99 (2H, m), 3.23 (2H, s), 2.98 (3H, d), 2.54 (2H, d), 2.18 (2H, m), 1.69 (2H, m), 1.21 (2H, m). 31P NMR (161 MHz, D2O): δ −9.65 (γP, d), −10.76 (αP, d), −22.36 (βP, t); ESIMS, m/z 864/784, M-2H2O/M-2H2O-80.
The stability of the boronic-acid-modified-TTP has been studied at 94°C for 2 h. No degradation was observed based on NMR and MS.
N-[2-(3, 4-Dihydroxy-phenyl)-ethyl]-acrylamide 13
To a suspension of dopamine hydrochloride (3 g, 16 mmol) in DCM (35 ml) was added triethylamine (6.7 ml). The mixture was stirred for 1 h then trimethylchlorosilane was added. After 4 h, additional triethylamine (2.5 ml) was added followed by acryloyl chloride in a dropwise fashion with an ice-bath cooling. After stirring the reaction for 12 h, the white precipitate was filtered, collected and re-dissolved in 30 ml DCM followed by the addition of 10% TFA/DCM. The reaction mixture was stirred over night at room temperature. The white precipitate product was filtered and collected (2.01 g, 64%). 1H NMR (300 MHz CDCl3) 6.69 (2H, m), 6.53 (1H, dd), 6.20 (1H, d), 6.18 (1H, d), 5.62 (1H, t, J = 6.0 Hz), 3.4 (2H, m), 2.66 (2H, t, J = 7.8 Hz). 13C NMR (100 MHz, CDCl3) δ 166.71, 144.87, 143.41, 130.71, 130.57, 125.10, 119.61, 115.10, 114.98, 41.00, 34.47.
Primer 21 nt: 5′-GCGTAATACGACTCACTATAG-3′
Primer 1: 5′-GCGTAATACGACTCACTATA-3′
Primer 2: 5′-TGTACGTTTCGGCCTTTCGG-3′
Template DNA: 3′-GCATTATGCTGAGTGATATCCGTTGGACTACTCCGGCTTTCCGGCTTTGCATGT-5′
32P labeling of the primers
A mixture of 10 µl of the primer DNA (100 µM), 2.0 μl of T4 polynucleotide kinase buffer (×10), 3.0 µl of water, 3.0 µl of T4 polynucleotide kinase (10 000 units/ml, Biolabs, Inc.) and 2.0 µl of γ-32P-ATP (from Perkin/Elmer) was incubated for 1 h at 37°C followed by heating in a water bath at 100°C for 5 min to denature the T4 polynucleotide kinase. Then the kinased DNA was precipitated with 2.2 µl of 3 M sodium chloride solution (3 M) and 66.6 µl of ethanol. The mixture was chilled at −20°C for 15 min and centrifuged at 14 000 rpm for 15 min. The supernatant was discarded and the pellet was re-dissolved in 8 µl of water (to obtain approximately 100 µM DNA solution, assuming an 80% recovery yield) and stored at −20°C.
Examples of DNA primer extension and time-course study with B-TTP (12)
Primer extensions were performed with the 5′-32P-labeled primer (21 nt, 5 µM) and the template (55 nt, 5 µM), Klenow (0.04 units/µl) and dNTPs (0.4 mM each). The reaction mixture was incubated at 37°C. Aliquots (5 µl) of the solution were taken at 0.5, 2, 5, 15 and 60 min and were put into an ice-bath to stop the reaction following the addition of 5 µl of denaturing dye solution (8 M urea) into each aliquot. These samples were analyzed later by electrophoresis and autoradiography.
Primer extension using boronic acid-labeled DNA as templates
Primer 1/template (5 µM), Klenow (0.04 units/µl), TTP (0.4 mM), B-TTP (0.4 mM) or M-TTP (0.4 mM), and three other dNTPs (0.4 mM each) were incubated at 37°C for an hour. The prepared DNAs were purified by membrane filtration for 15 min at 14 000 rpm, using Microcon centrifugal filter YM-3 from Millipore Corporation, to remove the labeled and non-labeled dNTPs and other low molecular weight molecules. 5′-32P-labeled Primer 2 was then added to the DNAs prepared using Primer 1, individually, and the mixtures were heated for 2 min at 95°C. The mixtures were cooled to room temperature over 10 min. The second run of the polymerizations on the labeled and non-labeled DNA templates was performed under the conditions of four dNTPs (0.4 mM each) and Klenow (0.04 units/μl) at 37°C for an hour. The resulting samples were analyzed by electrophoresis and autoradiography.
