Abstract
Plasmids replicating by the rolling circle mode usually possess a single site for binding of the initiator protein at the origin of replication. The origin of pMV158 is different in that it possesses two distant binding regions for the initiator RepB. One region was located close to the site where RepB introduces the replication-initiating nick, within the nic locus; the other, the bind locus, is 84 bp downstream from the nick site. Binding of RepB to the bind locus was of higher affinity and stability than to the nic locus. Contacts of RepB with the bind and nic loci were determined through high-resolution footprinting. Upon binding of RepB, the DNA of the bind locus follows a winding path in its contact with the protein, resulting in local distortion and bending of the double-helix. On supercoiled DNA, simultaneous interaction of RepB with both loci favoured extrusion of the hairpin structure harbouring the nick site while causing a strong DNA distortion around the bind locus. This suggests interplay between the two RepB binding sites, which could facilitate loading of the initiator protein to the nic locus and the acquisition of the appropriate configuration of the supercoiled DNA substrate.
INTRODUCTION
Replication of bacterial plasmids is generally initiated by a plasmid-encoded protein, termed Rep, which interacts with its cognate DNA at a specific region, the origin of replication. Binding of the Rep initiator to the origin is followed by recruitment of host-encoded proteins and generation of the replisome nucleoprotein complex. Settling of the initiation complex leads to opening of the DNA strands and loading of the replication machinery which performs DNA polymerase-mediated DNA synthesis from a newly acquired OH group (1). There are several mechanisms by which a free OH group is provided to the polymerases, one of them being the rolling-circle mode of replication (RCR), which is widespread among bacterial plasmids, single-stranded (ss) DNA coliphages and geminiviruses. Plasmids that use this mechanism for their replication are termed RCR plasmids, and they are found in bacteria, archaea and mitochondria (2,3). The number of sequenced RCR plasmids has been steadily increasing in the past years from around 20 (2) to nearly 300, making it now possible to develop accurate phylogenetic trees and to show that these plasmids are constructed by gene cassettes. According to homologies in their Rep proteins and in their double-strand origins of replication (dso), RCR plasmids have been grouped into several families (4). However, only a few of them have been studied in depth, namely the staphylococcal plasmids pT181/pC221 [(5) and references therein; (6)] and pC194/pUB110 (7), and the promiscuous plasmid pMV158 (1), originally isolated from Streptococcus agalactiae. These plasmids are now considered as the prototypes of three out of the seventeen families of RCR plasmids.
Initiation of RCR requires recognition of the plasmid dso by the cognate Rep protein. Upon binding to the DNA, Rep cleaves the phosphodiester bond of a specific di-nucleotide located in an unpaired region of the dso, leaving a free 3′-OH end that acts as the primer for the initiation of replication (1,2). Host-encoded proteins that participate in the initiation and elongation stages are, at least, a DNA helicase [the PcrA protein in gram-positive bacteria (8)], an ssDNA-binding protein, and DNA polymerases (9). The Rep-mediated nucleolytic cleavage is exerted by the OH group of a specific Tyr residue, so that the nature of the recognition and the reaction demands that the DNA substrate must be in an unpaired configuration. In addition to nicking the DNA, the Rep proteins are able to close the cleaved substrate, a reaction that takes place at the termination of replication stage (10,11).
The dso can be physically and functionally separated into two loci, termed bind (the binding region of the Rep protein) and nic (where Rep cleaves specifically at the nick site) [(12); see Figure 1]. The bind locus is replicon-specific and there is little or no cross-interaction between this region and heterologous Rep proteins belonging to plasmids of the same family. This is not the case for the nic region, whose nick sequence is conserved among plasmids of the same family (2,13). Thus, the Rep proteins encoded by different plasmids of the same family can perform in vitro the nicking-closing reaction on the dsos of all the plasmids belonging to the same family (13–16). The nic and the bind loci can be contiguous, as in the case of the plasmids of the pT181 family (17), or separated by up to 100 bp, as in the case of plasmids of the pMV158 family (18). Furthermore, whereas the bind locus is constituted by an inverted repeat (IRIII) in plasmids of the pT181 family, in the case of the pMV158 family it was reported to consist of two or three direct repeats (DR) whose length varied from 5 to 21 bp (12). In pMV158, three tandem 11-bp DR located 84 bp downstream from the nick site constitute the region (bind locus) where RepB binds with high affinity (18). RepB cleaves supercoiled DNA (but not linear duplex, ds, DNA) containing either the cognate or a related nic locus, as well as ss oligonucleotides harbouring the 9-mer nick sequence [5′-TACTACG/AC-3′, where / represents the nick site; (12); (15)]. The bind locus is not required for in vitro RepB-mediated cleavage of these ss or ds DNAs. These findings indicated that: (i) the reaction requires that the phosphodiester bond to be cleaved is on unpaired DNA; and (ii) on supercoiled DNA, a hairpin structure containing the nick sequence on its loop can be formed from an inverted repeat sequence (IR-I), as proven by sensitivity to endonuclease S1 (19) (Figure 1A). As yet unanswered questions on the in vivo function of the dso of pMV158 (and of other members of this family) are the role of a bind locus so distant from the nick site, and how binding of RepB to this region promotes initiation of plasmid replication (18,20).
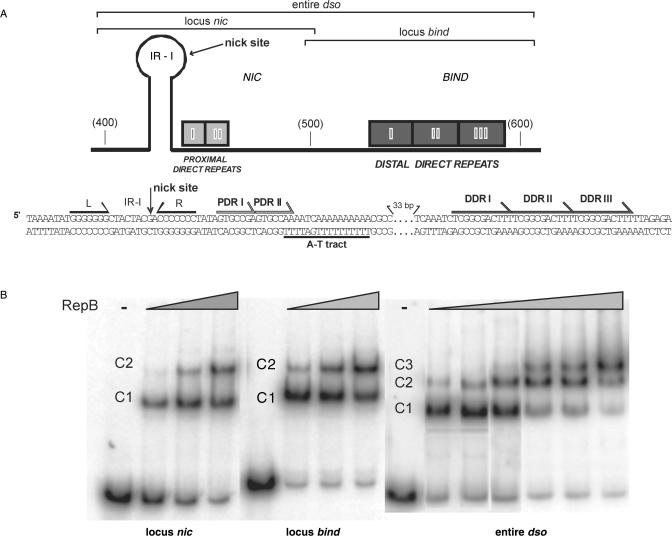
Figure 1.
The pMV158-dso and its interactions with RepB. (A) Schematic representation (top) and nucleotide sequence (bottom) of the pMV158-dso. This region is divided into two loci, a bind locus containing the DDR, and a nic locus harbouring the PDR (proximal direct repeats) and the IR-I element (L and R, left and right arms respectively) that generates the hairpin where the 5′-GpA-3′ dinucleotide cleaved by RepB would be placed in a ssDNA configuration. The three DNA fragments used in B are represented above the scheme. As a reference, some coordinates of the pMV158 sequence are indicated in parenthesis. The two repeats (I and II) constituting the PDR, and the three repeats (I, II and III) that compose the DDR are displayed. The A-T tract located between the PDR and the DDR is shown below the sequence. (B) Complexes between RepB and its DNA target. The protein was incubated with labelled DNA fragments containing the nic locus, the bind locus or the entire dso. RepB concentrations were 0 (-), 0.3, 0.4 and 0.5 µM for the reactions containing the nic or bind locus, and 0 (-), 0.3, 0.5, 0.7, 1.2, 1.5 and 1.7 µM for the entire-dso fragment. The different complexes are indicated.
Here we report that the dso of pMV158 contains not one but two regions for the specific binding of RepB: (i) the three 11-bp DR (hereafter termed distal direct repeats, DDR) previously reported to constitute the bind locus, and (ii) a region around the nick site, within the nic locus. Characterization of the relative affinity of the initiator protein for the bind and nic loci allowed us to show that the DDR constitute the primary binding site, whereas weak binding of RepB to the nic locus could be involved in recognition of the nick site during initiation of replication. Binding of RepB to the dso induces DNA bending, and electron microscopy (EM) of protein–DNA complexes indicates that the DNA winds through RepB. We propose a model in which, upon binding to its primary target, RepB could help to load new protein molecules in the catalytic binding site within the nic locus, a fact that would promote in turn the melting of the substrate nick sequence.
MATERIALS AND METHODS
Bacterial strains and plasmids
Escherichia coli BL21(DE3) (r−B m−B, gal, ompT, int::PlacUV5-T7 gene 1 imm 21 nin5; a gift of F.W. Studier) was used as the host for purification of RepB. Plasmid pGEMrepB1, containing repB under the control of promoter ϕ10 of phage T7 (18,21), was employed.