Preparation of the polyacrylamide gel containing catechol (19% acrylamide gels modified with 1% catechol)
Urea (12.6 g), N-[2-(3,4-dihydroxy-phenyl)-ethyl]-acrylamide (0.16 g), a 40% acrylamide solution (24 ml), ×5 TBE (Tris-borate-EDTA made from 108 g Tris base, 55 g boric acid, 9.3 g Na4EDTA in 1 l of water) (6 ml) and water (6 ml) were mixed and heated in a microwave for 30 s. After cooling, 20 µl of TEMED (N,N,N-tetramethyl thylenediamine) and 150 µl of APS (ammonium persulfate) were added before loading this solution into a gel cast.
Incorporation of B-TTP into DNA by PCR
Each 50 μl reaction was performed with 1.2 μM primers 1 and 2/template, 0.25 mM of each dNTPs, 0.25 mM of labeled-TTP (B-TTP) and 3.5 units of high fidelity DNA polymerase (Roche, Indianapolis, Ind.) under conditions of 1 cycle at 94°C for 2 min, 30 cycles at 94°C for 20 s, 59°C for 30 s, 72°C for 1 min and 1 cycle at 72°C for 7 min. (Primer 1: 5′-CGCCGCCCCCGCCGCG-3′ Primer 2: 5′-CGGCGGCCCGCGGGCG; Template DNA: 5′-CGCCGCCCCCGCCGCG-40N-CGCCCGCGGGCCGCCG-3′.) Ten microliters of each amplification product was separated by gel electrophoresis on 1.5% agarose, stained with ethidium bromide and visualized under UV light.
RESULTS AND DISCUSSION
In considering the eventual incorporation of the boronic acid into DNA, we focused on the strategy of covalently linking a boronic acid moiety to a nucleoside triphosphate, which can be used in DNA polymerization and amplification. It has been demonstrated in the literature that 5-position modification of deoxyuridine can be tolerated by polymerases and reverse transcriptases (17,34), though the situation with the attachment of a boronic acid moiety could be very different. One key factor that could lead to failure in this strategy with this type of modification is the strong Lewis acidity of the boronic acid moiety that can lead to tight interactions with Lewis bases commonly found on nucleic acids and enzymes. Such interaction could have several implications not common with the attachments of other organic functional groups at the 5-position of deoxyuridine including impeded incorporation and amplification, added secondary structures in the DNA products, enzyme binding and inhibition and even inter-strand interactions. Despite all these, 5-position modification on deoxyuridine represents the best chance for success.
For minimal interference of the polymerase reaction, we were interested in a long and somewhat linear linker, and for the ease of attachment we were interested in using a reaction that is mild, can be carried out in an aqueous solution, and does not interfere with other existing functional groups. For these reasons, we selected the Huisgen cycloaddition for the coupling of the boronic acid moiety with the nucleoside (35). Therefore, we have designed compound 12 as a monomeric building block for DNA polymerization. We chose an 8-quinoline boronic acid analog because of our prior experience with such compounds, its known affinity for various sugars and its water solubility. In addition, we were interested in using this somewhat large arylboronic acid as a model because its successful incorporation into DNA would probably mean that other smaller arylboronic acid analogs would have minimal problem to be incorporated.
With this strategy in mind, we were in need of a quinoline boronic acid analog with an azido group and a 5-modified deoxyuridine analog with an alkyne group. The synthesis of the quinoline boronic acid followed the procedures described in Scheme 1.
The synthesis of the quinoline boronic acid building block started with the commercially available 2-bromoaniline (1), which was converted to 2-methylquinoline (2) by refluxing with crotonaldehyde in 6N HCl (36–39) (Scheme 1). Bromination at methyl group gives compound 3, which was reacted with 40% methylamine aqueous solution in THF to yield 4. The amino group was first protected with Boc before borylation under the catalysis of dichloro-(bis-diphenylphospino)ferrocenyl)-palladium [Pd(dppf)Cl2] to give compound 6. Deprotection by TFA followed by amide formation with azido acetic acid generated the quinoline boronic acid (7).
For the synthesis of the final compound 12, there are two possible general approaches. The first approach is to attach the boronic acid first before triphosphorylation. The second approach is to triphosphorylate first before the attachment of the boronic acid moiety. In both approaches, the synthesis started with 5-iodo-2′-deoxyuridine (Scheme 2). By following literature procedures, an alkyne side chain was attached to the 5-position (34,40–42). Deprotection of the amino group and introduction of a terminal alkyne group gave intermediate 10. We first took the approach of attaching the boronic acid to nucleoside 10 followed by phosphorylation. Unfortunately, the incorporation of the boronic acid moiety made the phosphorylation very difficult (results not shown). As a consequence, we were unable to obtain the final product using this method. We suspect that the boronic acid moiety interferes with the phosphorylation because of the presence of two hydroxyl groups on the deoxyribose moiety or the difficulty in drying this highly polar and hygroscopic compound, though we did not analyze every component of the reaction mixture. Therefore, we turned to the second approach of preparing the triphosphate (11) before attaching the boronic acid moiety. The triphosphorylation of 10 allowed for the synthesis of 11 (M-TTP). The subsequent Huisgen cycloaddition allowed the tethering of the quinoline boronic acid moiety to give 12. The final product was purified by a Sephadex-DEAE A-25 followed by ion-exchange HPLC. Thermal stability studies using NMR under PCR conditions demonstrated that the boronic acid moiety does not present additional stability problems.
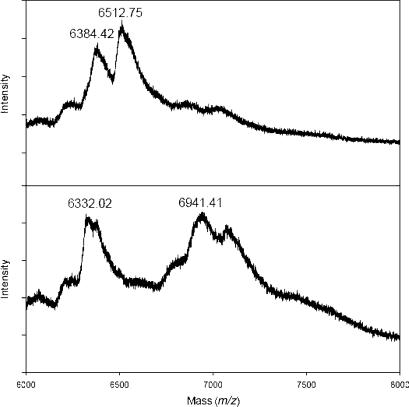
Since the goal is the incorporation of the boronic acid moiety into DNA, primer extension using B-TTP (12) was conducted using a short sequence of 21-nt template (5′-GGTTCCACCAGCAACCCGCTA-3′, molecular weight = 6336.2 Da) as the template and a 14-nt primer (5′-TAGCGGGTTGCTGG-3′, molecular weight = 4350.8 Da). The primer extension reaction using natural TTP yielded a DNA product with molecular weight of about 6512 a as determined using MALDI mass spectrometry (calculated molecular weight: 6518.2 a) (Figure 1). The same reaction using B-TTP yielded a DNA product with a molecular weight of about 6941 Da (calculated molecular weight: 6930.2 Da). Such results clearly demonstrated the successful incorporation of the boronic acid-labeled thymidine unit.
Figure 1.
The MALDI-TOF mass spectrometric analysis of primer extension products on a 21-nt template using TTP (top, showing a mixture of template and TTP product) and B-TTP (bottom: showing a mixture of template and B-TTP product). The mass difference of 418.6 reflects the incorporation of the boronic acid-labeled thymidine moiety.
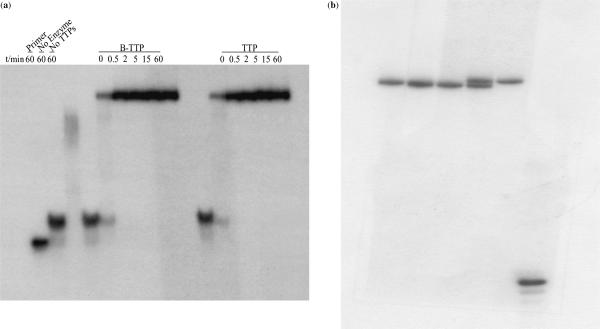
More detailed examination of the B-TTP incorporation was conducted through the extension of a 21-nt primer on a 55-nt template. This longer template has three As in the sequence allowing for the incorporation of three Ts or labeled Ts. We first studied the time-dependent incorporation of B-TTP compared with natural TTP using a 32P-labeled primer. Gel electrophoresis results showed that the full-length DNA was obtained from primer extension reactions (Figure 2a) that was confirmed by mass spectrometry. Furthermore, there was no noticeable difference in the rate of incorporation of natural TTP and B-TTP (12). For example, at 0.5 min, neither the B-TTP nor the natural TTP was incorporated significantly. From time 0 to 15 min, there was time-dependent incorporation in both cases. At 15 min, both reactions reached maximal incorporation. Control reactions with only the primer, without enzyme and without added TTP or labeled TTP showed no full-length DNA formation. (The smear in the third lane without TTP was from mismatch pairing and incomplete reaction.) All these indicate that the boronic acid-labeled base, B-TTP (12), was recognized by the Klenow fragment at approximately the same level as natural TTP. It is interesting to note that the B-TTP DNA and TTP-DNA were not well separated when using 19% acrylamide gel (Figure 2a), but were well resolved when using 15% acrylamide gel (Figure 2b).