DNA substrates
To obtain PCR-amplified DNA fragments for electrophoretic mobility shift assays (EMSA), the following oligonucleotides (oligos) were used. Their coordinates on the pMV158 sequence (accession number X15669) are given in parenthesis:
Oligo 1: (dso1) 5′-AGGGAGATGTTGTGGGGGAT-3′ (388-407)
Oligo 2: (dso2) 5′-CAGCTCTAAGGCTAAAGGCG-3′ (508-489)
Oligo 3: (dso3) 5′-CCTTAGAGCTGCAAGGGTTT-3′ (498-517)
Oligo 4: (dso4) 5′-GCATAACCGTGCACTCAATG-3′ (620-601)
Oligo 5: (DA) 5′-GATAACCCCATCTCTCTTGCC-3′ (734-714)
Oligo 1R (A): (1 repeat) 5′-AATCTCGGCGACTTTTTAGA-3′
Oligo 1R (B): (1 repeat) 5′- TCTAAAAAGTCGCCGAGATT-3′
The four DNA fragments used, namely entire dso (233 bp), nic locus (121 bp), bind locus (123 bp), and DA (347 bp), were amplified by PCR using oligos 1-4, 1-2, 3-4 or 1-5, respectively.
Amplification was done during 30 cycles using the DyNAZyME DNA polymerase (FINZYMES) and, as template, plasmid pLS1 DNA (22). For hydroxyl radical (HO•) or dimethyl sulphate (DMS) footprinting, synthesis of the DNA fragments by PCR was carried out after 5′-end labelling of one of the primers with [γ-32P]ATP and polynucleotide kinase (23). Fragment DA was internally labelled with [α -32P]dCTP during the PCR. It was then sequentially digested with HgaI and Fnu4HI, so that three fragments of 153, 119 and 75 bp, containing, respectively, the bind locus, the nic locus, and unspecific DNA, were generated and used in the relative affinity assays. They were separated on a native 5% polyacrylamide (PAA) gel and purified. DNA concentrations were calculated by counting the radioactivity incorporated in each fragment and correcting for their G+C content. A duplex oligonucleotide containing 1 repeat of the DDR was generated by annealing complementary oligos 1R(A) and 1R(B) (the repeat unit sequence is underlined). Double-stranded oligos with 2 or 3 DDR were generated with complementary oligos harbouring 2 or 3 repeat units in tandem and the same end sequences (non-underlined sequences) as oligos 1R(A) and 1R(B).
For DNA bending analysis, derivatives of plasmid pBEND2 (24) were constructed by inserting blunt-ended DNA fragments (carrying the bind or the nic locus), generated by PCR with the Pfu DNA polymerase, into the SalI site of the vector. The constructions were validated by sequencing of the cloned DNA. Nine different permutated DNA fragments were generated and purified from each of the two pBEND2 derivatives by digestion with BglII, XhoI, EcoRV, PvuII, SmaI, StuI, SspI, RsaI and BamHI. All restriction enzymes were purchased from New England Biolabs.
To prepare the linear DNA substrates for EM, DNA from pMV158 was digested with BstXI and StuI (which yields a 1825-bp DNA fragment containing the entire dso) or with BstXI and BanI (which results in a 700-bp DNA fragment also including the entire dso), and the desired fragments were gel purified.
Purification of RepB and EMSA
Protein RepB was purified as described (18,21) and stored at −80°C. Protein concentrations given in the text refer to protomers.
Purified RepB was mixed, at the indicated concentrations, with 32P-labelled DNA (0.5 nM) and 10 ng/µl of poly(dI-dC) in 10 µl of buffer B (20 mM Tris-HCl, pH 8.0, 1 mM EDTA, 5 mM DTT, 300 mM KCl). Reaction mixtures were incubated at 25°C for 30 min. Free and bound DNAs were separated by electrophoresis on native 5% PAA gels. Labelled DNA bands were detected by autoradiography and quantified with the storage phosphor technology, with the aid of a FLA-3000 (FUJIFILM) imaging system and the QuantityOne software (Bio-Rad).
RepB-DNA complexes stability assays
Dissociation experiments were performed by equilibrating RepB and 5′-end labelled DNA fragments (carrying the nic or the bind locus) under conditions in which the fraction of complex C1 was ∼0.8 and C2 was unpopulated, and at t = 0, adding a 300-fold molar excess of the respective unlabelled nic/bind DNA. Samples were taken at intervals and applied to running 5% native PAA gels. To test the competition conditions, RepB was added to a previously prepared mixture of unlabelled and labelled (molar ratio 300:1) specific DNA, and incubated in the same conditions. From these controls we calculated the background values, which were subtracted from the competition reactions. Labelled DNA bands were directly quantified as above. Data were analysed according to the integrated rate equation for a first-order process:
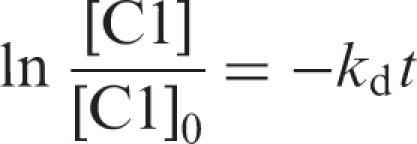 |
1 |
where [C1] represents the concentration of RepB-bind/nic complexes at time t and [C1]0 is its value at t = 0. Values of kd and half-life (given by ln2/kd) were estimated from the least-squares analysis of data.
Relative affinity assays
Competitive EMSA were used to determine the relative binding affinities of RepB for different DNA sequences. Various distinguishable-sized labelled DNA fragments were incubated simultaneously (at the indicated molar ratios) with increasing amounts of RepB protein. To identify correctly the different RepB–DNA complexes, binding reactions were also prepared for each separate fragment and loaded on the same gel. Labelled DNA bands were detected and quantified as above. The relative affinity of the protein for a pair of DNAs (D1 and D2) was determined as follows (25):
| 2 |
where Krel is the relative equilibrium binding constant of the protein to D1 with respect to D2. [P-D1] and [D1] are, respectively, the concentrations of bound and unbound D1. [P-D2] and [D2] are the concentrations of bound and unbound D2.
DNA bending assays
Purified permuted DNAs (10 nM) were equilibrated with RepB (protein to DNA molar ratio of 43:1) in 10 µl of buffer B. Samples were loaded on native 6% PAA gels (26). After electrophoresis, the gels were stained with ethidium bromide and the DNA bands were visualized with the aid of a Gel-Doc documentation system (Bio-Rad). Bending angles were determined by interpolating the relative mobility data with the standard curve built by Thompson and Landy (1988) (26).
Transmission EM
RepB–DNA complexes were formed in 4 µl of buffer B containing 3.6 nM of the 1825-bp DNA or 7 nM of the 700-pb DNA, at a protein to DNA molar ratio of 115:1 or 60:1, respectively. Following incubation for 10 min at 25°C, the complexes were fixed with 0.2% (v/v) glutaraldehyde for 10 min and, after a 20-fold dilution in 10 mM triethanolamine chloride, pH 7.5, and 10 mM MgCl2, adsorbed on to freshly cleaved mica, positively stained in 2% uranyl acetate, rotary shadowed with Pt/Ir, and covered with a carbon film as described (27). Micrographs of the carbon film replica were taken using a Philips CM100 (FEI Company, Hillsboro, Oregon) electron microscope at 100 kV and a Fastscan CCD camera (Tietz Video and Image Processing Systems GmbH, Gauting, Germany). To determine the contour length and to locate the RepB binding site, measurements were carried out on projections of 35-mm negatives using a digitizer (LM4, Brühl, Nuremberg, Germany).
Footprinting experiments on linear DNA
DNA fragments spanning the entire dso, the nic locus or the bind locus were 5′-end labelled, and 0.5 nM of each fragment was mixed with RepB (200 nM) in buffer B supplemented with 10 ng/µl of poly(dI-dC). For HO• probing, binding mixtures (50 µl) were incubated 30 min at 25°C, and footprinting reactions were initiated by the addition of 9 µl of the reactive mixture [Fe(II)-EDTA, H2O2 and sodium ascorbate] prepared as described (28). After 5 min of incubation at 25°C, reactions were stopped by addition of 15 µl of a solution containing 0.1 M thiourea and 20 mM EDTA. Bound and free DNAs were separated on native 5% PAA gels, and the purified DNAs were then applied to 8% PAA sequencing gels and run together with the sequencing chemical reactions (29) of the same fragment.
DMS footprinting was performed as reported (30,31). Binding reactions were prepared as above and electrophoresed in native 5% PAA gels. Free and bound DNAs were modified ‘in situ’ by soaking the gel in a 0.2% (v/v) DMS solution. Methylation reaction was allowed to proceed for 5 min at 25°C and stopped with a solution containing 0.5 M β-mercaptoethanol. The DNAs were eluted from the gel matrix, subjected to two sequential organic extractions and recovered by ethanol precipitation. The DNA pellets were dissolved in 50 µl of distilled water and digested with 10 M piperidine at 100°C. Digested DNAs were then applied to a sequencing gel as above.