Figure 2.
(a) Time-dependent primer extension using B-TTP and TTP: reaction was performed with 5 µM 5′5′-32P-primer/template, 0.4 mM of each dNTPs, 0.4 mM of B-TTP, and Klenow 0.04 units. Electrophoresis was conducted on 19% acrylamide gel. (b) Primer extension using B-TTP analyzed on 15% acrylamide gel: reaction was performed with 5 µM 5′5′-32P-primer/template, 0.4 mM of each dNTPs, 0.4 mM of B-TTP, and Klenow 0.04 units. From left to right, lane 1 (from left): M-TTP-DNA; lane 2: co-spot of M-TTP-DNA and TTP-DNA; lane 3: TTP-DNA; lane 4: co-spot of B-TTP-DNA, and TTP-DNA; lane 5 B-TTP-DNA, lane 6: primer.
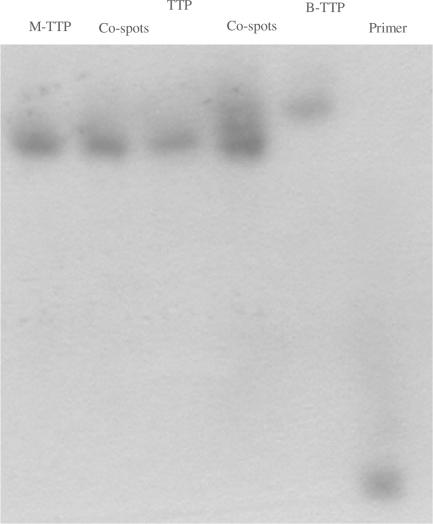
In order to allow for quick confirmation of boronic acid incorporation into DNA using electrophoresis, we have also developed a gel-shift method by using a low percentage of acrylamide (1%) modified with catechol, that was synthesized as shown in Scheme 3. Because catechol is known to form a tight complex with the boronic acid moiety (43–45), such gels are expected to exert extra retention power for boronic-acid-containing DNA and therefore allow for their separation from natural DNA of the same length and composition. Figure 3 shows the successful application of such a catechol-embedded acrylamide gel and its ability to differentiate the boronic acid-labeled DNA from that of the natural one. Specifically, when analyzed on the catechol-modified acrylamide gel, only the natural and non-boronic-acid-modified DNA (using M-TTP, 11) showed the same retention. The DNA labeled with boronic acid through the incorporation of B-TTP (12) moved more slowly compared with the other two as expected based on the known interaction between boronic acid and catechol (43–45). This was confirmed by co-loading these two different samples on the same lane (Figure 3).
Figure 3.
Gel-shifting experiments of full-length natural and boronic acid-labeled DNA using catechol-modified acrylamide gel: reaction was performed with 5 µM 5′-32P-primer/template, 0.4 mM of each dNTPs, 0.4 mM of B-TTP and Klenow 0.04 units for 1 h; the acrylamide gel was prepared with 19% acrylamide and 1% N-[2-(3,4-dihydroxyphenyl)-ethyl]-acrylamide co-loaded lane 1. From left to right: lane 1, M-TTP-derived DNA; lane 3, TTP-derived DNA; lane 5, B-TTP-derived DNA; lane 6, primer; lane 2, M-TTP and TTP-derived DNA co-loaded; lane 4, TTP and B-TTP-derived DNA co-loaded.
With the demonstration of incorporation of the boronic acid-labeled TTP, next it was important to examine whether the boronic acid-labeled DNA could serve as a template for further polymerization and amplification.
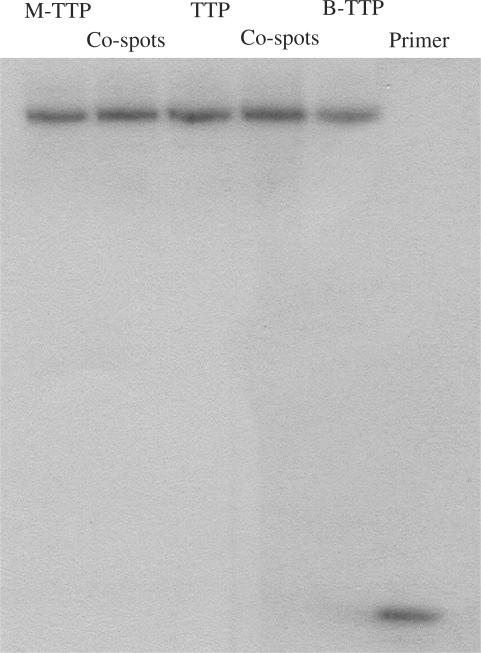
To demonstrate the recognition of boronic acid-labeled full-length DNA as templates by the Klenow fragment, two 20-nt primers for the 5′- and 3′-ends, respectively, were synthesized (Figure 4). The polymerase reactions using the boronic acid-labeled DNA as the template and with M-TTP (11), TTP or B-TTP (12) and the other three dNTPs as the monomers were carried out first with primer 1, which is complementary to the 3′-terminus of the full-length DNA. After primer extension, the full-length DNA obtained was purified by membrane filtration to remove the labeled and non-labeled dNTPs. Then further polymerization was conducted using natural dNTPs and 5′-32P-labeled primer 2, which is complementary to the 3′-terminus of the polymerized full-length DNA templates. Gel electrophoresis of the DNA products shows no noticeable differences between the experiments using natural and labeled full-length DNA as templates, indicating that all full-length DNA templates generated using M-TTP, TTP and B-TTP in the first primer extension were recognized with a similar efficiency by the polymerase (Figure 4). Such results are very critical for future work.
Figure 4.
Primer extension using the full-length DNA and boronic acid-labeled DNA as templates. Reaction was performed with 5 µM primer 1/template, 0.4 mM of each dNTPs, 0.4 mM of labeled-TTP (M-TTP and B-TTP), and Klenow 0.04 units for 1 h. After centrifugation–filtration, the reaction was performed with radio-labeled 5′-32P-primer 2 and 0.4 mM of each dNTPs. Co-spot 1: polymerization using M-TTP and TTP-derived DNA as templates, Co-spot 2: polymerization using B-TTP and TTP-derived DNA as templates. Primer 1: 5′-GCGTAATACGACTCACTATA-3′; Template DNA: 3′CGCATTATGCTGAGTGATATCCGTTGGACTACTCCGGCTT TCCGGCTTTGCATGT-5′; Primer 2: 5′-TGTACGTTTCGGCCTTTCGG-3′.
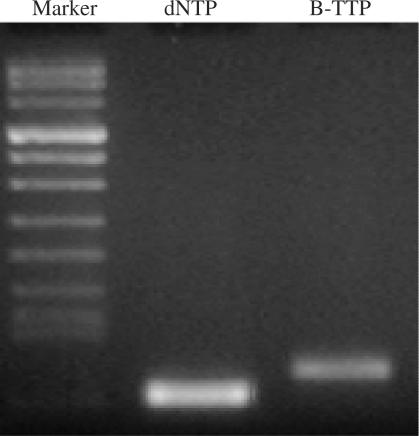
In order to confirm the general feasibility of incorporating the boronic acid-labeled TTP (B-TTP) into DNA, we have also carried out similar studies using a different template. The results again demonstrated the synthesis of the full-length DNA using B-TTP. Furthermore, using an agarose gel run for a longer time, the boronic acid-labeled DNA was differentiated from the natural DNA that is consistent with the increased molecular weight of the boronic acid-labeled DNA (Figure 5). It should be noted that the boronic acid has a pKa of about 9 (46) and is mostly charge neutral under the electrophoresis conditions (buffer pH 8.3).