Footprinting experiments on supercoiled plasmid DNA
DMS and KMnO4 footprinting assays were performed essentially as described (31). Binding reactions were prepared in 45 µl of buffer B, and RepB (0.18-4.8 µM) was allowed to bind to supercoiled DNA of plasmids pMV158 or pCGA7 (2 µg) at 25°C for 25 min. DMS reactions were started by adding 5 µl of the reactive solution (300 mM DMS in 10 mM Tris-HCl, pH 8). After 5 min at 25°C, reactions were stopped by adding 100 µl of stop solution (1 M β-mercaptoethanol, 3 M ammonium acetate, 20 mM EDTA), and DMS was carefully eliminated by ethanol precipitation. Methylated nucleotides were mapped by primer extension using T7 DNA polymerase and the dso1 primer labelled in its 5′-end with [γ-32P]ATP and polynucleotide kinase. About 300 fmol of modified supercoiled plasmid DNA (1 µg) was denatured with 0.2 M NaOH, neutralized, annealed with 1.5 pmol of the 5′-end-labelled primer, and incubated at 37°C in 10 µl containing 1.5 units of T7 DNA polymerase (USB), 0.2 mM of each dNTP, 50 mM Tris-HCl, pH 7.5, 45 mM NaCl, 10 mM MgCl2, and 16 mM DTT. Reactions were stopped by adding 7 µl of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). KMnO4 reactions were started by adding 5 µl of 100 mM KMnO4 (final concentration 10 mM). After exactly 2 min at 25°C, reactions were stopped by adding 50 µl of stop solution (3 M β-mercaptoethanol, 40 mM EDTA, 0.6 M sodium acetate, pH 4.8), and the DNA was ethanol-precipitated. Modified nucleotides were mapped by primer extension as described above.
RESULTS
Binding of the initiator protein to the bind and nic loci
RepB was obtained by a new method which yields high concentrations of pure protein (21). Analytical ultracentrifugation analyses of this protein preparation (not shown) revealed that RepB purified as a hexamer of identical subunits. It has been shown that RepB interacts with two distant DNA regions in the pMV158 dso (Figure 1A): it binds specifically to the bind locus and it initiates replication at the nick sequence within the nic locus (18,20). In spite of this dual interaction, only the complexes between RepB and the bind locus have been characterized regarding the contacts of the protein with the DNA backbone (18). To test whether RepB can also bind to the nic locus and to visualize the complexes that might result from the binding of the protein to different regions of the dso, EMSA were performed with DNA fragments harbouring the bind locus, the nic locus, or the entire dso, at different protein to DNA molar ratios (Figure 1B). Two retarded bands (C1 and C2) were observable upon binding of the protein to either the nic or the bind locus, C1 being the only one apparent at the lower RepB concentrations. More of the faster-migrating complex (C1) was observed with the bind fragment than with the nic fragment at the same RepB concentration, indicating that the affinity of RepB for the DDR is higher than for the DNA of the nic region (see below). With either fragment, complex C2 only became detectable when most of the DNA was in C1, indicating that generation of C2 from C1 is not cooperative. In fact, cooperative binding of RepB in a putative two-site system could not be deduced from the analysis of the fractions of free and C1- and C2-complexed DNA by applying the approach described in Supplementary Methods (data not shown). When a DNA fragment spanning the entire dso was assayed, three complexes were observed, the one with the lowest mobility, C3, accumulating at high RepB:DNA ratios (Figure 1B). Again, no cooperative binding of RepB could be deduced from the analysis of the pattern of formation of these complexes when considering a putative three-site system (see Supplementary Methods; data not shown). HO• footprinting experiments (below) showed that, whereas only the DDR were contacted by RepB in complex C1, both the nic locus and the bind locus were protected by the protein in C2. Preliminary results suggest that complex C3 would result from further binding of RepB to complex C2 through protein–protein or unspecific protein–DNA interactions, in a similar way to that proposed for generation of C2 from C1 in both the nic fragment and the bind fragment (see below).
High-resolution analysis of contacts between RepB and its target DNAs
Analysis of the contacts between RepB and its target DNAs were performed by HO• and DMS footprinting of complexes C1 and C2 generated by binding of the protein to the DNA fragments containing the nic locus, the bind locus, or the entire dso. The DNAs were 5′-end labelled (top or bottom strand) and modified either prior to (HO• assays) or after (DMS assays) separation of free and RepB-complexed DNAs. The results are summarized in Figure 2A and B.
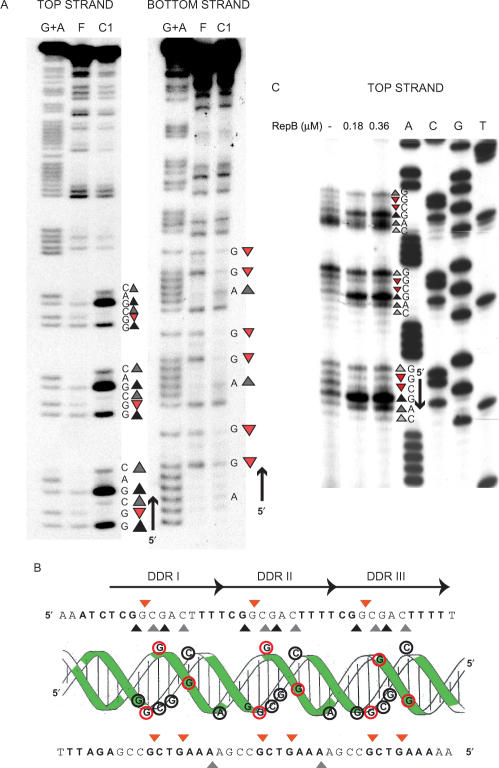
Figure 2.
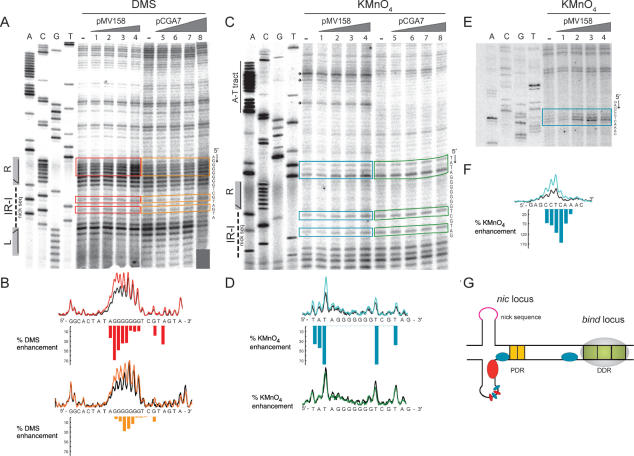
High resolution contacts of RepB with the entire dso (A) and with the nic locus (B). DNAs were 5′-end labelled and incubated with RepB. The complexes generated were treated with HO• (A and B) or with DMS (B) as described in ‘Materials and Methods’. Sequencing gels show the modification patterns of the 5′ regions of both strands. Absorption scans of naked DNAs (black lines) and complexes C1 (red lines) and C2 (blue lines) are displayed between the nucleotide sequence and the corresponding gel. To facilitate the detection of weak protections in B, the scans corresponding to the naked and complexed DNAs are shown superimposed around the footprints. PDR and DDR are shadowed. The IR-I element is displayed with its left (L) and right (R) arms. The dash shows the position of the nick site. Bases whose deoxyriboses are protected by RepB from HO• cleavage are indicated by triangles of different sizes (related to the extent of protection). In B, bases hyperexposed (black circles) or protected (open circles) by RepB against methylation with DMS are also shown.
When the entire dso-containing fragment was used, protections against HO• cleavage of the DNA in C1 showed that the DDR were fully occupied by RepB. In addition, some weak protections were observed in the nic locus (Figure 2A). The protection pattern observed in C2, although the same in the bind locus as that of C1, differed significantly from it in the nic locus (Figure 2A). In C2, the weak protections of the DNA backbone observed close to the nick site in C1 became stronger and larger (Figure 2A, top strand). These results, together with the footprinting pattern of the C1 complex arising from binding of RepB to the separate nic region (see below), suggested that while the RepB-dso C1 complex had the protein bound to only the bind locus, both the bind and the nic loci were occupied by RepB in C2. The low stability of the complexes between RepB and the nic locus (see below), led us to hypothesize that the weak protections found in the nic locus in the RepB-dso C1 complex could result from HO• reactivity of complexes harbouring RepB in both loci (complex C2), and subsequent dissociation of the protein from the nic locus previous to the electrophoretic separation of free and complexed DNAs. This would give rise to a fraction of molecules migrating as C1 but having the footprinting pattern corresponding to C2. Preliminary characterization of complex C3 of RepB-dso (not shown) did not reveal significant differences in the footprinting pattern as compared with C2, suggesting that C3 arises from binding of new RepB molecules to C2 through protein–protein interactions. Alternatively, C3 might arise from unspecific binding of RepB to the DNA of complex C2, a hypothesis that would also be consistent with the lack of new definite footprints in C3.
The interactions between RepB and the nic locus were also analysed in the fastest-migrating (C1) complex generated by binding of RepB to the DNA fragment that only contains this region (Figure 2B). On the top strand, two HO• footprints were located 3′ to the nick site (Figure 2B). These footprints were essentially identical to those observed within the nic locus in the RepB-dso C2 complex (Figure 2A). On the bottom strand of the separate nic locus, contacts of RepB with the DNA backbone were observed at the right arm of IR-I and in a region 5′ to it (Figure 2B). From the results on both strands, a picture of RepB bound to the nic locus emerges in which the protein only contacts the sugar-phosphate DNA backbone to the right of the nick site. One set of protections overlaps with the right arm of IR-I, and the other overlaps with a nearby region consisting of two 7-bp DR (5′-GTGCCPuA-3′; hereafter termed proximal direct repeats, PDR) (Figure 2B). When DMS footprinting was assayed, some G residues were slightly protected (Figure 2B), indicating specific interactions between RepB and these bases through the major groove of the DNA. On the top strand, the protected Gs were confined to the PDR, whereas on the bottom strand they were situated in the central sequence and in the right arm of the IR-I element. A clear hyperexposed band, corresponding to the G located precisely at the nick site, was also visualized on the top strand (Figure 2B). Enhanced methylation of this G residue might result from increased local hydrophobicity caused by the bound protein (32), indicating the proximity of a RepB region with these characteristics. The analysis of the RepB–DNA contacts in the nic region shows that the PDR direct repeats are not paralleled by a repeated pattern of either HO• or DMS footprints (Figure 2B).
The footprints mediated by RepB in the separate bind locus refine our previous finding that the DDR constitute the primary binding site of the initiator (18). Changes in the DMS reactivity profiles upon RepB binding were analysed for each strand of the DDR (Figure 3A and B). The results showed that RepB interacts through the major groove of the DNA with the same three Gs of each direct repeat, protecting them against methylation. Enhanced DMS reactivity of some Gs and Cs in each repeat constituting the DDR was also observed. Since DMS reactivity of C residues results from local DNA conformational changes such as unwinding or distortion of the double helix (32), we tested whether binding of RepB induced strand openings. To do this, the same RepB-bind C1 complex was subjected to KMnO4 probing, but no local melting of the DNA strands could be observed (not shown). The HO• and DMS reactivity profiles of the pMV158 bind locus are displayed on the DNA double helix in Figure 3B. They reflect that the interactions of RepB with the DDR involve only one face of the double helix, generating a significant distortion on it. Furthermore, a clear repeat profile was observed for the protein bound to the bind locus (Figure 3), which suggests that RepB (most likely in its hexameric form) employs three identical DNA-binding motifs to bind to the DDR. When, instead of linear DNA, supercoiled pMV158 DNA was used as the binding substrate, essentially the same DMS reactivity profile of the DDR region was observed (the methylation pattern of the top strand as deduced from stops in the extension of the bottom strand is shown in Figure 3C). This was expected as the DNA of the bind locus is predicted to have similar conformation in linear and supercoiled molecules.
Figure 3.
High resolution contacts of RepB with the bind locus. (A) DMS footprint of RepB bound to a linear DNA fragment containing the DDR. Bases hypermethylated (grey or black triangles, depending on whether there is a slight or a marked increase, respectively, in the DMS sensitivity) or protected (red triangles) in each strand by bound RepB are shown. To the right, the DNA sequence of the footprinted regions is displayed with its 5′→3′ directionality. Lanes: F, free DNA; C1, DNA of complex C1; G+A, Maxam and Gilbert sequencing ladder for purines. (B) Scheme of the B-DNA double helix of the DDR displayed with the nucleotide sequences of the top and bottom strands. Bases whose deoxyriboses are protected by RepB against HO• attack are shown in boldface letters or as green-shadowed regions on the double helix. Bases which become hypermethylated (grey/black triangles, black-lined encircled letters) or protected from methylation (red triangles, red-lined encircled letters) upon binding of RepB are indicated on the sequence and on the DNA double helix. The three repeats constituting the DDR are indicated by arrows. (C) DMS footprint of RepB bound to the DDR on supercoiled pMV158. The methylation sites on the top strand were mapped by primer extension using labelled primer dso4 corresponding to the bottom strand. The same primer was used for the control dideoxy sequencing ladder (lanes A, C, G, T). The methylation pattern of the bind locus was obtained in the absence of RepB or at the indicated concentrations of the protein. To the right, the DNA sequence at the footprinted regions is displayed with its 5′→3′ directionality. The same symbol code as in A was used.
Footprinting analysis of the RepB-nic or RepB-bind C2 complexes did not show additional protected regions compared to the corresponding C1 complexes (not shown), although the patterns of relative protections varied slightly. This suggests that new RepB molecules might bind, through protein–protein or protein–DNA interactions, to the region previously contacted by the protein in C1, thus generating the corresponding C2 complex.
Affinity of binding of RepB to the dso and stability of the RepB–DNA complexes
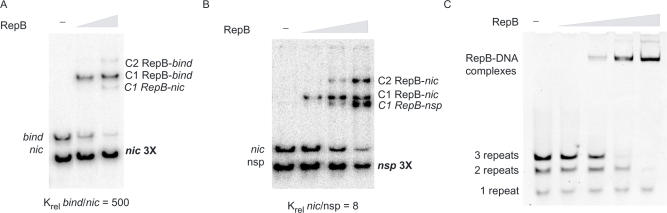
To measure the stability of binding of RepB to the nic or to the bind regions of the dso, the kinetics of dissociation of the corresponding C1 complex generated on either DNA (Figure 1B) was analysed. Dissociation of RepB from the labelled DNA was made irreversible by addition of a 300-fold excess of the respective unlabelled DNA. Samples were withdrawn at different time intervals and loaded on native PAA gels (Supplementary Figure S1). Plotting the logarithm of the fraction of the remaining undissociated C1 complex as a function of time allowed us to calculate the dissociation rate of RepB from either the nic or the bind locus. Whereas the half-life of the RepB-bind locus complex was around 18 min [kd = (0.64 ± 0.13) × 10−3 s−1] (Supplementary Figure S1), the dissociation rate of the complex between the protein and the nic locus was very high, so that its half-life (less than 1 min) could not be measured with accuracy (not shown). Thus, our results showed a significantly higher stability of the RepB–DNA complexes when the initiator protein bound to the DDR compared to when it bound by the replication initiation site.
We next measured the relative affinity of RepB for both DNA targets. Two labelled DNA fragments of different sizes, one harbouring the nic locus and the other the bind locus, were incubated simultaneously with various concentrations of RepB (Figure 4A). EMSA was performed and the fractions of free and complexed DNA were measured for each fragment. The relative affinities of RepB for the nic and bind loci were given by the relative equilibrium binding constant (Krel) (25). The results obtained (Figure 4A) showed that the affinity of binding of RepB to the DDR was about 500-fold higher than to the nic locus [Krel (bind/nic) ∼500]. The higher stability of the RepB–DDR complex compared to that of the protein bound to the nic locus (see above) could account for the higher affinity of RepB for the DDR. Binding of RepB to the nic locus seemed to be specific, as it showed about 10-fold higher affinity than the binding of the initiator protein to a non-specific DNA located outside of the pMV158-dso [Krel (nic/non-specific DNA) ∼8; Figure 4B]. A much higher affinity of binding of the initiator protein for its cognate bind locus than for the region where it specifically nicks the DNA has also been reported for the staphylococcal plasmid pC221, which belongs to the pT181 family (13). From the above set of results, we conclude that RepB has higher affinity for the DDR (that compose the bind locus) than for the nic locus, the former constituting the primary binding site of the protein.
Figure 4.
Relative affinity of RepB for different DNA sequences. The indicated fragments were mixed and incubated in the absence of RepB (-) or in the presence of increasing concentrations of the protein. The relative equilibrium binding constant (Krel) was calculated from Eq. 2. Relative affinities of RepB for the bind locus compared to the nic locus (A) and for the nic locus compared to a non-specific (nsp) DNA (B). A 3-fold molar excess of either the fragment containing the nic locus relative to the fragment harbouring the DDR (A), or of the nsp DNA relative to the nic fragment (B) was used. (C) Relative affinity of RepB for DNAs containing 1, 2 or 3 repeats of the DDR. The three duplex oligonucleotides were used at the same molar ratio. Note that the complexes corresponding to the three different DNAs could not be solved.
We have also compared, by EMSA, the binding affinities of RepB for double-stranded oligonucleotides comprising 1, 2 or all 3 of the DR that constitute the DDR (Figure 4C). The different complexes generated by binding of RepB to these oligonucleotides could not be properly solved upon electrophoresis. However, it was clear (from the decrease in the fraction of free DNA as the protein concentration increased) that RepB displayed the highest affinity for the DNA containing all three DR, and the lowest for the single repeat (Figure 4C).
RepB bends the DNA of the bind locus
Analysis of the interactions between RepB and the dso indicated that the protein recognizes the DDR as its primary binding site, inducing local conformational changes on this DNA. A possible explanation for the observed DNA distortion is that, upon binding, RepB strongly bends, or even wraps, the DNA. In fact, previous experiments indicated that an intrinsic bend might be located around this region (18), although neither the angle nor the centre of the curvature were determined.