Figure 5.
Primer extension using the full-length DNA and boronic acid-labeled DNA as templates. Each 50 µl reaction was performed with 1.2 µM primers 1 and 2/template, 0.25 mM of each dNTPs, 0.25 mM of labeled-TTP (B-TTP), and 3.5 units of high-fidelity DNA polymerase (Roche, Indianapolis, Ind.) under conditions of 1 cycle at 94°C for 2 min, 30 cycles at 94°C for 20 s, 59°C for 30 s, 72°C for 1 min and 1 cycle at 72°C for 7 min. Lane 1: Marker; lane 2: DNA synthesized using dNTPs; lane 3: DNA synthesized using B-TTP and the other three dNTPs. Primer 1: 5′-CGCCGCCCCCGCCGCG-3′ Primer 2: 5′-CGGCGGCCCGCGGGCG; Template DNA: 5′-CGCCGCCCCCGCCGCG-40N-CGCCCGCGGGCCGCCG-3′.
CONCLUSIONS
A modified TTP nucleotide containing a boronic acid functional group was synthesized using copper (I)-catalyzed Huisgen cycloaddition. This boronic acid-labeled nucleoside triphosphate can be incorporated into DNA with a similar efficiency as the natural TTP. Further DNA template efficiency studies indicated that the DNA sequences containing labeled nucleotides served as templates very well. PCR amplification of the DNA with the boronic acid-labeled TTP has also been achieved, indicating that B-TTP is stable under the elevated temperature. Such studies lay the foundation for future development of DNA-based boronolectins.
SUPPLEMENTARY DATA
NMR and MS spectra for compound 6–12 are available as Supplementary Data at NAR online.
ACKNOWLEDGEMENTs
Financial support from the National Institutes of Health (CA113917, CA123329, CA88343, NO1-CO-27184 and AI058051), the Georgia Cancer Coalition through a Distinguished Cancer Scientist Award and the Georgia Research Alliance through an Eminent Scholar endowment and an Eminent Scholar Challenge grant is gratefully acknowledged. Funding to pay the Open Access publication charge was provided by The National Institutes of Health.
Conflict of interest statement: None declared.
REFERENCES
- 1.Yang W, Gao X, Wang B. Boronic acid compounds as potential pharmaceutical agents. Med. Res. Rev. 2003;23:346–368. doi: 10.1002/med.10043. [DOI] [PubMed] [Google Scholar]
- 2.Yang W, Gao S, Wang B. In: Organoboronic Acids. Hall D, editor. New York: John Wiley and Sons; 2005. pp. 481–512. [Google Scholar]
- 3.Yan J, Fang H, Wang B. Boronolectins and fluorescent boronolectins-an examination of the detailed chemistry issues important for their design. Med. Res. Rev. 2005;25:490–520. doi: 10.1002/med.20038. [DOI] [PubMed] [Google Scholar]
- 4.Wang W, Gao X, Wang B. Boronic acid-based sensors for carbohydrates. Curr. Org. Chem. 2002;6:1285–1317. [Google Scholar]
- 5.James TD, Sandanayake KRAS, Shinkai S. Saccharide sensing with molecular receptors based on boronic acid. Angew. Chem. Int. Ed. Engl. 1996;35:1910–1922. [Google Scholar]
- 6.Davis CJ, Lewis PT, McCarroll ME, Read MW, Cueto R, Strongin RM. Simple and rapid visual sensing of saccharides. Org. Lett. 1999;1:331–334. doi: 10.1021/ol990105a. [DOI] [PubMed] [Google Scholar]
- 7.Manimala JC, Wiskur SL, Ellington AD, Anslyn EV. Tuning the specificity of a synthetic receptor using a selected nucleic acid receptor. J. Am. Chem. Soc. 2004;126:16515–16519. doi: 10.1021/ja0478476. [DOI] [PubMed] [Google Scholar]
- 8.Cao H, McGill T, Heagy MD. Substituent effects on monoboronic acid sensors for saccharides based on N-Phenyl-1,8-Naphthalenedicarboximides. J. Org. Chem. 2004;69:2959–2966. doi: 10.1021/jo035760h. [DOI] [PubMed] [Google Scholar]