To explore the possible RepB-induced bending, we cloned separately the nic and bind loci of the pMV158-dso into the test vector pBEND2 to conduct circular permutation assays (24). This technique uses DNA bending constructs which can be cleaved at different restriction sites, generating DNA fragments of uniform length but with the suspected bend site placed at different positions with respect to the ends of the fragments (33). If a protein induces a bend, it will affect the relative electrophoretic mobility of the fragments in a predictable manner that allows calculation of the bend angles (34). The DNA fragments obtained by restriction with different enzymes were incubated with RepB to perform EMSA under conditions in which 50% of the DNA was bound in the corresponding C1 complex, so that in the same assay we could calculate the electrophoretic mobility of the free and bound DNAs. To calculate the relative mobilities, the ratio of the mobility of the bound to the unbound DNA with the binding site in the middle of the fragment (µMB/µM) was divided by the same ratio for the DNA with the binding site at the end of the fragment (µEB/µE). This method cancels out the contribution of the intrinsic curvature of the DNA, although it only gives accurate estimations when the induced bend is large compared with the intrinsic curvature (26). In addition, the bends induced by RepB on the nic and bind loci, although probably located at the same place as the intrinsic curvatures of the corresponding fragments (plots in Figure 5), might not coincide with them in the directionality. This could complicate the deconvolution of the effects of the intrinsic and induced bends. For these reasons, we also estimated the intrinsic curvature of the unbound DNAs (from µM/µE), as well as the total bending angle of the RepB-complexed DNAs (from µMB/µEB). The results (Table 1) demonstrated that the DNA of the bind locus exhibited an intrinsic bend of about 50°, whereas the nic locus was essentially straight. RepB induced a bend of 120° (105° when the intrinsic curvature of the DNA was deducted) on the DDR-containing DNA, whereas an apparent bending angle of around 60° was measured as induced by the protein on the fragment harbouring the nic region. Since the method used underestimates bending angles sharper than 100° (26), we conclude that RepB induces a very strong bend (>100°) on the bind locus, while binding of the protein to the nic locus results in a moderate bending of the DNA.
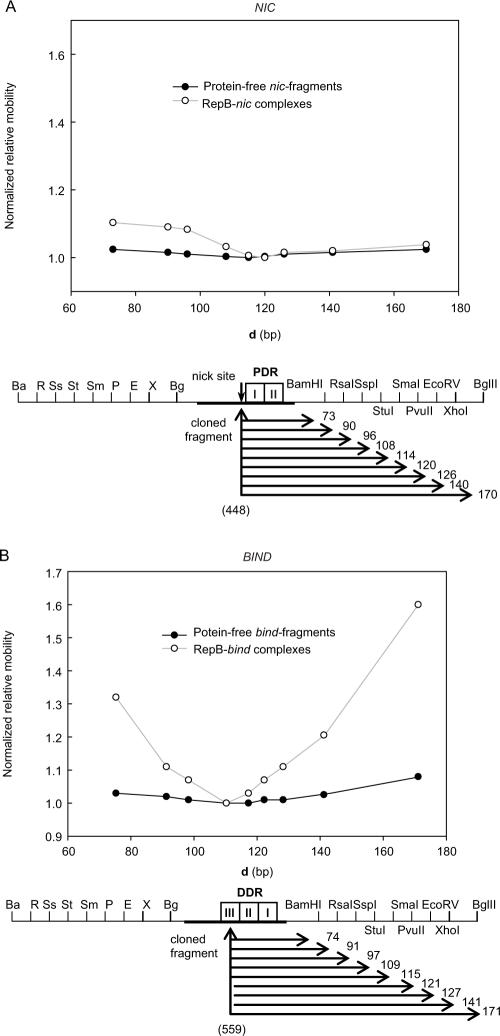
Figure 5.
RepB-induced DNA bending characterized by circular permutation assays. Both 241-bp nic- (A) and 243-bp bind- (B) permuted fragments were generated by restriction with the enzymes indicated in the scheme. Below, the distance (d, in bp) from the centre of the cloned pMV158 DNA (coordinate in brackets) to the right end of each fragment is displayed. The electrophoretic mobility of free (closed circles) or bound (open circles) DNA fragments, normalized to the mobility of the slowest same-sized DNA or complex, was plotted versus d. The apparent angles of both the intrinsic curvature and the RepB-induced bending on the nic and bind loci are shown in Table 1.
Table 1.
RepB induced DNA bending.
| Relative electrophoretic mobility (µM/µE)a StuI/BglII | Apparent bending angle α (°)b | |
|---|---|---|
| bind locus | 0.92 | 50 |
| RepB-bind | 0.61 | 120 |
| nic locus | 0.98 | 20 |
| RepB-nic | 0.88 | 62 |
aµM/µE determined in 6% polyacrilamide gels.
bα values were calculated from the electrophoretic mobilities as described by Thompson and Landy (26).
To investigate the location of the centre of the RepB-induced bending in either the nic or the bind locus, we used again circular permutation analysis. The bend centre was estimated by determining the coordinate of the pMV158 DNA located in the middle of the slowest-migrating fragment. In the 241-bp permuted fragments harbouring the nic locus, the centre of the cloned DNA corresponded to coordinate 448 of pMV158, exactly where the RepB nick site is located. The PvuII-ended nic-fragment (which has coordinate 448 located in the middle, that is about 120 bp from either end) exhibited the lowest mobility (Figure 5A), indicating that the bending site was centred on this fragment. This locates the bending site around coordinate 448 of the pMV158 DNA, within the protein secondary binding site (Figure 2B). In the 243-bp permuted bind-fragments, the centre of the cloned pMV158 DNA was located at coordinate 559, in the middle of repeat III of the DDR. The bind-fragment with the lowest mobility (the one with the StuI ends) has this coordinate 109 bp away from its right end, and the bend in the middle, that is, around 120 bp away from the same end (Figure 5B). This would locate the bend centre at the end of the DDR, within the region occupied by the protein bound to the bind locus.
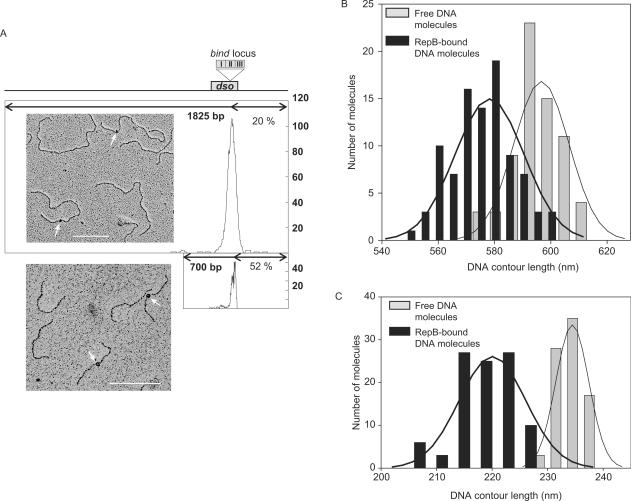
To further investigate the path of the DNA upon binding of RepB, we performed a set of electron microscopy (EM) assays to measure the contour length of bound and unbound DNAs. Two pMV158 DNA fragments of 1825 and 700 bp containing the entire dso were used, the former having the dso in an asymmetric position so that the nucleoprotein complex could be easily located from the EM images. The RepB–DNA complex was precisely located 365 bp from one of the ends in either fragment (Figure 6A). As both fragments share the BstXI end (coordinate 914 of the pMV158 DNA sequence), the nucleoprotein complex should be located around coordinate 549 of the plasmid, which is in the middle of repeat II of the DDR. These results confirm the specific binding of RepB to the bind locus of the dso. No indication of binding of the protein to its secondary site in the nic locus of the dso was found, perhaps because of the low stability of the complexes generated at this site. We measured the contour lengths of single 1825 and 700 bp DNA molecules, both free and bound to RepB, in the same deposition (Figure 6B and C; and Supplementary Table S1). A significant reduction in the average contour length of both fragments was observed upon binding of RepB. DNA length distribution showed two different populations, one of free DNA molecules, and the other of RepB-bound DNA molecules with a shortened contour length. The RepB-mediated shortening was estimated in about 50 bp (Figure 6B and C, and Supplementary Table S1), which matches the length of the DNA contacted by RepB in the bind locus. Shortening of complexed DNAs might reflect wrapping of it around the protein (35). Lacking further information on the structure of the RepB–DNA complexes, the fact that the path of the bind locus through the protein is longer than the measured contour length (traced as the shortest possible DNA path through the bound protein) could indicate DNA winding about RepB.
Figure 6.
EM of RepB-DNA complexes. (A) Distribution of the location of the RepB-DNA complexes from one end of the 1825-bp or 700-bp fragments. The length of the DNA fragments is represented by a thick line. RepB is located 20% (365 bp) from the closer end of the 1825-bp DNA, and 52% (365 bp) from one end of the 700-bp DNA. Since the bind locus is nearly centred in the 700-bp fragment, the peak observed is unsymmetrical. On top of the figure, a scale scheme indicates the position of the pMV158 dso, and the insert shows the bind locus. Alongside, micrographs of the 1825 bp and 700 bp fragments after incubation with RepB are shown. Arrows indicate RepB bound to the bind locus of the dso. Bar indicates 200 nm (about 600 bp). (B), (C) Contour length distribution of protein-free and RepB-bound DNA molecules. Histograms show the contour length of free (grey bars) and bound (black bars) DNA molecules of two different sizes: 1825 bp (B) and 700 bp (C). The Gaussian fit to the distribution is represented by the solid lines, and the statistics of the contour length determinations is shown in Table S1.
RepB–dso interactions on supercoiled plasmid DNA
At protein to DNA molar ratios up to 30:1, contacts of RepB with supercoiled pMV158 DNA were confined to the bind locus (Figure 3). Under these conditions, KMnO4 probing did not reveal the presence of unpaired or strongly distorted DNA regions within the plasmid dso. This excluded the possibility that the binding of RepB to its primary site contributes to the extrusion of the IR-I cruciform of the dso and thus, to the melting of the substrate nick sequence.
At higher RepB concentrations (protein to DNA molar ratio of 50:1 or higher), in addition to the footprints derived from the binding of RepB to the bind locus, DMS footprints consisting of hyperexposed bands were observed in the nic locus when analysing the methylation pattern of the bottom strand on supercoiled pMV158 DNA (Figure 7A and B). The relative intensity of the hyperexposed bands augmented as the protein concentration was increased, until a plateau was reached at a protein to DNA molar ratio of 150:1 (not shown). The hypermethylated residues (mostly Gs) lie on the right arm of IR-I and on the adjacent DNA region which is complementary to the nick sequence (Figure 7G) and may reflect the proximity of RepB and/or changes in the DNA conformation due to extrusion of the corresponding cruciform. As a control, DMS footprinting analysis was also performed on supercoiled DNA of plasmid pCGA7, a derivative of pC194 which bears the separate pMV158-nic locus (including IR-I and the PDR) (12,15). The presence of RepB also resulted in hypermethylation of the same nucleotides as in pMV158, although it was evident that much higher concentrations of the protein were required to obtain similar levels of enhancement of the DMS reactivity in pCGA7 (Figure 7A and B), with no plateau being reached at the RepB concentrations assayed. These results suggested that binding of RepB to its secondary site in the nic locus might be facilitated in supercoiled DNAs containing the entire dso.
Figure 7.
DMS and KMnO4 footprints of RepB bound to the nic locus on supercoiled DNAs. (A) Sequencing gels displaying DMS sensitivity of the bottom strand around the nic region in the absence and in the presence of RepB. As a control, pCGA7, a pC194 derivative which bears the nic region of the pMV158 dso was used. Supercoiled pMV158 or pCGA7 DNAs were incubated in the absence of RepB (lanes –) or with various protein concentrations, and treated with DMS as described under ‘Materials and Methods’. RepB concentrations were 0.36, 0.6, 1.2 and 1.8 µM (lanes 1–4) for the reactions containing pMV158 DNA, and 0.6, 1.2, 1.8 and 4.8 M (lanes 5–8) for the pCGA7 DNA. The methylation sites on the bottom strand were mapped by primer extension using a labelled primer corresponding to the top strand. The same primer was used for the control dideoxy sequencing ladder (lanes A, C, G, T). On the left, the location of IR-I, with its left (L) and right (R) arms and its central region harbouring the nick sequence, is shown. To the right, the DNA sequence of the bottom strand at the footprints is displayed with its 5′→3′ directionality. Bands whose intensity is modified in pMV158 in the presence of RepB are indicated with red boxes. The same bands in pCGA7 are in orange boxes. (B) Quantification of the experiment shown in A by PhosphorImager analysis using Quantity One software. Scans of lanes (–) and 4 corresponding to pMV158, and of lanes (–) and 8 of pCGA7 are drawn at the top and the bottom of the panel, respectively. The scans obtained with naked DNA (black line) and RepB-bound DNA (red line for pMV158 and orange line for pCGA7) are superimposed. Bases whose DMS sensitivity is modified in the presence of RepB are indicated with red (pMV158) or orange (pCGA7) bars. Bar heights are proportional to the percentage of DMS sensitivity enhancement. (C) Sequencing gels displaying KMnO4 sensitivity of the bottom strand around the nic region in the absence and in the presence of RepB. As a control, plasmid pCGA7 was used. Supercoiled pMV158 or pCGA7 DNAs were incubated in the absence of RepB (lanes –) or with various protein concentrations, and treated with DMS as described in ‘Materials and Methods’. RepB concentrations were 0.6 µM (lanes 1 and 5), 1.8 µM (lanes 2 and 6), 2.4 µM (lanes 3 and 7) and 4.8 µM (lanes 4 and 8). The modification sites on the bottom strand were mapped by primer extension using a labelled primer corresponding to the top strand. The same primer was used for the control dideoxy sequencing ladder (lanes A, C, G, T). On the left, the location of the central region and right arm (R) of IR-I is displayed. To the right, the DNA sequence of the bottom strand at the distorted DNA regions is shown with its 5′→3′ directionality. Bands whose intensity is modified in pMV158 in the presence of RepB are within blue boxes. The same bands in pCGA7 are within green boxes. The arrows point to sites of high RepB-independent KMnO4 reactivity. (D) Quantification of the experiment shown in C. Scans of lanes (–) and 4 corresponding to pMV158, and of lanes (–) and 8 corresponding to pCGA7 are drawn at the top and the bottom of the panel, respectively. The scans obtained with naked DNA (black line) and RepB-bound DNA (blue line for pMV158 and green line for pCGA7) are superimposed. Bases whose KMnO4 sensitivity is modified in the presence of RepB are indicated with blue (pMV158) or green (pCGA7) bars. Bar heights are proportional to the percentage of enhancement in the KMnO4 reactivity. (E) Sequencing gels displaying KMnO4 sensitivity of the bottom strand close to the bind region, in the absence and in the presence of RepB. Supercoiled pMV158 DNA was incubated without RepB (–) or in the presence of 0.6, 1.8, 2.4 and 4.8 µM of the protein (lanes 1–4, respectively). KMnO4 reaction and mapping of the modification sites was as in C. A dideoxy sequencing ladder (lanes A, C, G, T) was included as a control. To the right, the DNA sequence of the bottom strand at the DNA region distorted upon binding of RepB to the nic locus is displayed with its 5′→3′ directionality. Bands whose intensity is modified in the presence of RepB are within a blue box. (F) Quantification of the experiment shown in ahown in E. Scans of lanes (–) (black line) and 4 (blue line) are drawn superimposed. Bases that increase their KMnO4 sensitivity in the presence of RepB are indicated with blue bars whose heights are proportional to the percentage of enhancement of the KMnO4 reactivity. (G) Scheme of the bottom strand sites of the pMV158 dso whose DMS or KMnO4 sensitivity is modified upon binding of RepB to the nic locus. Once RepB is bound to its high affinity target, the bind locus, the protein would bind to its lower affinity target in the nic locus, then rendering some sites highly sensitive to DMS (red ellipses) or to KMnO4 (blue ellipses). Whereas most of the sites that increase their KMnO4 sensitivity are within the nic locus, the most distorted site was found close to the DDR. The IR-I cruciform of the pMV158 dso is represented since binding of RepB to the nic locus promotes its extrusion.
To test whether the interaction of the initiator protein with the nic locus caused melting of DNA within this region, analyses of the KMnO4 reactivity of the bottom strand were performed on supercoiled pMV158 and pCGA7 DNAs (Figure 7C and D). KMnO4 preferentially oxidizes thymines in severely distorted or denatured regions of DNA or at their junction with native duplex DNA (36). In pMV158, increased reactivity of two thymines located between both IR-I arms as well as of three bases (5′-TAT-3′) 5′-adjacent to the right arm of IR-I was observed at RepB:DNA molar ratios above 50:1 (Figure 7). Although the KMnO4 hypersensitivity of the two T bases in the central region of the IR-I element only proved that the DNA of this region was melted, extrusion of the IR-I cruciform could be deduced from the increased reactivity of the bases (5′-TAT-3′) contiguous to the IR-I right arm; high KMnO4 reactivity caused by disruption of the DNA base stacking is to be expected in nucleotides located at the junctions between intra- and inter-stranded duplexes of a cruciform structure (see the scheme in Figure 7G). High KMnO4 reactivity of the same T bases (two in the IR-I central region, and the two thymines of the 5′-TAT-3′ sequence contiguous to the IR-I) was observed in the unbound supercoiled pCGA7 DNA, and no enhancement of this reactivity was obtained in the presence of RepB, even at protein concentrations at which binding of RepB was indicated by DMS hypersensitivity of the G residues in the stem of the hairpin (Figure 7A, B, C and D). In fact, the KMnO4 reactivity pattern of the unbound supercoiled pCGA7 DNA coincides with that of the RepB-bound pMV158 DNA at the plateau. These results show that the frequency of spontaneous extrusion of the IR-I hairpin is higher in pCGA7 than in pMV158, which might arise from pCGA7 lacking possible secondary structures able to compete with IR-I in releasing the torsional stress of the supercoiled DNA. The fact that binding of RepB to the supercoiled pCGA7 DNA enhanced methylation of the Gs located at the IR-I hairpin stem without further promoting the cruciform extrusion indicated that the variation in the DMS reactivity pattern arose from the proximity of the protein to the hypersensitive bases, rather than from any RepB-promoted strong distortion of the DNA.
Unexpectedly, the major change observed in the KMnO4-sensitivity pattern upon binding of RepB to the pMV158 supercoiled DNA was the great increase (up to about 150%) in the reactivity of a region located 11 pb upstream of the bind locus, in the intervening DNA between the nic and bind loci, indicating a strong distortion or even base unpairing of the DNA close to the bind locus. However, such deformation of the helix was only observed at RepB concentrations where the protein is bound also to the nic locus (Figure 7E, F and G).