- 9.Badugu R, Lakowicz JR, Geddes CD. Fluorescence sensors for monosaccharides based on the 6-methylquinolinium nucleus and boronic acid moiety: potential application to ophthalmic diagnostics. Talanta. 2005;65:762–768. doi: 10.1016/j.talanta.2004.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexeev VL, Sharma AC, Goponenko AV, Das S, Lednev IK, Wilcox CS, Finegold DN, Asher SA. High ionic strength glucose-sensing photonic crystal. Anal. Chem. 2004;75:2316–2323. doi: 10.1021/ac030021m. [DOI] [PubMed] [Google Scholar]
- 11.Dowlut M, Hall DG. An improved class of sugar-binding boronic acids, soluble and capable of complexing glycosides in neutral water. J. Am. Chem. Soc. 2006;128:4226–4227. doi: 10.1021/ja057798c. [DOI] [PubMed] [Google Scholar]
- 12.Yoon J, Czarnik AW. Fluorescent chemosensors of carbohydrates. A means of chemically communicating the binding of polyols in water based on chelation-enhanced quenching. J. Am. Chem. Soc. 1992;114:5874–5875. [Google Scholar]
- 13.Adams J, Kauffman M. Development of the Proteasome Inhihitor Veleade(TM) (Bortezomib) Can. Invest. 2004;22:304–311. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- 14.Kabalka GW, Smith GT, Dyke JP, Reid WS, Desmond-Longford CP, Roberts TG, Reddy NK, Buonocore E, Hübner KF. Evaluation of Fluorine-18-BPA-Fructose for Boron Neutron Capture Treatment Planning. J. Nuclear Med. 1997;38:1762–1767. [PubMed] [Google Scholar]
- 15.Smith BD. Sugar separation using liquid membranes and boronic acid carriers. Chem. Sep. Liq. Memb. 1996;642:194–205. [Google Scholar]
- 16.Valigra L. First aptamer-based drugs take on tough diseases-tools to identify cell signaling mechanisms yield better targets and drug leads with high binding affinity and specificity for previously untargeted diseases. Drug Discovery Dev. 2005;8:61–64. [Google Scholar]
- 17.Eaton BE, Gold L, Hicke BJ, Janjic N, Jucker FM, Sebesta DP, Tarasow TM, Willis MC, Zichi D. Post-SELEX combinatorial optimization of aptamers. Bioorg. Med. Chem. 1997;5:1087. doi: 10.1016/s0968-0896(97)00044-8. [DOI] [PubMed] [Google Scholar]
- 18.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 19.Ellington AD, Szostak JW. In Vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 20.Lato SM, Ozerova NDS, He KZ, Sergueeva Z, Shaw BR, Burke DH. Boron-containing aptamers to ATP. Nucleic Acids Res. 2002;30:1401–1407. doi: 10.1093/nar/30.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw BR, Dobrikov M, Wang X, Wan J, He KZ, Lin JL, Li P, Rait V, Sergueeva ZA, et al. Reading, writing, and modulating genetic information with boranophosphate mimics of nucleotides, DNA, and RNA. Therap. Oligonucl. Ann. NY Acad. Sci. 2003;1002:12–29. doi: 10.1196/annals.1281.004. [DOI] [PubMed] [Google Scholar]
- 22.Eaton BE, Gold L, Hicke BJ, Janjic N, Jucker FM, Sebesta DP, Tarasow TM, Willis MC, Zichi DA. Post-SELEX combinatorial optimization of aptamers. Bioorg. Med. Chem. 1997;5:1087–1096. doi: 10.1016/s0968-0896(97)00044-8. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z, Szostak JW. Evolution of aptamers with a new specificity and new secondary structures from an ATP Aptamer. RNA. 2003;9:1456–1463. doi: 10.1261/rna.5990203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 25.Eaton BE. The joys of in vitro selection: chemically dressing oligonucleotides to satiate protein targets. Curr. Opin. Chem. Biol. 1997;1:10–16. doi: 10.1016/s1367-5931(97)80103-2. [DOI] [PubMed] [Google Scholar]