In addition to the abovementioned sites, which increased their sensitivity to KMnO4 upon binding of RepB to the nic locus, several thymines within an A–T-rich region contiguous to the PDR (Figure 1A) were hyperexposed to KMnO4 on the naked supercoiled pMV158 DNA, and did not increase their reactivity in the presence of the protein (Figure 7C). These results reveal that the DNA of this A–T tract located between the PDR and the DDR is strongly distorted on the supercoiled DNA of pMV158. Although also present in pCGA7, the A–T tract exhibits a much lesser degree of distortion on the supercoiled DNA of this plasmid (Figure 7C).
DISCUSSION
The results presented here demonstrate the generation of a nucleo-protein complex at the dso of plasmid pMV158 promoted by the binding of the RepB initiator to its target DNA. Upon binding to its primary site, the three DDR, RepB would be loaded to the nic locus, to which it binds with weaker affinity. This positioning would facilitate RepB-mediated extrusion of the IR-I hairpin and, as a consequence, the protein would be placed in position to initiate replication by introducing the nick at the 5′-GpA-3′ dinucleotide. Although in vivo replication of pMV158 requires the presence of the DDR, in vitro RepB-mediated nicking/closing of the DNA has been observed on single-stranded oligonucleotides lacking both the DDR and the PDR, as well as on supercoiled plasmid DNAs either lacking the DDR or harbouring non-cognate PDR and DDR (15). Thus, it would appear that the only requirement for the nicking activity of RepB is that the sequence around the nick site has an ssDNA configuration, which in supercoiled DNA would be accomplished by IR-I cruciform extrusion. Binding of RepB to the nic locus would promote cruciform extrusion. It has previously been shown that the cruciform involving IR-I (see Figure 1A) is one of the three main secondary structures extruded on the supercoiled DNA of a pMV158 derivative at 10°C. In contrast, at 37°C, only extrusion of a secondary structure located upstream of the plasmid dso (about coordinate 170 of pMV158) could be clearly detected (19). Thus, at the physiological temperature for the pMV158 host (37°C), the IR-I cruciform seems to exhibit a low frequency of extrusion, so that cruciform extrusion would require the binding of RepB to the nic locus. As a consequence, initiation of replication would take place only when specific binding of RepB occurs. It can also be argued that only when RepB is bound to the bind and nic loci, the structure generated could facilitate the assembly of the host-encoded proteins [PcrA helicase (8), ssDNA-binding protein, and DNA polymerase I (37)] that constitute the replisome. In addition, contacts of RepB with the nic locus span DNA sequences that would be located in the base or within the stem-loop of the IR-I hairpin, where the cleavage activity of the protein is exerted (Figures 1A, 2B and 7G). Thus, this secondary RepB binding site would be the region expected to be bound by the catalytic RepB molecule. Interestingly, footprinting patterns of the initiator protein of plasmids belonging to the pT181 family have demonstrated contacts of the protein with both the 3′ side of the DNA hairpin that contains the nick site and an adjacent inverted repeat sequence constituting the specific bind region (38,39). Therefore, the secondary RepB binding site within the pMV158-nic locus has the same arrangement relative to the nick site as the specific bind locus of plasmids of the pT181 family. In these plasmids, binding of the initiator to the bind locus also promotes extrusion of the cruciform structure containing the nick site (40). However, the main difference between pT181 and pMV158 is that in the former, the initiator is placed directly in the vicinity of the nick site, whereas RepB of pMV158 binds with high affinity to a distant bind locus and then it has to reach the low-affinity site located close to the nick site, within the nic locus. This might be achieved through the interplay between the primary and secondary binding sites of RepB, a hypothesis that fits well with our unpublished observation that the in vivo replication of pMV158 requires the DDR to be in-phase with the nic locus.
Interplay between both loci of the pMV158-dso might be mediated by protein–protein interactions between two RepB hexamers, each bound to a different locus, as, at least in vitro, a protein hexamer seems to be unable to bind simultaneously to the bind and nic loci (results not shown). Although on linear DNA RepB bound to the bind locus does not seem to cooperate positively in binding to the nic locus, indications of positive cooperativity on supercoiled DNA came from the observation that binding of RepB to the nic locus required lower protein concentrations when the high-affinity bind locus was present in the same molecule than when it was absent (Figure 7A). Protein–protein interactions, which are suggested to generate RepB-nic or RepB-bind C2 complexes as well as the RepB-dso C3 complex, may account for the apparent positive cooperativity of binding of the protein to the nic region on supercoiled plasmid DNA bearing the entire dso. In fact, analytical ultracentrifugation experiments have revealed that, at high protein concentrations (>10 µM), RepB hexamers can interact leading to higher order aggregation states (JA Ruiz-Masó, unpublished data), which might account for the observed footprinting pattern of complexes C2 generated on the separate nic and bind loci.
It is relevant to point out the great distortion of the DNA helix in the region close to the bind locus (Figure 7E, F and G) which is observed when RepB is bound to both loci of the pMV158 dso. This could be an indication of the generation of a DNA loop on supercoiled DNA, as reported for AraC (41) or LacI (42). Although we have not found any consensus sequence for binding of IHF protein at the dso region, contribution of host factors, such as architectural proteins, to the in vivo interaction between the two RepB binding sites cannot be ruled out. In fact, the pMV158 DNA sequence between PDR and DDR has a large A–T tract, conserved in other plasmids of the family (see below), which could provide a binding site for architectural proteins recognizing distorted DNA, such as HU and HNS (43). Some thymines in the bottom strand of this A–T tract exhibit a marked RepB-independent reactivity to KMnO4 in the supercoiled pMV158 DNA, indicating that this DNA region really is distorted (Figure 7C). A host-factor-mediated strong bending of the DNA intervened between the PDR and the DDR might bring together the primary and secondary RepB binding sites in the dso, thus facilitating the loading of the protein on the catalytic nic locus.
A comparative analysis of the dso regions of the members of the pMV158 family (Supplementary Results) showed that these replicons share a similar organization: a highly conserved nick sequence and, downstream of it, two DR clusters, the PDR and the DDR. The PDR have been identified in all these plasmids. The DDR, which have been found in most members of the pMV158 family, are nearly in phase with the DNA helical repeat.
Why have plasmids of the pMV158 family evolved to create a high-affinity binding site for the initiator that is apart from the cleavage site? While other RCR-plasmid initiators are monomeric [RepA of pC194; (44)] or dimeric [RepC-pT181, RepD-pC221 (10)], RepB purifies as a hexamer. The interaction of the initiator with its cognate dso could be envisaged as a dynamic and sequential process requiring the oligomerization ability of RepB. First, RepB would bind to its high affinity site, favouring its loading into the nic region. As a consequence, the next stage would be extrusion of the IR-I hairpin, and finally, the protein would cleave at the initiation site. This loading of the initiator could be followed by the assembly of the other replisome proteins. Thus, successive nucleo-protein complexes would be generated in which, as in the case of the AAV (adeno-associated virus) and papillomavirus initiator proteins [containing RCR domains (45,46)], specific protein–protein and protein–DNA interactions would be essential for replication.
SUPPLEMENTARY DATA
Supplementary data is available at NAR online.
ACKNOWLEDGMENTS
Thanks are due to M.T. Alda for purification of RepB, to members of our lab for helpful discussions, and to A.M.L. Jones for correcting the English throughout the manuscript. J. A. R-M. was a recipient of a FPI fellowship from the Ministerio de Educación y Ciencia. Research financed by Ministerio de Educación y Ciencia (grant BFU2004-00687/BMC to G.d.S. and Acción Especial BMC2002-11562-E to M.E.).
REFERENCES
- 1.del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan SA. Plasmid rolling-circle replication: recent developments. Mol. Microbiol. 2000;37:477–484. doi: 10.1046/j.1365-2958.2000.02001.x. [DOI] [PubMed] [Google Scholar]
- 3.Novick RP. Contrasting lifestyles of rolling-circle phages and plasmids. Trends Biochem. Sci. 1998;23:434–438. doi: 10.1016/s0968-0004(98)01302-4. [DOI] [PubMed] [Google Scholar]
- 4.Osborn M. Database of plasmid replicons. 2002. http://www.essex.ac.uk/bs/staff/osborn/DPR_home.htm.