- 26.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 27.Shimkus M, Levy J, Herman T. A chemically cleavable biotinylated nucleotide: usefulness in the recovery of protein–DNA complexes from avidin affinity columns. Proc. Natl. Acad. Sci. U.S.A. 1985;82:2593–2597. doi: 10.1073/pnas.82.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rye HS, Yue S, Wemmer DE, Quesada MA, Haughland RP, Mathies RA, Glazer AN. Stable fluorescent complexes of double-stranded DNA with bis-intercalating asymmetric cyanine dyes: properties and applications. Nucleic Acids Res. 1992;20:2803–2812. doi: 10.1093/nar/20.11.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pohl NL. Array methodology singles out pathogenic bacteria. Nature Chem. Biol. 2006;2:125–126. doi: 10.1038/nchembio0306-125. [DOI] [PubMed] [Google Scholar]
- 30.Zheng T, Peelen D, Smith LM. Lectin arrays for profiling cell surface carbohydrate expression. J. Am. Chem. Soc. 2005;127:9982–9983. doi: 10.1021/ja0505550. [DOI] [PubMed] [Google Scholar]
- 31.Mahal LK, Pilobello K, Krishnamoorthy L. Development of a lectin microarray for the glycomic profiling of cells. Glycobiology. 2004;14:1203. [Google Scholar]
- 32.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem. 2005;6:985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 33.Hsu K-L, Pilobello KT, Mahal LK. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nature Chem. Biol. 2006;2:153–157. doi: 10.1038/nchembio767. [DOI] [PubMed] [Google Scholar]
- 34.Lee SE, Vyle JS, Williams DM, Grasby TA. Novel synthesis of (Z)-Alkene and Alkane base-modified nucleosides. Tetrahedron Lett. 2000;41:267–270. [Google Scholar]
- 35.Huisgen R. In: 1,3-Dipolar Cycloaddition Chemistry. Padwa A, editor. New York: John Wiley; 1984. pp. 1–176. [Google Scholar]
- 36.Leir CM. An improvement in the Doebner-Miller synthesis of quinaldines. J. Org. Chem. 1977;42:911–913. [Google Scholar]
- 37.Matsugi M, Tabusa F, Minamikawa J-I. Doebner-Miller synthesis in a two-phase system: practical prepration of Quinolines. Tetrahedron Lett. 2000;41:8523–8525. [Google Scholar]
- 38.Cohn EW. A modification of the skraup synthesis of Quinoline. J. Am. Chem. Soc. 1930;52:3685–3688. [Google Scholar]
- 39.McNaughton BR, Miller BL. A mild and efficient one-step synthesis of Quinolines. Org. Lett. 2003;5:4257–4259. doi: 10.1021/ol035333q. [DOI] [PubMed] [Google Scholar]
- 40.Lee SE, Sidorov A, Grasby JA, Williams DM. Enhancing the Catalytic Repertoire of Nucleic Acids: A Systematic Study of Linker Length and Rigidity. Nucleic Acids Res. 2001;29:1565–1573. doi: 10.1093/nar/29.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hobbs FW. Palladium-catalyzed synthesis of Alkynylamino Nucleosides. A universal linker for nucleic acids. J. Org. Chem. 1989;34:3420–3422. [Google Scholar]
- 42.Sakthivel K, Barbras CF. Expanding the potential of DNA for binding and catalysis: Highly functionalized dUTP derivatives that are substrates for thermostable DNA polymerases. Angew. Chem. Int. Ed. 1998;37:2872–2875. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2872::AID-ANIE2872>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 43.Springsteen G, Wang B. A detailed examination of boronic acid-diol complexation. Tetrahedron. 2002;58:5291–5300. [Google Scholar]
- 44.Yan J, Springsteen G, Deeter S, Wang B. The relationship among pKa, pH, and Binding constants in the interactions between Boronic Acids and Diols – It is not as simple as it appears. Tetrahedron. 2004;60:11205–11209. [Google Scholar]
- 45.Yang W, Gao X, Springsteen G, Wang B. Catechol pendant polystyrene for solid phase synthesis. Tetrahedron Lett. 2002;43:6339–6342. [Google Scholar]
- 46.Yang W, Springsteen G, Yan J, Deeter S, Wang B. A novel type of fluorescent boronic acid that shows large fluorescence intensity changes upon binding with a diol in aqueous solution at physiological pH. Bioorg. Med. Chem. Lett. 2003;13:1019–1022. doi: 10.1016/s0960-894x(03)00086-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.