- 5.Khan SA. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid. 2005;53:126–136. doi: 10.1016/j.plasmid.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Thomas CD, Balson DF, Shaw W. In vitro studies of the initiation of staphylococcal plasmid replication. Specificity of RepD for its origin (oriD) and characterization of the Rep-ori tyrosil ester. J. Biol. Chem. 1990;265:5519–5530. [PubMed] [Google Scholar]
- 7.Noirot-Gross MF, Bidnenko V, Ehrlich SD. Active site of the replication protein in the rolling circle plasmid pC194. EMBO J. 1994;13:4412–4420. doi: 10.1002/j.1460-2075.1994.tb06761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang T-L, Naqvi A, Anand SP, Kramer MG, Munshi R, Khan SA. Biochemical characterization of the Staphylococcus aureus PcrA helicase and its role in plasmid rolling circle replication. J. Biol. Chem. 2002;277:45880–45886. doi: 10.1074/jbc.M207383200. [DOI] [PubMed] [Google Scholar]
- 9.Khan SA. DNA-protein interactions during the initiation and termination of plasmid pT181 rolling-circle replication. Prog. Nucleic Acid Res. Mol. Biol. 2003;75:113–137. doi: 10.1016/s0079-6603(03)75004-1. [DOI] [PubMed] [Google Scholar]
- 10.Rasooly A, Wang P, Novick R. Replication-specific conversion of the Staphylococcus aureus pT181 initiator protein from an active homodimer to an inactive heterodimer. EMBO J. 1994;13:5245–5251. doi: 10.1002/j.1460-2075.1994.tb06856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao AC, Khan SA. Sequence requirements for the termination of rolling-circle replication of plasmid pT181. Mol. Microbiol. 1997;24:535–544. doi: 10.1046/j.1365-2958.1997.3641730.x. [DOI] [PubMed] [Google Scholar]
- 12.del Solar G, Moscoso M, Espinosa M. In vivo definition of the functional origin of replication (ori(+)) of the promiscuous plasmid pLS1. Mol. Gen. Genet. 1993;237:65–72. doi: 10.1007/BF00282785. [DOI] [PubMed] [Google Scholar]
- 13.Thomas CD, Nikiforov TT, Connolly BA, Shaw WV. Determination of sequence specificity between a plasmid replication initiator protein and the origin of replication. J. Mol. Biol. 1995;254:381–391. doi: 10.1006/jmbi.1995.0625. [DOI] [PubMed] [Google Scholar]
- 14.Dempsey L, Birch P, Khan S. Uncoupling of the DNA topoisomerase and replication activities of an initiator protein. Proc. Natl. Acad. Sci. USA. 1992;89:3083–3087. doi: 10.1073/pnas.89.7.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moscoso M, del Solar G, Espinosa M. In vitro recognition of the replication origin of pLS1 and of plasmids of the pLS1 family by the RepB initiator protein. J. Bacteriol. 1995;177:7041–7049. doi: 10.1128/jb.177.24.7041-7049.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zock JM, Birch P, Khan SA. Specificity of RepC protein in plasmid pT181 DNA replication. J. Biol. Chem. 1990;265:3484–3488. [PubMed] [Google Scholar]
- 17.Jin R, Novick RP. Role of the double-strand origin cruciform in pT181 replication. Plasmid. 2001;46:95–105. doi: 10.1006/plas.2001.1535. [DOI] [PubMed] [Google Scholar]
- 18.de la Campa AG, del Solar G, Espinosa M. Initiation of replication of plsmid pLS1. The initiator protein RepB acts on two distant regions. J. Mol. Biol. 1990;213:247–262. doi: 10.1016/S0022-2836(05)80188-3. [DOI] [PubMed] [Google Scholar]
- 19.Puyet A, del Solar G, Espinosa M. Identification of the origin and direction of replication of the broad-host-range plasmid pLS1. Nucl. Acids Res. 1988;16:115–133. doi: 10.1093/nar/16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moscoso M, del Solar G, Espinosa M. Specific nicking-closing activity of the initiator of replication protein RepB of plasmid pMV158 on supercoiled or single-stranded DNA. J. Biol. Chem. 1995;270:3772–3779. doi: 10.1074/jbc.270.8.3772. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Maso JA, Lopez-Zumel C, Menendez M, Espinosa M, del Solar G. Structural features of the initiator of replication protein RepB encoded by the promiscuous plasmid pMV158. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics. 2004;1696:113–119. doi: 10.1016/j.bbapap.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Lacks SA, López P, Greenberg B, Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J. Mol. Biol. 1986;192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T, editors. Molecular Cloning. A Laboratory Manual. 2nd edn. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Kim K, Zwieb C, Wu C, Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989;85:15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- 25.Man T-K, Stormo GD. Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucl. Acids Res. 2001;29:2471–2478. doi: 10.1093/nar/29.12.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JF, Landy A. Empirical estimation of protein-induced DNA bending angles: applications to l site-specific recombination complexes. Nucl. Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiess E, Lurz R. Electron microscopic analysis of nucleic acids and nucleic acid-protein complexes. Methods Microbiol. 1988;20:293–323. [Google Scholar]
- 28.del Solar GH, Perez-Martin J, Espinosa M. Plasmid pLS1-encoded RepA protein regulates transcription from repAB promoter by binding to a DNA sequence containing a 13-base pair symmetric element. J. Biol. Chem. 1990;265:12569–12575. [PubMed] [Google Scholar]
- 29.Maxam AH, Gilbert W. Sequencing end-labelled DNA with base-specific chemical cleavages. Methods Enzymol. 1980:65. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 30.Papavassiliou AG. Localisation of DNA-protein contact points by DMS resistance of complexes resolved in gel retardation assays. Nucl. Acids Res. 1993;21:757–758. doi: 10.1093/nar/21.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasse-Dwight S, Gralla JD. Footprinting Protein-DNA Complexes in Vivo. Methods Enzymol. 1991;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- 32.Shaw PE, Stewart F. In: DNA-Protein Interactions: Principles and Protocols. Kneale GG, editor. Vol. 30. Totowa, NJ: Humana Press Inc.; 1994. pp. 79–87. [Google Scholar]
- 33.Wu HM, Crothers DM. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 34.Krüger R, Rakowski AA, Filutowicz M. Isomerization and apparent DNA bending by Pi, the replication protein of plasmid R6K. Biochem. Biophys. Res. Comm. 2004;313:834–840. doi: 10.1016/j.bbrc.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 35.Beerens N, Hoeijmakers JHJ, Kanaar R, Vermeulen W, Wyman C. The CSB Protein Actively Wraps DNA. J. Biol. Chem. 2005;280:4722–4729. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- 36.Schlax PJ, Capp MW, Record MT. Inhibition of transcription initiation by lac repressor. J. Mol. Biol. 1995;245:331–350. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- 37.Díaz A, Lacks SA, López P. Multiple roles for DNA polymerase I in establishment and replication of the promiscuous plasmid pLS1. Mo.l Microbiol. 1994;14:773–783. doi: 10.1111/j.1365-2958.1994.tb01314.x. [DOI] [PubMed] [Google Scholar]
- 38.Koepsel RR, Murray RW, Khan SA. Sequence-specific interaction between the replication initiator protein of plasmid pT181 and its origin of replication. Proc. Natl. Acad. Sci. USA. 1986;83:5484–5488. doi: 10.1073/pnas.83.15.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas CD, Balson DF, Shaw W. Identification of the tyrosine residue involved in bond formation between replication origin and the initiator protein of plasmid pC221. Biochem. Soc. Trans. 1988;16:758–759. [Google Scholar]
- 40.Jin R, Fernandez-Beros M-E, Novick RP. Why is the initiation nick site of an AT-rich rolling circle plasmid at the tip of a GC-rich cruciform? EMBO J. 1997;16:4456–4466. doi: 10.1093/emboj/16.14.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schleif R. DNA looping. Annu. Rev. Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 42.Swigon D, Coleman BD, Olson WK. Modeling the Lac repressor-operator assembly: The influence of DNA looping on Lac repressor conformation. Proc. Natl. Acad. Sci. USA. 2006;103:9879–9884. doi: 10.1073/pnas.0603557103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004;14:28. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Noirot-Gros M-F, Bidnenko V, Ehrlich SD. Active site of the replication protein of the rolling circle plasmid pC194. EMBO J. 1994;13:4412–4420. doi: 10.1002/j.1460-2075.1994.tb06761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F. The nuclease domain of adeno-associated virus Rep coordinates replication initiation using two distinct DNA recognition interfaces. Molecular Cell. 2004;13:403. doi: 10.1016/s1097-2765(04)00023-1. [DOI] [PubMed] [Google Scholar]
- 46.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 47.Senear DF, Brenowitz M. Determination of binding constants for cooperative site-specific protein-DNA interactions using the gel mobility-shift assay. J. Biol. Chem. 1991;266:13661–13671. [PubMed] [Google Scholar]
- 48.Moscoso M, del Solar G, Espinosa M. In vitro recognition of the replication origin of pLS1 and of plasmids of the pLS1 family by the RepB initiator protein. J. Bacteriol. 1995;177:7041–7049. doi: 10.1128/jb.177.24.7041-7049.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SW, Jeong EJ, Kang HS, Tak JI, Bang WY, Heo JB, Jeong JY, Yoon GM, Kang HY, Bahk JD. Role of RepB in the replication of plasmid pJB01 isolated from Enterococcus faecium JC1. Plasmid. 2006;55:99–113. doi: 10.1016/j.plasmid.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 50.de la Campa AG, del Solar G, Espinosa M. Initiation of replication of plsmid pLS1. The initiator protein RepB acts on two distant regions. J. Mol. Biol. 1990;213:247–262. doi: 10.1016/S0022-2836(05)80188-3. [DOI] [PubMed] [Google Scholar]